Abstract
The naturally occurring plasmid ColE1 was found to localize as a cluster in one or both of the cell poles of Escherichia coli. In addition to the polar localization of ColE1 in most cells, movement of the plasmid to the midcell position was observed in time-lapse studies. ColE1 could be displaced from its polar location by the p15A replicon, pBAD33, but not by plasmid RK2. The displacement of ColE1 by pBAD33 resulted in an almost random positioning of ColE1 foci in the cell and also in a loss of segregational stability, as evidenced by the large number of cells carrying pBAD33 with no visible ColE1 focus and as confirmed by ColE1 stability studies. The addition of the active partitioning systems of the F plasmid (sopABC) or RK2 (OB1 incC korB) resulted in movement of the ColE1 replicon from the cell pole to within the nucleoid region. This repositioning did not result in destabilization but did result in an increase in the number of plasmid foci, most likely due to partial declustering. These results are consistent with the importance of par regions to the localization of plasmids to specific regions of the cell and demonstrate both localization and dynamic movement for a naturally occurring plasmid that does not encode a replication initiation protein or a partitioning system that is required for plasmid stability.
Recent studies using fluorescent in situ hybridization (FISH) of plasmids in fixed cells or tagging of plasmids with a fluorescent protein in living cells have shown that these extrachromosomal elements exist in bacterial cells as discrete clusters at specific cellular locations. In Escherichia coli, low-copy-number plasmids such as F (13, 33), P1 (13), and R27 (24) were located at quarter- or midcell positions, while R1 (18) was localized near the cell pole. The first suggestion that multicopy plasmids also occurred as clusters in distinct cellular locations was provided by a study in which DAPI (4′,6′-diamidino-2-phenylindole) was used to stain a high-copy-number derivative of the plasmid R100 or a derivative of pBR322 (7). In this study, the R100 derivative was evenly spaced throughout the cell (which formed elongated cells or filaments due to the cultural conditions used), while the pBR322 derivative was localized to the cell poles (7). The localization of a multicopy plasmid by tagging with a fluorescent protein was reported in a study using plasmid RK2 (37). This naturally occurring broad-host-range plasmid was found as a cluster at midcell and quarter-cell positions in several gram-negative bacteria. In that same study, plasmid pAFS52, a derivative of the commonly used high-copy-number cloning vector pUC19, was also found as discrete foci in E. coli at either the midcell position in those cells with only one focus or at quarter positions in cells with two foci (37).
It has been a basic tenet of plasmid biology that intermediate- or high-copy-number plasmids do not require an active partitioning (par) system for stable maintenance, as random distribution is sufficient to ensure segregation of plasmid molecules to the two daughters at the time of cell division. However, the clustering of plasmids would seem to act against random distribution, particularly because, for all plasmids studied, the number of plasmid clusters is always less than the number of plasmid copies per cell (14, 37, 47). Some moderate-copy-number plasmids such as RK2, with a copy number of ∼5 to 8 per chromosome, do encode an active partitioning system, while for others, such as ColE1 with an estimated 10 to 15 copies per chromosome (6), there is no evidence for an active partitioning system. The purpose of this study was to determine the location inside E. coli cells of ColE1, a naturally occurring 6.6-kb, intermediate-copy-number plasmid. ColE1 replication utilizes host proteins, including PolI, but does not require a plasmid-encoded protein (4, 5, 22, 44). Stable maintenance of ColE1 in E. coli is dependent on a multimer resolution system consisting of host-encoded proteins acting at the plasmid carried cer site (43).
By using a fluorescent fusion protein to tag plasmid molecules, ColE1 was found to be stably maintained as a cluster located in one or both of the cell's poles. ColE1 was displaced from its polar location by the presence of plasmid pBAD33 in the same cell. The presence of pBAD33 in the cell also caused ColE1 instability. The addition of the partitioning system of the F plasmid to ColE1 increased ColE1 plasmid stability in the presence of pBAD33, while the addition of an RK2 par region did not. Notably, the presence of either par system on ColE1-tetO resulted in the movement of the ColE1 derivative from the cell pole into the nucleoid.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strain MG1655 (2) or a derivative, MGTRY (C. Verheust, unpublished data), was used as the bacterial host. MGTRY expresses a TetR-enhanced yellow fluorescent protein (EYFP) fusion protein from the chromosome under control of the arabinose-inducible PBAD promoter.
Plasmids used in this study are listed in Table 1. As ColE1 has no selectable marker, the aphAI gene encoding kanamycin resistance was isolated as an EcoRI fragment from plasmid pUC4K (45) and inserted into the unique EcoRI site of ColE1, yielding plasmid pSY1. The EcoRI site in ColE1 is located at the 3′ end of the gene encoding the colicin E1 protein.
TABLE 1.
