Abstract
In this study, we report the isolation of colony morphology variants from Streptococcus pneumoniae serotype 3 biofilms. The colony variants differed in colony size (large, medium, and small) and their mucoid appearance on blood agar. The small nonmucoid variant (SCV) emerged during the initial attachment stage of S. pneumoniae biofilm formation and dominated over the course of biofilm growth. Mucoid variants appeared at later biofilm developmental stages. The reduction in colony size/mucoidy correlated with a decrease in capsule production and an increase in initial attachment. The large mucoid variant formed flat unstructured biofilms, failed to aggregate in liquid culture, and adhered poorly to solid surfaces. In contrast, SCVs autoaggregated in liquid culture, hyperadhered to solid surfaces, and formed biofilms with significant three-dimensional structure, mainly in the form of microcolonies. The variants showed similar antibiotic resistance/susceptibility based on a modified Kirby-Bauer test and when grown as biofilms. However, antimicrobial treatment of S. pneumoniae biofilms altered the colony variant's distribution and mainly affected the most interior areas of biofilm microcolonies. To further explore the nature of the variants, the capsule biosynthetic operon (cps3DSUM) was explored in greater detail. The genetic analysis indicated that the emergence of nonmucoid variants was due to a deletion comprising cps3DSU as well as additional genes upstream of the cps3 operon. Overall, our findings suggest that in vitro biofilm formation of S. pneumoniae serotype 3 coincides with the emergence of colony variants with distinct genotypic and phenotypic characteristics.
Streptococcus pneumoniae, a gram-positive, alpha-hemolytic bacterium, causes pneumococcal infections of the upper respiratory tract leading to pneumonia, as well as infections such as acute sinusitis, otitis media, conjunctivitis, meningitis, osteomyelitis, septic arthritis, endocarditis, peritonitis, pericarditis, cellulitis, and brain abscess. The major virulence determinant of S. pneumoniae is the polysaccharide capsule (3, 13), which renders the organism resistant to opsonophagocytosis, the major mechanism of clearance, in hosts lacking type-specific antibody of sufficient quantity or avidity (15, 26). Expression of antiphagocytic capsular polysaccharide (CPS), however, has been shown to inhibit adherence of pneumococci to host cells, a critical step in carriage and possibly in the later aspects of the pathogenesis of disease (25). Expression of capsule and/or key surface structures must be varied at different stages of invasive infection, allowing for both adhesive interactions with host cells and resistance to humoral clearance mechanisms, for adaptation to various ecological niches in the human host (9, 10, 20, 36). In S. pneumoniae, the reversible expression or phase variation of surface structures, resulting in a detectable change in colony morphology, has been correlated with differences in rates of autolysis, production of capsular polysaccharide and teichoic acid, and adherence (15, 27, 39). Several studies have suggested a role for the capsule in attachment by demonstrating that the rate of attachment of pneumococci to eukaryotic cells is inversely proportional to the presence and size of the polysaccharide capsule (1, 30, 32). Furthermore, Weiser et al. described two distinct colony morphology variants that switched between a transparent form, facilitating adherence and carriage, and an opaque form that poorly adhered but was better adapted to evade the host immune response during inflammation or invasive infection (16, 37). The opaque form was shown to produce two to six times more capsular polysaccharide than the transparent form (15). Differences in the production of capsular polysaccharide between opaque and transparent variants were accentuated by changes in the environmental concentration of oxygen; decreased oxygen levels correlated with an increase in CPS and expression of cps-encoded proteins (38).
Changes in the environmental concentration of oxygen are commonly noted in biofilms, sessile communities composed of aggregates of cells that are encased in an extracellular polymeric matrix. Waite et al. (33) demonstrated that S. pneumoniae serotype 3 biofilms grown in a Sorbarod reactor gave rise to small revertable acapsular colony variants. The high-frequency phase variation generated during biofilm growth was attributed to random tandem duplications within the cps3D gene (33).
The formation of colony variants during biofilm growth has also been described in other pathogenic bacteria, including Pseudomonas aeruginosa. Unlike the colony variants reported for S. pneumoniae (33), the phenotype of these laboratory-derived, spontaneous variants has been shown to be heritable, suggesting that genetic changes produced them (5, 6, 11, 12, 17). Webb et al. (34, 35) reported the emergence of a small colony variant (SCV) during P. aeruginosa biofilm development that exhibited enhanced attachment, accelerated biofilm development, and accelerated biofilm detachment. Boles et al. discovered two variants, termed “mini” and “wrinkly,” that arose spontaneously from biofilms at a high frequency (5). The wrinkly variant exhibited reduced detachment rates but increased attachment and cell cluster formation rates, while the mini variant exhibited a hyperdetaching phenotype by a mechanism requiring the biosurfactant rhamnolipid (5, 6). Furthermore, colony variants such as SCVs have been shown to provide a selective advantage under biofilm conditions by displaying biofilm-related phenotypes, including hyperadherence, autoaggregation, increased hydrophobicity, and reduced swimming and twitching motilities (5, 7, 17). Interestingly, similar colony variants have been detected in sputum and deep throat swab samples from cystic fibrosis patients (11, 12).
Overall, the ability of biofilm bacteria to produce colony variants with specialized functions has been suggested to be a survival strategy, ensuring tolerance to a wide variety of environmental conditions (5). Here, we demonstrate the isolation of at least three colony morphology variants, including an SCV, from aging S. pneumoniae biofilms that differ in colony size, capsule production, and adhesion. The small colony variant displayed biofilm-related phenotypes, including autoaggregation, hyperadherence, and hyper-cluster formation. The molecular basis of the SCV acapsular phenotype was attributed to an ∼7-kbp deletion located within the cps3DSU operon.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All strains used in this study are listed in Table 1. All Streptococcus pneumoniae strains were grown in Todd-Hewitt broth (Acumedia; Neogen Corporation, Lansing, MI) or on Trypticase soy agar (TSA II) supplemented with 5% sheep's blood (Becton Dickinson, Sparks, MD) at 37°C in 5% CO2. The laboratory-derived colony morphology variants were generated, as described below, in a continuous flow tube reactor.
TABLE 1.