Relevant plasmids
| Plasmid | Description | Reference(s) |
|---|---|---|
| ColE1 | Naturally occurring E. coli plasmid | 40 |
| pSY1 | ColE1 with Kmr | This work |
| pAS2 | pSY1 with tetO repeats and Gmr | This work |
| pAS4 | pAS2 with sopABC from F plasmid | This work |
| pAS6 | pAS2 with OB1incC korB from RK2 | This work |
| pFX234 | pBAD24 expressing TetR-EYFP | 14, 23 |
| pBAD33 | p15A-based expression vector utilizing PBAD promoter | 15 |
| pCV234 | pBAD33 expressing TetR-EYFP; derived from pFX234 | This work |
| pLAU44 | Source of tetO repeats and Gmr gene | 23 |
| RK2 | Naturally occurring broad-host-range plasmid | 36 |
To construct plasmid pAS2, a PvuII fragment containing a Gmr-tetO operator array was isolated from pLAU44, a derivative of pUC18 with an ∼ 9-kb DNA insert consisting of a stable genetic array of 120 copies of the 19-bp tetO binding site separated by 10-bp random spacer sequence flanking either side of a gene encoding gentamicin (Gm) resistance (23). The ∼9.3-kb PvuII fragment was inserted into EcoRV-digested pSY1. pSY1 has two EcoRV sites ∼1 kb apart, both located in the gene encoding the colicin E1 protein.
Derivatives of pAS2 carrying the partitioning region either of the F plasmid or of RK2 were constructed as follows. An ∼3.7-kb XbaI-HindIII fragment from pRR53 (38) containing sopABC from plasmid F was inserted into XbaI-HindIII-digested pAS2, yielding pAS4. pAS6 has OB1 incC korB of RK2 isolated as a HindIII-PvuII fragment from pCV126 cloned into HindIII-SmaI digested pAS2. pCV126 (C. Verheust, unpublished data) is pUC19 with OB1 incC korB isolated from a derivative of pRK214.1 (9).
pFX234 carries the gene for expression of a TetR-EYFP fusion protein (23). This plasmid is derived from pBAD24, a ColE1-based replicon. The gene encoding the fusion protein was transferred to p15A-based pBAD33 as follows. Plasmids pFX234 and pBAD33 were digested with MluI and HindIII. MluI has a unique site, between araC and PBAD, in both pBAD24 (the vector for pFX234) and pBAD33. HindIII is unique in pBAD33 and pFX234. In pFX234 the HindIII site is after the stop codon for the fusion protein. The desired fragments were isolated and ligated to yield plasmid pCV234, which is ∼6 kb in size and encodes chloramphenicol resistance (C. Verheust, unpublished data).
Plasmid localization studies.
For localization of tetO-containing plasmids tagged by TetR-EYFP in E. coli MGTRY, cultures were grown overnight in LB with 10 μg/ml Gm at 30°C. The culture was then diluted 1:100 into M63 with 0.2% glycerol and 10 μg/ml Gm and incubated at 30°C with aeration to an absorbance at 600 nm of 0.1 over the inoculum. TetR-EYFP expression was then induced by adding l-arabinose to a final concentration of 0.6%. Aliquots of the culture were examined at various time points by fluorescence microscopy. Foci typically became visible ∼1 h after induction of TetR-EYFP. Where indicated, 200 μg/ml cephalexin was added after dilution of the overnight culture. Localization of pAS2 in E. coli MGTRY in the presence of RK2 or pBAD33 was carried out as described above except for the inclusion of 100 μg/ml ampicillin or 20 μg/ml chloramphenicol, respectively, in all media.
For localization of tetO-containing plasmids in E. coli strain MG1655 expressing TetR-EYFP from plasmid pCV234, cultures were grown overnight in LB supplemented with 0.2% glucose (which is essential to suppress expression of the EYFP-TetR protein from the PBAD promoter present on the moderate-copy-number plasmid pCV234 as opposed to the chromosome in MGTRY), 10 μg/ml Gm, and 20 μg/ml chloramphenicol, followed by 1:10 dilution into M63 containing 0.2% glycerol, 10 μg/ml Gm, and 20 μg/ml chloramphenicol the next morning. Approximately 1.5 h after inoculation, l-arabinose was added to a final concentration of 0.2%. Starting at 20 min after induction of TetR-EYFP, aliquots of the culture were examined by fluorescence microscopy.
Foci were visualized with a BX60 fluorescence microscope (Olympus), and images were captured with a C-5050 digital camera (Olympus). For visualization of nucleoids, 10 μl of cells stained with 5 μg/ml DAPI was added to slides without fixation. Cells and foci were measured using ImageJ public-domain software available through the National Institutes of Health. For the purposes of this study, polar location was defined as from 0 to 0.2 cell length, quarter-cell location as from 0.2 to 0.4 cell length, and midcell location as from 0.4 to 0.5 cell length in every case that focus location was measured.
For time-lapse microscopy, overnight cultures were diluted 1:100 in 20 ml M63 containing 0.2% glycerol and 10 μg/ml Gm. Cells were incubated at 30°C with aeration for 3 h before addition of l-arabinose to a final concentration of 0.6%. When foci were visible, ∼1 h later, an aliquot of the culture was concentrated 20×, and 10 μl was placed on an agarose slab (0.8% agarose prepared in M63 containing 0.2% glycerol and 0.6% l-arabinose) and observed at 15-min intervals for up to 4 h at 30°C (27, 28, 37).
Stability assays.
To assay the stability of plasmid pAS2 or pAS4 in E. coli MG1655 and of pAS6 in E. coli MGTRY, colonies were picked from a fresh overnight plate of LB with 10 μg/ml Gm, resuspended in 5 ml LB broth with 10 μg/ml Gm, and then incubated overnight with shaking at 30°C. At time zero, 50 μl of the stationary-phase culture was transferred to 5 ml M63 with 0.2% glycerol without Gm, and incubation with shaking was continued. The culture was similarly diluted at 24 h. At 0, 8, 24, and 48 h, an aliquot of the cultures was diluted and plated on LB agar. After overnight incubation at 30°C, 100 colonies from the LB plate were patched to LB with 10 μg/ml Gm and to LB to determine the percentage of cells still maintaining Gm resistance.
The stability of pAS2, pAS4, and pAS6 in the presence of pBAD33 or RK2 was similarly assayed except that either 20 μg/ml chloramphenicol for pBAD33 or 100 μg/ml ampicillin for RK2 was added to all liquid cultures in LB or M63 with 0.2% glycerol.
RESULTS
Localization of ColE1-tetO.
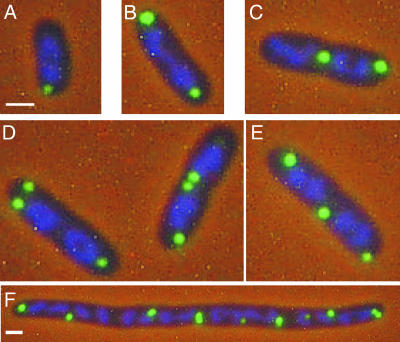
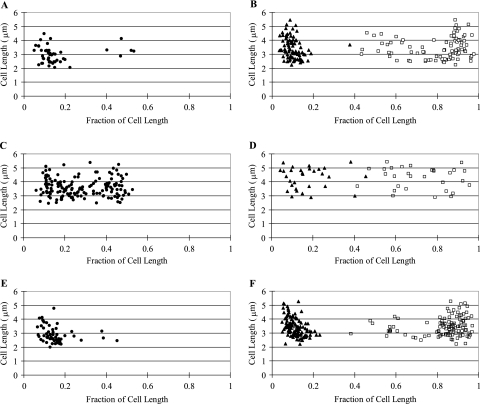
A reporter system consisting of a TetR-EYFP fusion protein and an array of repeating units of the tetO binding site was used to localize plasmid ColE1 in E. coli cells. A derivative of ColE1 containing an ∼9.3 kb insert with an array of ∼240 tetO binding sites was constructed. This plasmid, pAS2, was transformed into E. coli strain MGTRY, which carries the gene for a TetR-EYFP fusion protein on the chromosome. In MGTRY, expression of the TetR-EYFP fusion protein is dependent on the l-arabinose-inducible PBAD promoter. As shown in Fig. 1, discrete foci of ColE1-tetO were observed in cells after induced expression of the TetR-EYFP protein. The majority of cells (66%) had either one focus or two foci, and about 30% had three or more foci (Table 2). The position of foci in these cells was measured as a function of cell length (Fig. 2A and B). In cells with only one focus, the single focus was primarily in the polar region (from 0 to 0.2 fractional cell length) (Fig. 1A and 2A). When there were two foci per cell, 99% of the cells had one polar focus, and in ∼66% of these cells the second focus was also polar (Fig. 1B and 2B). In the remaining cells, the position of the second focus was variable (Fig. 2B), with 14% of cells having the second focus in the midcell position (Fig. 1C).
FIG. 1.
Localization of the ColE1 derivative pAS2 in E. coli. Cells of E. coli MGTRY(pAS2) with one (A), two (B and C), or three (D and E) foci or after treatment with cephalexin (F) are shown. In all panels, the nucleoid has been stained with DAPI. The bars in panels A and F correspond to 1 μm. Cells shown in panels B to D are at the same magnification as those in panel A.
TABLE 2.
Number of foci in E. coli cells carrying ColE1 with or without a second replicon
| Strain | % of cells with the following no. of focia:
|
||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ≥4 | |
| MGTRY(pAS2) | 4 | 18 | 48 | 24 | 7 |
| MGTRY(pAS2)(pBAD33) | 59 | 33 | 7 | <1 | 0 |
| MGTRY(pAS2)(RK2) | 4 | 8 | 50 | 27 | 12 |
The numbers of cells counted were 285 for MGTRY(pAS2), 787 for MGTRY(pAS2)(pBAD33), and 379 for MGTRY(pAS2)(RK2).
FIG. 2.
Location of pAS2 foci in E. coli in the presence or absence of a second replicon. The position of foci in cells of MGTRY(pAS2) (A and B), MGTRY(pAS2)(pBAD33) (C and D), or MGTRY(pAS2)(RK2) (E and F) was determined as a fraction of cell length. Each point represents a single focus. The position of pAS2 in those cells with only a single focus (•) is shown in panels A, C, and E. The position of pAS2 foci in cells with two foci is shown in panels B, D, and F (▴, first focus; □, second focus).
The location of plasmid foci in MGTRY(pAS2) relative to the nucleoid was determined by visualizing cells stained with DAPI. As shown in Fig. 1A, when one polar focus was present in the cell, the focus was located outside of the nucleoid. In cells with two polar foci, both foci were located outside of the nucleoid (Fig. 1B). In two-focus cells with one polar focus and one nonpolar focus, the nonpolar focus was in most cases located on the edge of the nucleoid or in a space between two nucleoids (Fig. 1C). A similar positioning of foci outside the nucleoid was observed in cells with three foci (Fig. 1D and E). In MGTRY(pAS2) cells treated with cephalexin, foci were evenly spaced at positions outside the nucleoid (Fig. 1F). This antibiotic prevents septal cell wall synthesis, resulting in cell filamentation but not cell death during the course of the assay.
The question of whether the observed localization is actually an artifact due to aggregation of plasmid elements is always a concern when using fluorescently tagged proteins. The facts that foci were evenly spaced in cephalexin-treated cells (Fig. 1F) and that foci showed dynamic and directed movement (Fig. 3) (see below) argue against aggregation.