Streptococcus pneumoniae strains and colony morphology variants used in this study
| Strain or colony variant | Capsule serotype | Colony appearance on blood agar | Colony diameter (mm) | Source or reference |
|---|---|---|---|---|
| ATCC 6303, WT | 3 | Large mucoid | 4-5 | 2 |
| BS68 | 9 | Small mucoid | 1-1.5 | 2 |
| BS69 | 14 | Small mucoid | 1-1.5 | 2 |
| BS70 | 11 | Small mucoid | 1-1.5 | 2 |
| BS71 | 3 | Large mucoid | 4-5 | 2 |
| BS72 | 23 | Small mucoid | 1-1.5 | 2 |
| BS73 | 6 | Small mucoid | 1-1.5 | 2 |
| BS74 | 18 | Small mucoid | 1-1.5 | 2 |
| BS75 | 19 | Small mucoid | 1-1.5 | 2 |
| LMV | Large mucoid | 4-5 | This study | |
| MMV | Medium mucoid | 2.5-3 | This study | |
| SMV | Small mucoid | 1-1.5 | This study | |
| SCV | Small nonmucoid | 1 | This study |
Initial attachment in a 96-well microtiter dish assay.
Initial biofilm formation was measured by using the microtiter dish assay system, as described previously (31). Briefly, microtiter wells were inoculated with 20 μl of an S. pneumoniae culture grown in Todd-Hewitt broth to mid-logarithmic phase (turbidity of ∼0.5 at 600 nm). The cells were grown for 3, 6, and 12 h before they were stained with crystal violet and quantified.
Biofilm formation using a continuous flow tube reactor.
S. pneumoniae serotype 3 (ATCC 6303 and clinical isolate BS71) (Table 1) biofilms were grown in 0.2×-diluted Todd-Hewitt medium (6 g/liter) in a once-through continuous flow tube reactor system as previously described (2). Biofilms were incubated at 37°C in 5% CO2 for 1, 3, 6, and 9 days under flowing conditions. The biofilm architecture was studied by using an inverted confocal laser scanning microscope (CLSM) from Zeiss (Heidelberg, Germany) as described previously (2, 28). For visualization, S. pneumoniae serotype 3 (ATCC 6303) biofilms were stained using the Live/Dead BacLight stain from Invitrogen (Carlsbad, CA). Quantitative analysis of epifluorescence microscopic images obtained at the 3- and 6-day time points was performed with COMSTAT image analysis software (14). To ensure reproducibility, each experiment was performed in triplicate and COMSTAT analysis was carried out from a minimum of six image stacks.
Evaluation of colony variance during Streptococcus pneumoniae biofilm development.
A once-through reactor, as described above, was used to evaluate and quantitate the emergence of colony morphology variants associated with S. pneumoniae serotype 3 biofilm formation (ATCC 6303 and clinical isolate BS71 [Table 1]). Biofilm biomass was harvested from a tube biofilm reactor, resuspended in saline (total volume of 1 ml), homogenized for 30 s to disrupt cell clusters, serially diluted, and plated on blood agar. Colonies were scored as either mucoid or nonmucoid based on their appearance on blood agar and categorized according to diameter as large, medium, or small variants (Table 1). Experiments for each time point were repeated at least three times. To determine whether the variants were stable mutants, well-isolated colony morphology variants (large mucoid variant [LMV], medium mucoid variant [MMV], small mucoid variant [SMV], and SCV) were restreaked on blood agar (and/or subcultured in Todd-Hewitt medium and subsequently plated onto blood) and incubated for 24 h. This was repeated six times, and reversion with respect to colony size and mucoidy was monitored. The experiments were repeated for the wild type (WT; ATCC 6303).
Capsule quantification of S. pneumoniae serotype 3 colony variants.
The relative amount of capsule for each colony variant was determined using the Stains-All reagent as described by Schrager (29). Absorbance was measured at 640 nm, and values were subsequently compared with a standard curve generated using known concentrations of pneumococcal serotype 3 polysaccharide (ATCC, Manassas, VA). In addition, the total protein concentration of the original bacterial suspension was determined by the modified method of Lowry (23) using reagents from Sigma. Bovine serum albumin was used as the standard.
Microbial adhesion to hydrocarbon (MATH) test.
To compare the relative hydrophobicities of S. pneumoniae wild type and colony variants, cell adherence to hexadecane was determined as described by Deziel et al. (7). Briefly, exponential-phase cultures grown in Todd-Hewitt medium were washed twice with saline and resuspended in saline. The resulting suspension was diluted to a turbidity of 0.5 (600 nm), and 2-ml aliquots were mixed with increasing volumes (100 to 800 μl) of xylene, vortexed for 120 seconds, and incubated at room temperature for 30 min, after which time the turbidity of the aqueous phase was recorded at 600 nm and the percent hydrophobicity was calculated.
Disk susceptibility tests of planktonic cells.
Susceptibilities of planktonic cells were determined by the Kirby-Bauer technique as described by Bauer et al. (4), except that TSA II agar supplemented with 5% sheep's blood was used. Triplicate sets of inoculated plates, containing various antibiotic-impregnated filter paper disks, were prepared by the Kirby-Bauer technique. The following antibiotics were tested: amoxicillin (30 μg/disk), tetracycline (30 μg/disk), tobramycin (10 μg/disk), bacitracin (10 U/disk), polymyxin B (300 U/disk), nalidixic acid (30 μg/disk), gentamicin (10 μg/disk), erythromycin (15 μg/disk), and rifampin (5 μg/disk).
Biofilm susceptibility test.
To determine whether colony variance is affected by antimicrobial treatment, S. pneumoniae wild-type (ATCC 6303), LMV, and SCV biofilms were grown in tube reactors as described above for 1 and 6 days and subsequently treated with tetracycline (10 μg/ml) for 6 and 12 h, respectively. Tetracycline was chosen because S. pneumoniae was found to be susceptible to tetracycline. This was done by exposing biofilms to tetracycline in the same reactor in which they were grown by switching the flow from Todd-Hewitt medium to Todd-Hewitt medium containing tetracycline (flow rate of 0.0348 ml/min). After 6 or 12 h, biofilms were removed from the biofilm reactor, and total CFU were determined as described above. Experiments for each time point were repeated at least three times. Untreated biofilms were used as controls. To ensure that tetracycline was stable under the conditions tested, disk diffusion assays using Escherichia coli were performed. Tetracycline-containing medium obtained prior to and upon completion of the experiment (6 and 12 h of incubation time at 37°C) was used in the diffusion assays to impregnate filter paper disks. No significant difference was detected (not shown). Furthermore, S. pneumoniae wild-type (ATCC 6303) biofilms were grown in once-through flow cells as described above to monitor the effect of antimicrobial treatment on biofilm architecture and to visualize the zone of killing. Biofilms were grown for 6 days, after which time the biofilms were exposed to tetracycline (10 μg/ml) or tobramycin (10 μg/ml) as described above. After 12 h, biofilms were stained using the Live/Dead BacLight stain from Invitrogen (Carlsbad, CA) and viewed by CLSM. Biofilms were removed from the flow cells, homogenized, serially diluted, and plated for total CFU. Untreated biofilms were used as controls. Experiments for each time point were repeated at least three times.
Analysis of the cps3DSUM operon.
To determine whether the molecular basis of colony variants was based on random insertion or duplication within the cps3D gene, the cps3D gene was amplified under standard conditions and the internal primer pair cps3Dint (5′-TACTCCGACTAATTATGATGTAG, 5′-TTACCTCGCTATATGTATCTATCT) as previously described (33). Genomic DNA for cps3D PCR analysis was obtained using the DNA isolation kit from MO BIO Laboratories, Inc. (Carlsbad, CA). To map the deletion within the cps3DSUM operon of SCVs, PCR was used. SCV genomic DNA obtained from four independent experiments was purified as described above. PCR was conducted using the following parameters: 94°C for 5 min; 35 cycles of 94°C for 25 s, 41 to 58°C for 30 s, and 72°C for 50 s; followed by a final extension step at 72°C for 5 min. The following primer pairs (based on the sequence for U15171) were used: cps3-promoter (5′-CACTTTTTGACAGAG, 5′-GACCTTAACTTCATG; 1,083 bp), cps3-promoter2 (5′-CAAAGCTGATACTAAG, 5′-CTTTATAAACGTGTGC, 284 bp), cps3D (5′-GGTATACTAAAGAAG, 5′-CCAAGACAATTATTG, 423 bp), cps3Dint (5′-TACTCCGACTAATTATGATGTAG, 5′-TTACCTCGCTATATGTATCTATC; 440 bp), cps3DS (5′-AGTAACACTTACTTGG, 5′-AGGACTTATATCTGAC; 715 bp), cps3S (5′-GTTCTGTAATTATCCCTGTCGTG, 5′-CTAAAAGCAATTGTTCGACCAGG; 502 bp), cps3U (5′-GGCAAAAGAAATGCTTCC, 5′-GTTGCGTTGTAATCATCC; 390 bp), cps3M (5′-GACACCTATGAATTGT, 5′-GTAGGCTTTCAAAAAC; 368 bp). Genomic DNA isolated from S. pneumoniae WT (6303), LMV, MMV, and SMV was used as controls. To determine whether the deletion was identical in all SCVs obtained from four independent biofilm experiments, PCR was performed using primers that amplified a fragment located between dexB and cps3M (see U15171; dexB, 5′-TATTGTCTCAATCTGG; cps3M-del, 5′-GTATCAATAGCATCTG). The same primers were used to characterize the deletion in two independent SCVs obtained from clinical isolate BS71 in vitro biofilms.
The identity of the PCR product was confirmed by DNA sequencing using the BigDye Terminator sequencing kit (Applied Biosystems) and the ABI Prism sequencer (Applied Biosystems).
RESULTS
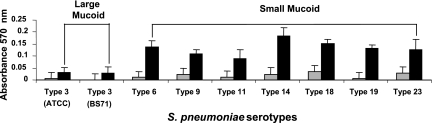
We recently characterized biofilm development of 14 different Streptococcus pneumoniae strains representing 10 unique serotypes and categorized them into three distinct biofilm architectural groups (2). However, the distinction was not apparent when initial attachment was analyzed using a 96-well plate assay. Both S. pneumoniae serotype 3 strains, ATCC 6303 and BS71, showed significantly reduced levels of attachment compared to the other pneumococcal strains (Fig. 1). Based on our findings, we hypothesized that the difference in initial attachment was based on capsule amount, since S. pneumoniae serotype 3 colonies produced large mucoid colonies on blood agar compared to the other tested serotypes (Table 1).
FIG. 1.
Initial biofilm attachment of different S. pneumoniae capsule serotypes. Various S. pneumoniae capsule serotypes were allowed to form biofilms in polystyrene microtiter plates for 6 (gray bars) and 12 h (black bars), after which time the attached biomass was stained with crystal violet, solubilized with ethanol, and subsequently measured at 570 nm. The capsule serotypes are grouped as large mucoid or small mucoid according to their appearance on blood agar.
Biofilm growth of S. pneumoniae serotype 3 produces colony morphology variants.
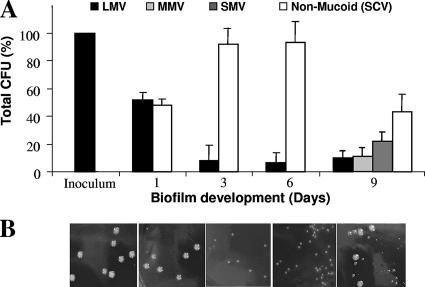
We therefore analyzed the colony morphologies of S. pneumoniae serotype 3 (ATCC 6303) biofilms soon after attachment and over the course of biofilm development. While the inoculum appeared to be composed of uniform mucoid colonies roughly 5 mm in size, S. pneumoniae biomass harvested from a tube biofilm reactor produced two prevalent colony morphologies: the wild-type-like colony morphology, with its mucoid appearance, and a small, nonmucoid colony morphology variant (SCV) (Fig. 2). On solid growth medium, the SCVs formed colonies that were roughly 1 mm in size and appeared nonmucoid. SCVs were detectable within 1 day of biofilm growth and represented approximately 50% of the bacterial population. However, in some instances (3 out of 10), SCVs accounted for less than 20% of the total biofilm population or were absent in some of the harvested biofilms. Following 3 and 6 days of biofilm growth, up to ∼80% of the biofilm population was composed of SCVs (Fig. 2). Interestingly, the later biofilm developmental stage (9 days) was characterized by a diverse population of mucoid variants discernible based on colony diameter: LMV, MMV, and SMV (Table 1). In some instances, these mucoid variants were detectable as early as 1 day of biofilm growth (not shown). Similar results were obtained for the clinical S. pneumoniae serotype 3 isolate, BS71 (data not shown).