FIG. 3.
Time-lapse photography of single cells of MGTRY(pAS2), showing movement of polar foci (A) or cell division in the absence of focus duplication (B). The time after transfer of the culture to an agarose pad for microscopic observation is noted in the top left corner of each picture. The bars correspond to 1 μm.
Movement of pAS2 foci.
The movement of pAS2 foci in MGTRY was observed by examining cells at 15-min intervals over a 4-h time course. A polar focus was often observed to split into two foci, with subsequent movement of one of the two foci to a midcell location (Fig. 3A, 0- to 45-min time points). Occasionally the polar focus was observed to move to the midcell position without splitting (not shown). After division, the midcell focus becomes located in the polar position of one of the two daughter cells (Fig. 3A, 90- to 120-min time points). Also commonly observed were cells with two polar foci that did not duplicate prior to cell division, with each focus again located in the polar position of the daughter cells (Fig. 3B, 0- to 90-min time points). The position of pAS2 in MGTRY was not static, showing apparent movement over short distances at the polar (Fig. 3B, 45- to 60-min time points) or midcell (Fig. 3A, 30- to 45-min time points) position. Occasionally, a single focus appeared to split into two foci, which after a period of time appeared to rejoin as a single focus (Fig. 3B, 120- to 180-min time points). It is of course possible that rather than showing movement, the plasmid clusters observed are actually dissolved and reformed at another position. It should, however, be noted that plasmids with par systems have been shown to move along cellular tracks (29; see references 10 and 11 for reviews).
Localization of ColE1 in the presence of a second plasmid.
The observed polar localization of pAS2 was not in agreement with a previous study using the ColE1-type replicon pAFS52. This derivative of pUC had been localized to quarter-cell or midcell positions in E. coli (37). Localization of pAFS52 involved the binding of lacO repeats present on pASF52 by a LacI-green fluorescent protein fusion protein expressed from a second plasmid in the cell. This second plasmid, pGAP60 (37), was a derivative of the expression vector pBAD33 (15).
To determine if the difference between the polar localization of pAS2 and the quarter-cell localization of pAFS52 was due to the presence of the second replicon in the cell, we localized pAS2 in MGTRY carrying pBAD33. In this two-plasmid system, 59% of the cells had no visible focus, while the rest of the cells had either one focus (33%) or two foci (7%) (Fig. 4A and Table 2). Very few cells were observed to have more than three foci (Table 2). In those cells with one pAS2 focus, there was a fairly random distribution in the location of the single focus (Fig. 2C). Forty percent of the cells had the focus in a polar location (defined as 0 to 0.2 cell length), 32% had a quarter-cell location (from 0.2 to 0.4 cell length,) and 28% had a midcell location (from 0.4 to 0.5 cell length). A similar random distribution was observed in cells with two foci (Fig. 2D). Almost all pAS2 foci, including those that had a quarter-cell or midcell location, appeared to be outside of the nucleoid stained with DAPI (Fig. 4A).
FIG. 4.
Localization of ColE1 in the presence of a second plasmid. Plasmid pAS2 was localized in E. coli MGTRY in the presence of plasmids pBAD33 (A) or RK2 (B) or in E. coli MG1655 in the presence of plasmid pCV234 (C). In panel A, cells were visualized for pAS2 and the nucleoid was stained with DAPI. In panels B and C, only foci are shown. The bar in each panel corresponds to 1 μm.
To see if the essentially random positioning of pAS2 observed in the two-plasmid system was specific to pBAD33, we localized pAS2 in MGTRY containing the plasmid RK2. RK2 is unrelated to either ColE1 or p15A (the parent replicon of pBAD33). With the pAS2/RK2 two-plasmid system, only 4% had no foci and the majority (89%) had two or more foci (Fig. 4B and Table 2). In the presence of RK2, pAS2 was again primarily in a polar location in cells with either one focus (Fig. 2E) or two foci (Fig. 2F).
Lastly, to duplicate the conditions used in the earlier study (37), we localized pAS2 in a strain expressing the TetR-EYFP fusion protein from pBAD33 (E. coli MG1655 carrying plasmid pCV234) and not from the chromosome. In this two-plasmid system, 47% of cells had no foci. In those cells that did have a focus, the position was random (Fig. 4C). Thus, the effect on ColE1 positioning by a second plasmid in the cell seemed to be specific to pBAD33 and its derivative pCV234 (which express the TetR-EYFP fusion protein) and not simply due to a second replicon in the cell.
Stability of replicons in the two-plasmid system.