FIG. 2.
Emergence of colony morphology variants over the course of S. pneumoniae serotype 3 biofilm development. (A) Distribution of colony variants was determined from total CFU, colony size, and mucoidy on blood agar. (B) Appearance of colony variants on solid medium (from left, inoculum, planktonic growth conditions after 1, 3, 6, and 9 days of biofilm growth).
All variants showed similar growth rates (data not shown). The colony variants were found to be stable genotypic variants; no revertants were detected in liquid or on solid medium, not even after six consecutive passages. Similar results were found upon subculturing the S. pneumoniae wild-type strain. To exclude the possibility that the variants were already present in the wild-type population, wild-type cells grown planktonically were serially diluted and plated, and the colony morphology was evaluated. Under the conditions tested, S. pneumoniae wild type was found to only give rise to large mucoid colonies (Table 1). The finding suggests that variants do not form in liquid culture inoculated with the wild type but only when grown as biofilms.
The SCV has a hyper- and autoaggregating phenotype.
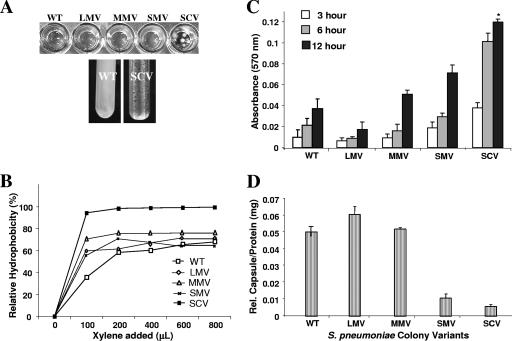
The emergence of SCVs within 1 day of biofilm growth indicated a role in initial attachment. This was further supported by a strong aggregative phenotype in liquid culture of SCVs, including autoaggregation and hyperadherence to both hydrophilic and hydrophobic surfaces (Fig. 3A). Similar observations were made for SCVs obtained from the clinical isolate BS71, although the variants' aggregative phenotype was less pronounced (not shown). The aggregative phenotype was not due to increased sedimentation and/or increased S. pneumoniae chain length, since no difference in chain length between LMVs and SCVs grown in liquid was observed by microscopy (data not shown). Interestingly, we observed small clusters formed by SCV that were >50 μm in diameter. The cohesiveness of SCV cells suggested an increase in hydrophobicity, and so relative hydrophobicity of SCV was evaluated using the MATH test (7). As shown in Fig. 3B, the SCV phenotype was more hydrophobic than that of the WT (P < 0.05). In contrast, no significant difference in the relative hydrophobicity was detected between the mucoid variants and the wild type.
FIG. 3.
Liquid culture phenotype, relative hydrophobicity (MATH), microtiter plate adhesion assay, and relative capsule concentration of S. pneumoniae WT and colony variants. (A) Hyperadherence and hyperautoaggregation of SCV in liquid cultures. The WT showed homogeneous growth in shaken liquid culture, as opposed to autoaggreation, seen for SCV. (B) Adherence to hexadecane as a measure of relative hydrophobicity (MATH) of colony morphology variants. Each datum point is based on the results from three replicate measurements. SCVs were significantly more hydrophobic than WT and the mucoid variants (P < 0.05). (C) Microtiter plate adhesion assay. SCV showed significantly greater biofilm adhesion than did WT, LMV, or MMV (P < 0.01). Initial attachment of S. pneumoniae colony variants was analyzed after 3, 6, and 12 h. (D) Relative capsule concentration of S. pneumoniae WT and colony variants as determined by using the Stains-All reagent.
SCV was also characterized by its hyperadherence to a hydrophobic surface compared to the wild type and mucoid variants. In a microtiter plate adhesion assay, the SCV showed a four- and sixfold increase in crystal violet-stainable biofilm biomass compared to the WT and LMV, respectively (P < 0.05). MMVs were able to attach more efficiently than LMVs but less than SMVs (Fig. 3C).
The increase in initial attachment correlated with a decrease not only in the colony diameter but also in the relative amount of capsule present on the cell surface of each colony variant (Fig. 3D). Among the colony morphology variants, the LMVs were found to produce the highest relative amount of capsule, followed by MMV and SMV. Interestingly, the amount of capsule produced by SMV was similar to that produced by S. pneumoniae serotype 19, which forms small mucoid colonies on blood agar (Fig. 3D; Table 1). The least amount of capsule was produced by SCV. Overall, the findings indicate that the rate of initial attachment was inversely proportional to the colony diameter (Fig. 3C) and, thus, the amount of capsule produced (Fig. 3D), confirming the hyperadhering and autoaggregating phenotype associated with the SCV.
Colony variants and biofilm architecture.
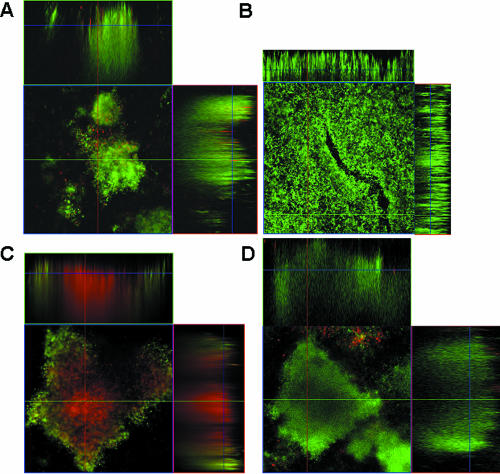
To further explore differences between the colony variants, the in vitro biofilm architecture of two colony variants, SCV and LMV, was examined after 3 days of biofilm growth using a flow cell biofilm reactor system. SCV formed biofilms composed of only large microcolonies (>40 μm) after 3 days of growth (Fig. 4A). The microcolonies were characterized by having defined borders and being separated by an extensive network of channels. Almost no cells were detectable at the substratum surrounding the microcolonies, indicating the tendency of SCVs to stay near the point of initial attachment in a biofilm. The finding suggests that SCVs may be involved in microcolony and cell cluster formation. In contrast, biofilms formed by LMV appeared unstructured and composed of a thick uniform layer of cells (Fig. 4B). Further quantitative analysis using COMSTAT (14) indicated that the biomass and average thickness were found to be similar in both variant biofilms (Table 2). However, SCV biofilms differed from LMV biofilms with respect to maximum thickness and roughness coefficient (a measure of biofilm heterogeneity) (Table 2). These findings are consistent with the distinct three-dimensional architecture composed of large microcolonies and defined channels observed for the SCV colony variant biofilm (Fig. 4A) and the decreased prevalence of channels and the poorly defined, unstructured architecture in LMV biofilms (Fig. 4B). Interestingly, wild-type biofilms of the same age (3 days old) were characterized by the presence of small cell clusters (∼10 μm), low biomass accumulation, sparsely covered substratum, and heterogeneity with respect to biofilm height (Table 2). At day 6, S. pneumoniae wild-type biofilms were composed of large microcolonies similar to the ones seen in 3-day-old SCV biofilms (Fig. 4D). The maximum height was comparable to that seen for SCV biofilms at day 3 (Fig. 4A; Table 2), while the average biofilm thickness and biomass were twofold higher than in either colony variant biofilm. S. pneumoniae wild-type biofilms (6 days) were also characterized by a lower roughness coefficient compared to SCV biofilms. The difference in biofilm architecture between the two variants and the wild-type biofilms at the 3-day time point suggested that biofilm development in the colony variants may be accelerated. The data also suggested that both colony variants may contribute to the overall biofilm architecture of S. pneumoniae in vitro.
FIG. 4.
Biofilm architecture of colony variants LMV and SCV and S. pneumoniae WT biofilms before and after treatment with tetracycline. (A) Three-day-old biofilm of SCV; (B) 3-day-old biofilm of LMV; (C) 6-day-old S. pneumoniae WT biofilm after treatment with tetracycline (10 μg/ml) for 12 h; (D) untreated 6-day-old S. pneumoniae WT biofilm. The CLSM images show the x-y and x-z planes. Flow cell experiments were performed in triplicate as described in Materials and Methods.
TABLE 2.
Quantitative analysis of biofilm structure formed by Streptococcus pneumoniae wild type and colony variants
| Strain | Biomass (μm3/μm2) | Thickness (μm)
|
Roughness coefficient | |
|---|---|---|---|---|
| Avg | Maximum | |||
| WT (3 days)a | 1.531 (2.0) | 4.62 (2.5) | 19.2 (3.2) | 1.46 (0.1) |
| WT (6 days)b | 78.96 (2.2) | 88.54 (4.8) | 119.88 (5.2) | 0.52 (0.09) |
| LMVa | 30.4 (5.05) | 43.1 (6.1) | 61.9 (6.6) | 0.36 (0.22) |
| SCVa | 26.4 (9.2) | 33.3 (4.9) | 159.6 (4.7) | 1.65 (0.16) |
Values are means of data from six z-series image stacks for each strain/variant taken at day 3. Values in parentheses indicate standard deviations.
Values are means of data from six z-series image stacks for each strain/variant taken at day 6. Values in parentheses indicate standard deviations.
Distribution of colony variants in biofilms is altered by treatment with antimicrobial agents.
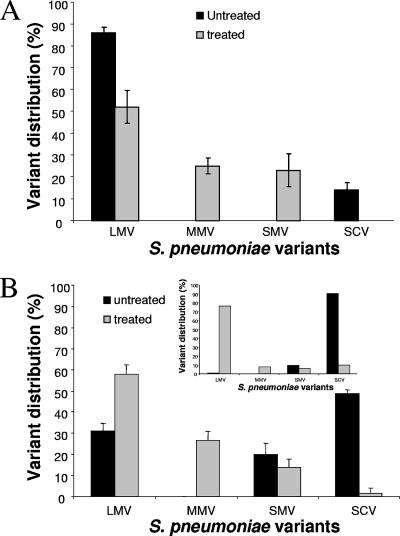
To further explore differences in biofilm characteristics among colony morphology variants, we determined whether colony variants were differentially affected by antimicrobial treatment. No differences in susceptibility were observed for variants grown on solid agar medium using a modified Kirby-Bauer test (not shown). Similarly, no significant difference was observed in susceptibility of variants when grown under biofilm growth conditions. Treatment of 1-day-old biofilms of both LMV and SCV with tetracycline for 6 h under once-through flow conditions resulted in a 1-log reduction of the viable counts. No revertants were detected in treated LMV and SCV biofilms (not shown). However, upon treatment of 1-day-old wild-type biofilms with tetracycline, we observed an alteration in the distribution of colony variants. While untreated biofilms were composed of LMV and SCV (Fig. 5A), the biofilm population after treatment with tetracycline for 6 h was comprised of only mucoid variants (52% LMV [±7.5%], 25% MMV [±3.7%], and 23% SMV [±7.5]) (means ± standard deviations). Interestingly, no SCVs were detected (Fig. 5A). Furthermore, treatment of 6-day-old wild-type biofilms with tetracycline for 12 h resulted in biofilms in which less than 2% were SCVs (Fig. 5B). This is in contrast to the untreated 6-day-old biofilm population, in which more than 50% was comprised of SCVs (Fig. 5B). Similar results were obtained after treatment with tobramycin (data not shown).
FIG. 5.
Colony variant distribution in S. pneumoniae biofilms is affected by treatment with tetracycline. Biofilms were grown in tube reactors for 1 day before treatment with tetracycline (10 μg/ml) for 6 h (A) or for 6 days before treatment with tetracycline (10 μg/ml) for 12 h (B). Inset: distribution of colony variants in 6-day-old S. pneumoniae WT biofilm grown in flow cells before and after treatment with tetracycline. Distribution of colony variants was assessed by total CFU, colony size, and mucoidy on blood agar. Experiments were performed in triplicate.