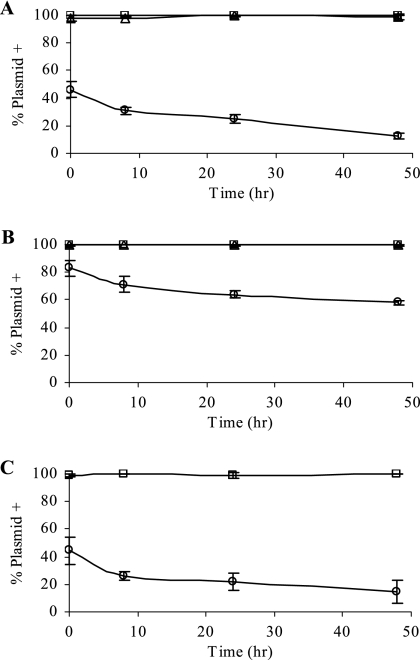
In addition to the almost random distribution of the ColE1 replicon pAS2 in E. coli cells carrying pBAD33 was the apparent instability of pAS2 as indicated by the 59% of cells with no visible focus (Table 2). To establish that the absence of a focus was in fact due to loss of pAS2, stability studies were performed using MG1655 carrying either pAS2, pAS2 and pBAD33, or pAS2 and RK2. As shown in Fig. 5A, pAS2 was stable in E. coli MG1655 and its stability was not influenced by the presence of RK2, as measured by the percentage of cells maintaining Gm resistance in the absence of selection. However, there was a significant loss of pAS2 in MG1655 carrying pBAD33. Similar results were obtained when E. coli MGTRY was the host (data not shown).
FIG. 5.
Influence of a second replicon on stability of ColE1 derivatives in E. coli. The stabilities of ColE1 derivative pAS2 in MG1655 (□) (A), of pAS4 (pAS2 + sopABC) in MG1655 (□) (B), and of pAS6 (pAS2 + OB1 incC korB) in MGTRY (□) (C) were determined as the percentage of cells maintaining Gm resistance in the absence of selection, as described in Materials and Methods. Stability was also determined for pAS2 (A) and pAS4 (B) in the presence of pBAD33 (○) or RK2 (▵) and for pAS6 in the presence of pBAD33 (C) (○). Results shown are the averages and standard deviations from two or three experiments.
This result was surprising, as plasmids derived from ColE1 and p15A (the parent of pBAD33) have previously been reported to be compatible even though the origins of replication of these two plasmids have several regions of extensive DNA homology (3, 41). These compatibility studies were performed in RecA− E. coli strains. Many of the experiments with localization of pAFS52 were also performed in RecA− E. coli (37). The studies presented here, however, were carried out in the RecA+ hosts MG1655 and MGTRY, which may account for the discrepancy. To examine this possibility, the stability of pAS2 was measured, in the presence or absence of pBAD33, in the RecA− E. coli strain DH5. The results were that pAS2 was unstable during the 48 h of the assay if pBAD33 was present and selected for in the host (∼42% loss of Gm resistance after 48 h), while no loss of pAS2 was observed if pBAD33 was not present (data not shown). These results suggest a replication incompatibility between pAS2 and pBAD33.
Effect of par genes on positioning of ColE1 foci.
The low-copy-number plasmids P1, F, and R27 are located at quarter- or midcell positions in E. coli. Deletion or inactivation of the partitioning regions from these plasmids not only reduces or eliminates their segregational stability, it also results in mislocalization (F [33] and P1 [8]) or in random distribution of plasmid foci throughout the cell (R27 [24]). Conversely, mini-oriC plasmids localize to the cell poles; however, the addition of the partitioning system of the F plasmid, sopABC, will result in localization of mini-oriC to the quarter-cell positions (34).
Even though ColE1 is extremely stable despite the absence of any known partitioning function (43), we wanted to determine if a heterologous par system would influence its cellular location. The F plasmid sopABC genes were inserted into pAS2, generating pAS4. To test for functionality of the inserted genes, we first determined their ability to stabilize the plasmid.
As shown above, even though pAS2 alone is stably maintained in E. coli, introduction of pBAD33 into the cell results in pAS2 instability (Fig. 5A) and mislocalization (Fig. 2). The addition of F par to pAS2 (plasmid pAS4) improved plasmid stability in MG1655 in the presence of pBAD33 (compare pAS4 with pBAD33 in Fig. 5B to pAS2 with pBAD33 in Fig. 5A). This suggested that the heterologous partitioning system was functional.
We then examined the cellular location of pAS4 in MGTRY. As shown in Fig. 6, multiple foci of pAS4 were detected throughout the nucleoid. This repositioning did not result in destabilization (Fig. 5B) but did result in an increase in the number of plasmid foci. To see if these effects on ColE1-tetO were specific to the F par system or if another partitioning system would also affect plasmid positioning, the OB1 incC korB partition system of plasmid RK2 was introduced into pAS2, yielding pAS6. Once again there was a random distribution and partial declustering, of plasmid pAS6 molecules in MGTRY (Fig. 6). However, pAS6 did not have improved stability relative to pAS2 in the presence of pBAD33 (compare Fig. 5C with 5A), though it was stable when present in the cell alone (Fig. 5C). To determine if the increase in the number of foci observed in MGTRY cells carrying pAS4 or pAS6 relative to the number of foci in MGTRY(pSY2) was due to an actual increase in plasmid copy number, the relative copy numbers of the ColE1 derivatives were estimated as described previously (50). The average relative copy numbers of pAS4 and pAS6 were found to be less than that of pAS2 by 4% and 15%, respectively. Therefore the increased number of foci was not due to any increase in the number of plasmid molecules per cell but is most likely due to partial declustering.
FIG. 6.
Localization of ColE1 derivatives carrying heterologous partition systems. (A to C) MGTRY(pAS4) (A, foci only; B, DAPI staining of nucleoid; C, merge of A and B); (D to F) MGTRY(pAS6) (D, foci only; E, DAPI staining of nucleoid; F, merge of D and E). The bar in panel A corresponds to 1 μm.