To determine the effect of antimicrobial treatment on the population distribution and to visualize which area of the biofilms was most affected, 6-day-old S. pneumoniae wild-type biofilms were treated with tetracycline for 12 h, stained using the Live/Dead stain, and subsequently viewed by CLSM. As shown in Fig. 4C, treatment with tetracycline affected the inside of biofilms, as indicated by the interior portion of microcolonies being stained red (an indication for dead cells), while the outer layer of the biofilms was unaffected (as indicated by live cells stained green). In contrast, only a few dead cells stained red were detectable in untreated 6-day-old biofilms (Fig. 4D). Similar results were obtained after treatment with tobramycin (data not shown). To correlate the results obtained by Live/Dead staining with CFU results, treated and untreated biofilms were harvested from the flow cells upon acquisition of image stacks, homogenized, serially diluted, and plated on blood agar. Treatment of wild-type biofilms with tetracycline resulted in a ∼2-log reduction of the viable counts. Furthermore, while more than 90% of the untreated biofilm population was composed of SCVs, only 1.5 to 3% of the treated biofilm population was composed of SCVs (Fig. 5B, inset). The remaining population was mainly composed of LMV (70 to 90%) as well as MMV and SMV (Fig. 5B, inset). Similar results were obtained after treatment with tobramycin (data not shown).
The results suggest that antimicrobial treatment of wild-type biofilms for 12 h mainly affected the interior of microcolonies and resulted in the reduction of SCV from 50 to 90% of the entire population in untreated to less than 3% in treated biofilms. However, susceptibility of SCV biofilms was similar to that of LMV biofilms, and no revertants were detected in variant biofilms upon treatment.
Colony variance is independent of phase variation within the cps3D gene.
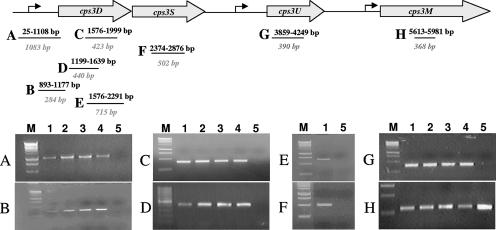
Previous reports indicated that the molecular basis for the formation of acapsular colonies or small colony variants obtained under biofilm conditions was due to random duplications within cps3D (33). We therefore analyzed the cps3D locus of the colony variants for random duplications or insertions by PCR using primers described by Waite et al. (33). Interestingly, none of the mucoid variants was found to carry an insertion within the cps3D gene (Fig. 6D). In addition, no PCR product was detectable for the SCV cps3D locus under the conditions tested (Fig. 6D), suggesting that the molecular basis for the SCV was due to a cps3D deletion rather than a random insertion. To further pursue this possibility, PCR was used to determine the extent to which the cps locus was deleted in all SCV clones obtained from four independent experiments. A total of eight oligonucleotide pairs were used to amplify regions within the cps3DSUM operon as well as the promoter region (Fig. 6). Genomic DNAs obtained from the WT and the mucoid variants were used as controls. By doing so, we found that the SCV not only lacked cps3D but also both the cps3S and cps3U genes as well as a 1-kbp fragment comprising the cap3 promoter region (Fig. 6A to G). However, we were able to detect cps3M (Fig. 6H). The identity of the PCR products was confirmed by DNA sequencing. Our finding of the cps3DSU deletion is consistent with the observation that SCVs do not revert to the capsular phenotype upon subculturing in liquid culture and on solid media.
FIG. 6.
Mapping of the cps3DSU deletion in the genome of SCV by PCR. Genomic DNA of WT and mucoid variants was used as a control. The PCR product was amplified using the following primers: (A) cps3-promoter (1,083-bp PCR product); (B) cps3-promoter2 (284 bp); (C) cpsD (342 bp); (D) cps3Dint (440 bp); (E) cps3DS (715 bp); (F) cps3S (502 bp); (G) cps3U (390 bp); (H) cps3M (368 bp). Genomic DNAs isolated from S. pneumoniae wild type (6303), LMV, MMV, and SMV were used as controls. Lanes: 1, S. pneumoniae wild type (6303); 2, LMV; 3, MMV; 4, SMV; 5, SCV; M, DNA ladder (for PCR products shown in panels A to C and E to H, a 1-kb DNA ladder was used; for PCR products shown in panel D, a 100-bp DNA ladder was used).
Emergence of SCV during biofilm formation is based on a >7-kb deletion.
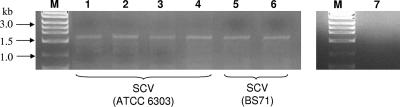
To determine whether the deletion in SCVs was a random or precise event, we isolated DNA from SCVs obtained from four independent biofilm experiments and PCR amplified a DNA fragment located between dexB and cps3M. The total length of the PCR product in the wild type was calculated to be ∼9.5 kb (see accession no. U15171). However, a PCR product of only ∼1.8 kb was obtained for all four SCVs (Fig. 7), indicating that the deletion consistently comprised a >7-kb region of the SCV chromosome. The identity of the PCR product was confirmed by DNA sequencing using the cps3M-del primer. It is worth noting that amplification of the region located between dexB and cps3M using DNA obtained from three independent BS71 SCV isolates yielded PCR products of similar size (Fig. 7). In contrast, no PCR product was obtained for wild-type genomic DNA under the conditions tested (Fig. 7, lane 7). The lack of a PCR product in the wild type suggests that SCV was not present in the S. pneumoniae wild-type population.
FIG. 7.
Survey of SCVs obtained by PCR from independent biofilm growth experiments. Genomic DNA from SCVs obtained from four independent S. pneumoniae ATCC 6303 biofilm experiments (lanes 1 to 4) and from two independent S. pneumoniae BS71 biofilm experiments (lanes 5 and 6) was amplified using dexB and cps3M-del primers. Genomic DNA of S. pneumoniae ATCC 6303 grown planktonically was used as a control (lane 7). Lane M, 1-kb DNA ladder.
DISCUSSION
In this study, we report that in vitro biofilm growth of S. pneumoniae serotype 3 selects for colony morphology variants differing in colony diameter, mucoidy, autoaggregation, initial attachment, hydrophobicity, capsule production, and biofilm formation (Fig. 3 and 4). Colony morphology variants similar to the parental strain (LMV) were found to decrease soon after initial attachment. After 3 days of biofilm growth, ∼10% of the biofilm population consisted of LMVs. The LMV population remained at this low level even after extended biofilm growth. Although the LMV appeared on solid medium to be similar to the WT, LMV isolated from biofilms showed phenotypic characteristics that were distinct from the WT, including reduced initial attachment, increased capsule production, and tendency to spread over surfaces and, thus, to only form flat unstructured biofilms (Fig. 3 and 4). Interestingly, the dominating colony variant to emerge under in vitro once-through biofilm growth flow conditions was SCV (Fig. 2). The acapsular variant was characterized by increased adherence in initial attachment assays and hyper-/autoaggregation in liquid, its tendency to stay near the point of initial attachment at the substratum in a biofilm, and its ability to form large microcolonies under biofilm growth conditions (Fig. 3 and 4A; Table 2). SCV formation coincided with a deletion comprising the cps3DSU operon necessary for capsule formation (Fig. 6 and 7). While it is unclear how SCVs arise, the variant was not detectable in the wild-type population (Fig. 7).
Why do SCVs arise in a biofilm community? Certainly, under in vivo conditions, loss of capsule would be considered a double-edged sword for the pneumococcus during intimate contact with host cells. A reduction in the amount of capsular material might strongly enhance adherence and uptake, but the reduced amount of capsule might convert the pneumococcus into a more apathogenic state in terms of its ability to evade the immune system (10, 18). One possibility is that the colony variants (SCVs) successfully compete in a particular biofilm niche. Rainey and Travisano (24) noted the appearance of colony morphology variants in standing liquid cultures of Pseudomonas fluorescens but not in shaken liquid cultures. They proposed and ultimately demonstrated that these variants were adapted to specific niches in the standing liquid culture. Kirisits et al. isolated sticky variants in standing liquid cultures and in biofilms but not in a shaken liquid culture inoculated with WT, suggesting that the variant may have a competitive advantage in a particular niche of a structured system, such as a biofilm (17). Similarly, we did not isolate SCVs from liquid cultures inoculated with WT but only from biofilms. Furthermore, biofilms are characterized by high cell densities containing both chemical and physical gradients that may affect variations in capsule production or contribute to increased mutation rates. Interestingly, changes in availability of oxygen have been shown to accentuate differences in capsular polysaccharide expression in S. pneumoniae (38), and endogenous hydrogen peroxide production has been demonstrated to influence the frequency of spontaneous mutations in pneumococcal genes (21, 22).
Two other in vitro biofilm growth conditions have been shown to give rise to SCVs. Waite et al. (33) reported that S. pneumoniae serotype 3 biofilms grown in a Sorbarod reactor gave rise to small revertable acapsular colony variants with tandem sequence duplications in cps3D. In contrast to the tube reactors or flow cells used in this study, Sorborad filters provide a high surface-to-bulk liquid ratio and are characterized by reduced cell densities and chemical and physical gradients. McEllistrem et al. (19) recently reported the emergence of phase variants upon extended growth (4 to 7 days) in colony biofilms. Colony biofilms have been described as bacterial colonies behaving like planktonic cells “stranded” on a surface (8). This type of biofilm also differs from Sorbarod filters and the ones used in this study by the lack of flowing conditions and the use of a surface that allows for diffusion. Interestingly, most acapsular phase variants isolated from colony biofilms had mutations in the cps3D gene, the first gene of the capsular operon. While the variants were phenotypically indistinguishable, 11 had single nucleotide polymorphisms (SNPs) in the cps3D gene, 1 had an SNP in the −10 promoter, and 3 had large deletions in cps3D gene (19). It is worth noting that reversion was only observed for variants that were based on SNPs (19). While the molecular mechanisms of phase variation are not well understood, it is apparent from the above-mentioned studies that growth conditions play a significant role in the emergence of colony variants in in vitro biofilms. This is further supported by the drastic change observed in the colony variants distribution in biofilms upon treatment with antimicrobial agents (Fig. 5). Upon treatment, the SCV population was drastically reduced. SCV biofilms were not more susceptible to antimicrobial agents than LMV biofilms, and antimicrobial treatment did not result in the emergence of variants in both LMV and SCV biofilms. The findings indicate that the reduction of SCV in in vitro wild-type biofilms upon treatment for 12 h is not explained by reversion or selective killing of SCVs. We hypothesize that the change in colony variants distribution in biofilms upon treatment may be based on genetic transfer of the capsule biosynthetic operon to SCVs.
Overall, the findings support the notion that SCV may have a competitive advantage in a particular niche within a biofilm. The competitive advantage of SCV under in vitro biofilm growth conditions may be due to its hyper-/autoaggregating phenotype and its ability to form large cellular aggregates or microcolonies (Fig. 3 and 4A; Table 2). These characteristics may provide SCV with a selective advantage compared to mucoid variants under flowing conditions, in the absence of antimicrobial agents.
In conclusion, we demonstrated that in vitro S. pneumoniae biofilm formation correlated with the formation of colony variants, suggesting that biofilm growth may select for distinctive subpopulations with their own specific biofilm-related phenotypes, with colony morphology variation being just one example of a distinctive subpopulation. However, whether the appearance of these variants in in vitro biofilms correlates with the establishment of chronic pneumococcal infections remains to be determined.
Acknowledgments
This work was supported by grants from the National Institutes of Health (RO1 DC005659, subcontract) and the National Science Foundation (DBI-0321046).