DISCUSSION
ColE1-tetO exhibited specific localization in E. coli cells, with a cluster of plasmid molecules present in one or both of the cell poles (Fig. 1). The polar location of ColE1-tetO was not fixed; a polar focus was often observed to split into two foci, with subsequent movement of one of the two foci to a midcell location (Fig. 3). The midcell focus was located outside the nucleoid or between two nucleoids in longer cells, a position that upon cell division would become a new cell pole. The majority of cells had two foci, though a significant number of smaller cells had only one focus (Table 2 and Fig. 2).
Consistent with these localization results for ColE1-tetO are the results from earlier studies utilizing minicell-producing strains of E. coli. In minicell mutants of E. coli there is aberrant placement of the cell septum near a cell pole. The resulting asymmetric cell division gives rise to one daughter, the minicell, containing no chromosomal DNA but, depending on the replicon, possibly containing plasmid DNA. In these previous studies, ColE1 was found to segregate into and replicate in E. coli minicells (17). Derivatives of the F plasmid were not found in minicells (16), though copy-up or par− derivatives of F were found in minicells (16). It has now been shown that derivatives of F containing the F partition region, sopABC, localize at quarter- or midcell positions in E. coli (13, 33) while par− mutants are distributed randomly in the cell, including the polar region. Plasmid R100 (related to R1, which is located in the cell pole [18]) and a derivative of pBR322 (a ColE1-based replicon) were also found in minicells by DAPI staining, but plasmids P1Cm (which is found at quarter- or midcell positions [13]) and F′lac were not (7). That study noted not only the polar localization of the pBR322 derivative but also that the number of DAPI-labeled foci observed was significantly lower than the estimated plasmid copy number (7).
Attempts were also made to localize pAS2 in E. coli by using FISH. Fluorescent foci were observed in the polar region; however, they were very large and indistinct. The inability to obtain clear results using FISH may be because the small size of ColE1 makes fixation of the plasmid in the cell difficult.
The polar localization of ColE1-tetO reported here was in contrast to the midcell or quarter-cell localization previously reported for pAFS52, a high-copy-number ColE1-based plasmid (37). Localization of pAFS52 was carried out in the presence of a second plasmid, pGAP60, a derivative of pBAD33, which carried the gene for the fluorescent fusion protein. The introduction of pBAD33 into a cell carrying the ColE1 derivative pAS2 resulted in the movement of pAS2 clusters out of the cell pole to essentially random positions outside the nucleoid (Fig. 2). The presence of pBAD33 also resulted in a considerable decrease in the stability of pAS2 (Fig. 5).
The instability of pAS2 was specific to pBAD33 and not just due to a second replicon in the cell (Fig. 5). The cause of the instability is not obvious. Pogliano et al. (37) reported observing a significant loss of pAFS52 during growth that they attributed to the clustering of the plasmid. The results presented here suggest that it is due not to clustering but rather to an incompatibility between ColE1 and derivatives of pBAD33, since instability was not observed in the presence of RK2. Previous studies have reported that plasmids derived from ColE1 and p15A (the parent of pBAD33) are compatible in RecA− E. coli (3, 41). However, ColE1 and p15A do share significant regions of DNA homology, and it is possible that the incompatibility observed in this study is expressed only in RecA+ hosts such as MGTRY. Though it is possible that a contributing factor to ColE1 instability in the presence of pBAD33 is the colocalization of the two plasmids at the cell pole (unpublished observations), this is unlikely because for other plasmid pairs that have been examined (e.g., ColE1 and pACYC184 [unpublished observations]), colocalization at the cell pole does not result in instability or in displacement of a plasmid from its normal cellular location.
Naturally occurring low-copy-number plasmids typically have an active partitioning mechanism to ensure efficient distribution of plasmid molecules to each daughter cell at the time of cell division. ColE1, an intermediate-copy-number plasmid, has not been found to encode an active partitioning system. The stability of ColE1 has been shown to be dependent on a very efficient multimer resolution system, which ensures that the plasmid exists as monomeric units inside the cell (42). The addition of F par to ColE1-tetO (plasmid pAS4) increased ColE1 plasmid stability in the presence of pBAD33 (Fig. 5). The addition of RK2 par (plasmid pAS6) did not lead to an increase in stability in the presence of pBAD33 (Fig. 5). However, the RK2 par region used, OB1 incC korB, contained only one of three KorB binding sites present in RK2 par, and this site, OB1, had previously been shown not to be as effective as OB3 in stabilizing RK2 derivatives (39, 48).
Interestingly, in cells containing only the ColE1-derived plasmid (i.e., without pBAD33), the presence of either the F or the RK2 par region on the ColE1 replicon resulted in the movement of the ColE1 replicon from the cell pole to within the nucleoid region (Fig. 6). This repositioning of the ColE1 derivative did not result in destabilization (Fig. 5) but did result in an increase in the number of plasmid foci observed in the cell. These results are consistent with the importance of par regions to the localization of plasmids to specific regions of the cell. Aberrant localization has previously been observed for derivatives of plasmids F (33), P1 (8), R27 (24), and R1 (18) in which the par region has been deleted or inactivated. In these cases, plasmid foci are distributed throughout the cell, though outside the nucleoid (8, 18, 33), and the plasmid is not stably maintained. However, in these cases, the repositioning of foci does not result in partial declustering as found for ColE1.