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Adamou, J. E., T. M. Wizemann, P. Barren, and S. Langermann. 1998. Adherence of Streptococcus pneumoniae to human bronchial epithelial cells (BEAS-2B). Infect. Immun. 66:820-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allegrucci, M., F. Z. Hu, K. Shen, J. Hayes, G. D. Ehrlich, J. C. Post, and K. Sauer. 2005. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 188:2325-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austrian, R. 1981. Some observations on the pneumococcus and the current status of pneumococcal disease and its prevention. Rev. Infect. Dis. 3:S1-S17. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, A. W., D. M. Perry, and W. M. M. Kirby. 1959. Single disc antibiotic sensitivity testing of staphylococci. Arch. Int. Med. 104:208-216. [DOI] [PubMed] [Google Scholar]
- 5.Boles, B. R., M. Thoendel, and P. K. Singh. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA 101:16630-16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boles, B. R., M. Thoendel, and P. K. Singh. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 57:1210-1223. [DOI] [PubMed] [Google Scholar]
- 7.Déziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammerschmidt, S., A. Muller, H. Sillman, M. Muhlenhoff, R. Borrow, et al. 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol. Microbiol. 20:1211-1220. [DOI] [PubMed] [Google Scholar]
- 10.Hammerschmidt, S., S. Wolff, A. Hocke, S. Rosseau, E. Muller, and M. Rohde. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect. Immun. 73:4653-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Häußler, S., B. Tümmler, H. Weißbrodt, M. Rohde, and I. Steinmetz. 1999. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29:621-625. [DOI] [PubMed] [Google Scholar]
- 12.Häußler, S., I. Ziegler, A. Löttel, F. V. Götz, M. Rohde, D. Wehmhöhner, S. Saravanamuthu, B. Tümmler, and I. Steinmetz. 2003. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J. Med. Microbiol. 52:295-301. [DOI] [PubMed] [Google Scholar]
- 13.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heydorn, A., B. K. Ersboll, M. Hentzer, M. R. Parsek, M. Givskov, and S. Molin. 2000. Experimental reproducibility in flow-chamber biofilms. Microbiology 146:2409-2415. [DOI] [PubMed] [Google Scholar]
- 15.Kim, J., and J. Weiser. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177:368-377. [DOI] [PubMed] [Google Scholar]
- 16.Kim, J. O., S. Romero-Steiner, U. B. Sorensen, J. Blom, M. Carvalho, S. Barnard, G. Carlone, and J. N. Weiser. 1999. Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae. Infect. Immun. 67:2327-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirisits, M. J., L. Prost, M. Starkey, and M. R. Parsek. 2005. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71:4809-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magee, A. D., and J. Yother. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect. Immun. 69:3755-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McEllistrem, M. C., J. V. Ransford, and S. A. Khan. 2006. Characterization of in vitro biofilm-associated pneumococcal phase variants of a clinically relevant serotype 3 clone. J. Clin. Microbiol. 45:97-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Overweg, K., C. D. Pericone, G. G. Verhoef, J. N. Weiser, H. D. Meiring, A. P. De Jong, R. DeGroot, and P. W. M. Hermans. 2000. Differential protein expression in phenotypic variants of Streptococcus pneumoniae. Infect. Immun. 68:4604-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pericone, C. D., D. Bae, M. Shchepetov, T. McCool, and J. N. Weiser. 2002. Short-sequence tandem and nontandem DNA repeats and endogenous hydrogen peroxide production contribute to genetic instability of Streptococcus pneumoniae. J. Bacteriol. 184:4392-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pericone, C. D., K. Overweg, P. W. Hermans, and J. N. Weiser. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 68:3990-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson, G. L. 1977. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83:346-356. [DOI] [PubMed] [Google Scholar]
- 24.Rainey, P. B., and M. Travisano. 1998. Adaptive radiation in a heterogeneous environment. Nature 394:69-72. [DOI] [PubMed] [Google Scholar]
- 25.Ring, A., J. N. Weiser, and E. I. Tuomanen. 1998. Pneumococcal penetration of the blood-brain barrier: molecular analysis of a novel re-entry path. J. Clin. Investig. 102:347-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romero-Steiner, S., D. Musher, M. Cetron, L. Pais, J. Groover, A. Fiore, B. Plikaytis, and G. Carlone. 1999. Reduction in functional antibody activity against Streptococcus pneumoniae in vaccinated elderly individuals highly correlates with decreased IgG antibody avidity. Clin. Infect. Dis. 29:281-288. [DOI] [PubMed] [Google Scholar]
- 27.Saluja, S. K., and J. N. Weiser. 1995. The genetic basis of colony opacity in Streptococcus pneumoniae: evidence for the effect of box elements on the frequency of phenotypic variation. Mol. Microbiol. 16:215-227. [DOI] [PubMed] [Google Scholar]
- 28.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrager, H. M., J. G. Rheinwald, and M. R. Wessels. 1996. Hyaluronic acid capsule and the role of streptococcal entry into keratinocytes in invasive skin infection. J. Clin. Investig. 98:1954-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selinger, D. S., and W. P. Reed. 1979. Pneumococcal adherence to human epithelial cells. Infect. Immun. 23:545-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Southey-Pillig, C. J., D. G. Davies, and K. Sauer. 2005. Characterization of biofilm-specific protein production patterns in Pseudomonas aeruginosa and their influence on biofilm formation. J. Bacteriol. 187:8114-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talbot, U. M., A. W. Paton, and J. C. Paton. 1996. Uptake of Streptococcus pneumoniae by respiratory epithelial cells. Infect. Immun. 64:3772-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waite, R. D., J. K. Struthers, and C. G. Dowson. 2001. Spontaneous sequence duplication within an open reading frame of the pneumococcal type 3 capsule locus causes high-frequency phase variation. Mol. Microbiol. 42:1223-1232. [DOI] [PubMed] [Google Scholar]
- 34.Webb, J. S., M. Lau, and S. Kjelleberg. 2004. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa. J. Bacteriol. 185:8066-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Giskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacertiol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiser, J. N. 1993. Relationship between colony morphology and the life cycle of Haemophilus influenzae: the contribution of lipopolysaccharide phase variation to pathogenesis. J. Infect. Dis. 168:672-680. [DOI] [PubMed] [Google Scholar]
- 37.Weiser, J. N., R. Austrian, P. K. Sreenivasan, and H. R. Masure. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62:2582-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiser, J. N., D. Bae, H. Epino, S. B. Gordon, M. Kapoor, L. A. Zenewicz, and M. Shchepetov. 2001. Changes in availability of oxygen accentuate differences in capsular polysaccharide expression by phenotypic variants and clinical isolates of Streptococcus pneumoniae. Infect. Immun. 69:5430-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiser, J. N., Z. Markiewicz, E. I. Tuomanen, and J. H. Wani. 1996. Relationship between phase variation in colony morphology, intrastrain variation in cell wall physiology, and nasopharyngeal colonization by Streptococcus pneumoniae. Infect. Immun. 64:2240-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]