The analysis of the ColE1 plasmid pAS2 presented in this study shows that an intermediate-copy-number plasmid can be found in E. coli as clusters targeted to the cell pole despite the absence of a plasmid-specified replication initiation protein. While plasmid initiation proteins may play a role in clustering through their binding to the plasmid origin of replication for those plasmids that do encode an initiation protein, that clearly is not a factor in clustering and localization in the ColE1 plasmids. For plasmids such as ColE1, the pole may simply be the default position. Alternatively, there may be host-encoded factors that are responsible for targeting.
The cellular location of the machinery for ColE1 replication is not known. A midcell location for the initiation of chromosomal DNA replication in E. coli has been proposed (1, 23). The time-lapse studies showed movement of some ColE1 clusters to the midcell position, and it is possible that movement of the plasmid cluster to the region of the midcell is a requirement for replication. If that is the case, it raises the question of the nature of the cellular and ColE1 plasmid elements responsible for this movement and repositioning of plasmid clusters. This is of particular interest since ColE1 has not been shown to encode a partition system similar to the ones shown to be responsible for movement of either R1 (30) or F (29).
It is not clear whether the polar position of ColE1 is a default position or whether there are plasmid sequences and host factors that target this plasmid to the poles. A polar location has been demonstrated for plasmid R1 in E. coli (31) and several naturally occurring plasmids of Agrobacterium tumefaciens and Sinorhizobium meliloti (20). In addition, a cell pole location has been shown for the chromosomal origins of replication of E. coli (12, 32), Bacillus subtilis (26), and Caulobacter crescentus (19). In E. coli, a 25-bp sequence within the origin was identified as playing a role in the bipolar migration of a newly replicated oriC region (49). However, virtually nothing is known about the cellular components that may interact with plasmid or oriC nucleotide sequences for targeting to the polar position. In addition to the localization of the replication machinery at the midcell position in B. subtilis (25) and E. coli (21), subunits of the E. coli (35) and C. crescentus (46) replisome have been shown to be present at the poles. Thus, the replication complex conceivably may be playing a role in polar positioning. It is clear that the identification of macromolecular structures at the cell poles that play an essential role in fixing the chromosomal origin of replication and plasmids, such as ColE1, to this region of the cell is critical to understanding the targeting of replicating elements in bacteria.
Acknowledgments
This work was supported by National Institutes of Health research grant AI-07194.
We thank Celine Verheust for providing E. coli strain MGTRY and plasmids pCV126 and pCV234 in advance of publication.
Footnotes
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Bates, D., and N. Kleckner. 2005. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121:899-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 3.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clewell, D. B. 1972. Nature of Col E1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J. Bacteriol. 110:667-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clewell, D. B., and D. R. Helinski. 1972. Effect of growth conditions on the formation of the relaxation complex of supercoiled ColE1 deoxyribonucleic acid and protein in Escherichia coli. J. Bacteriol. 110:1135-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clewell, D. B., and D. R. Helinski. 1970. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 9:4428-4440. [DOI] [PubMed] [Google Scholar]
- 7.Eliasson, A., R. Bernander, S. Dasgupta, and K. Nordstrom. 1992. Direct visualization of plasmid DNA in bacterial cells. Mol. Microbiol. 6:165-170. [DOI] [PubMed] [Google Scholar]
- 8.Erdmann, N., T. Petroff, and B. E. Funnell. 1999. Intracellular localization of P1 ParB protein depends on ParA and ParS. Proc. Natl. Acad. Sci. USA 96:14905-14910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figurski, D., R. Meyer, D. S. Miller, and D. R. Helinski. 1976. Generation in vitro of deletions in the broad host range plasmid RK2 using phage Mu insertions and a restriction endonuclease. Gene 1:107-119. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes, K., J. Moller-Jensen, G. Ebersbach, T. Kruse, and K. Nordstrom. 2004. Bacterial mitotic machineries. Cell 116:359-366. [DOI] [PubMed] [Google Scholar]
- 11.Gitai, Z. 2006. Plasmid segregation: a new class of cytoskeletal proteins emerges. Curr. Biol. 16:R133-R136. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, G. S., R. P. Shivers, and A. Wright. 2002. Polar localization of the Escherichia coli oriC region is independent of the site of replication initiation. Mol. Microbiol. 44:501-507. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, G. S., D. Sitnikov, C. D. Webb, A. Teleman, A. Straight, R. Losick, A. W. Murray, and A. Wright. 1997. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90:1113-1121. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, S., J. Rech, D. Lane, and A. Wright. 2004. Kinetics of plasmid segregation in Escherichia coli. Mol. Microbiol. 51:461-469. [DOI] [PubMed] [Google Scholar]
- 15.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan, J. E., B. C. Kline, and S. B. Levy. 1982. Regions on the F plasmid which affect plasmid maintenance and the ability to segregate into Escherichia coli minicells. Plasmid 8:36-44. [DOI] [PubMed] [Google Scholar]
- 17.Inselburg, J. 1970. Segregation into and replication of plasmid deoxyribonucleic acid in chromosomeless segregants of Escherichia coli. J. Bacteriol. 102:642-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen, R. B., and K. Gerdes. 1999. Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. EMBO J. 18:4076-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen, R. B., S. C. Wang, and L. Shapiro. 2002. Dynamic localization of proteins and DNA during a bacterial cell cycle. Nat. Rev. Mol. Cell Biol. 3:167-176. [DOI] [PubMed] [Google Scholar]
- 20.Kahng, L. S., and L. Shapiro. 2003. Polar localization of replicon origins in the multipartite genomes of Agrobacterium tumefaciens and Sinorhizobium meliloti. J. Bacteriol. 185:3384-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koppes, L. J., C. L. Woldringh, and N. Nanninga. 1999. Escherichia coli contains a DNA replication compartment in the cell center. Biochimie 81:803-810. [DOI] [PubMed] [Google Scholar]
- 22.Kues, U., and U. Stahl. 1989. Replication of plasmids in gram-negative bacteria. Microbiol. Mol. Biol. Rev. 53:491-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau, I. F., S. R. Filipe, B. Soballe, O.-A. Okstad, F.-X. Barre, and D. J. Sherratt. 2003. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol. 49:731-743. [DOI] [PubMed] [Google Scholar]
- 24.Lawley, T. D., and D. E. Taylor. 2003. Characterization of the double-partitioning modules of R27: correlating plasmid stability with plasmid localization. J. Bacteriol. 185:3060-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemon, K. P., and A. D. Grossman. 1998. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282:1516-1519. [DOI] [PubMed] [Google Scholar]
- 26.Lewis, P. J., and J. Errington. 1997. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol. Microbiol. 25:945-954. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y., and S. Austin. 2002. The P1 plasmid is segregated to daughter cells by a ‘capture and ejection’ mechanism coordinated with Escherichia coli cell division. Mol. Microbiol. 46:63-74. [DOI] [PubMed] [Google Scholar]
- 28.Li, Y., B. Youngren, K. Sergueev, and S. Austin. 2003. Segregation of the Escherichia coli chromosome terminus. Mol. Microbiol. 50:825-834. [DOI] [PubMed] [Google Scholar]
- 29.Lim, G. E., A. I. Derman, and J. Pogliano. 2005. Bacterial DNA segregation by dynamic SopA polymers. Proc. Natl. Acad. Sci. USA 102:17658-17663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moller-Jensen, J., J. Borch, M. Dam, R. B. Jensen, P. Roepstorff, and K. Gerdes. 2003. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol. Cell 12:1477-1487. [DOI] [PubMed] [Google Scholar]
- 31.Møller-Jensen, J., R. B. Jensen, J. Löwe, and K. Gerdes. 2002. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 21:3119-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niki, H., and S. Hiraga. 1998. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev. 12:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niki, H., and S. Hiraga. 1997. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell 90:951-957. [DOI] [PubMed] [Google Scholar]
- 34.Niki, H., and S. Hiraga. 1999. Subcellular localization of plasmids containing the oriC region of the Escherichia coli chromosome, with or without the sopABC partitioning system. Mol. Microbiol. 34:498-503. [DOI] [PubMed] [Google Scholar]
- 35.Onogi, T., K. Ohsumi, T. Katayama, and S. Hiraga. 2002. Replication-dependent recruitment of the β-subunit of DNA polymerase III from cytosolic spaces to replication forks in Escherichia coli. J. Bacteriol. 184:867-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncPα plasmids: compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 37.Pogliano, J., T. Q. Ho, Z. Zhong, and D. R. Helinski. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts, R. C., R. Burioni, and D. R. Helinski. 1990. Genetic characterization of the stabilizing functions of a region of broad-host-range plasmid RK2. J. Bacteriol. 172:6204-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosche, T. M., A. Siddique, M. H. Larsen, and D. H. Figurski. 2000. Incompatibility protein IncC and global regulator KorB interact in active partition of promiscuous plasmid RK2. J. Bacteriol. 182:6014-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth, T. F., and D. R. Helinski. 1967. Evidence for circular DNA forms of a bacterial plasmid. Proc. Natl. Acad. Sci. USA 58:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selzer, G., T. Som, T. Itoh, and J. I. Tomizawa. 1983. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 32:119-129. [DOI] [PubMed] [Google Scholar]
- 42.Summers, D. 1998. Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol. Microbiol. 29:1137-1145. [DOI] [PubMed] [Google Scholar]
- 43.Summers, D. K., and D. J. Sherratt. 1984. Multimerization of high copy number plasmids causes instability: ColE1 encodes a determinant essential for plasmid monomerization and stability. Cell 36:1097-1103. [DOI] [PubMed] [Google Scholar]
- 44.Tomizawa, J. I., Y. Sakakibara, and T. Kakefuda. 1975. Replication of Colicin E1 plasmid DNA added to cell extracts. Proc. Natl. Acad. Sci. USA 72:1050-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 46.Wang, S. C., and L. Shapiro. 2004. The topoisomerase IV ParC subunit colocalizes with the Caulobacter replisome and is required for polar localization of replication origins. Proc. Natl. Acad. Sci. USA 101:9251-9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weitao, T., S. Dasgupta, and K. Nordstrom. 2000. Plasmid R1 is present as clusters in the cells of Escherichia coli. Plasmid 43:200-204. [DOI] [PubMed] [Google Scholar]
- 48.Williams, D., D. Macartney, and C. Thomas. 1998. The partitioning activity of the RK2 central control region requires only incC, korB and KorB-binding site O(B)3 but other KorB-binding sites form destabilizing complexes in the absence of O(B)3. Microbiology 144:3369-3378. [DOI] [PubMed] [Google Scholar]
- 49.Yamaichi, Y., and H. Niki. 2004. migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J. 23:221-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao, S., A. Toukdarian, and D. R. Helinski. 2006. Inhibition of protein and RNA synthesis in Escherichia coli results in declustering of plasmid RK2. Plasmid 56:124-132. [DOI] [PubMed] [Google Scholar]