Abstract
Haemophilus somnus can be either a commensal of bovine mucosal surfaces or an opportunistic pathogen. Pathogenic strains of H. somnus are a significant cause of systemic disease in cattle. We report the genome sequence of H. somnus 129Pt, a nonpathogenic commensal preputial isolate, and the results of a genome-wide comparative analysis of H. somnus 129Pt, Haemophilus influenzae Rd, and Haemophilus ducreyi 35000HP. We found unique genes in H. somnus 129Pt involved in lipooligosaccharide biosynthesis, carbohydrate uptake and metabolism, cation transport, amino acid metabolism, ubiquinone and menaquinone biosynthesis, cell surface adhesion, biosynthesis of cofactors, energy metabolism, and electron transport. There were also many genes in common among the three organisms. Our comparative analyses of H. somnus 129Pt, H. influenzae Rd, and H. ducreyi 35000HP revealed similarities and differences in the numbers and compositions of genes involved in metabolism, host colonization, and persistence. These results lay a foundation for research on the host specificities and niche preferences of these organisms. Future comparisons between H. somnus 129Pt and virulent strains will aid in the development of protective strategies and vaccines to protect cattle against H. somnus disease.
Haemophilus somnus (Histophilus somni), a member of the family Pasteurellaceae (47), can be either a commensal of bovine mucosal surfaces or an opportunistic pathogen. Commensal H. somnus strains colonize and reside relatively harmlessly in the upper respiratory and reproductive tracts, while pathogenic strains spread systemically and cause diseases such as pneumonia, thrombotic meningoencephalitis, myocarditis, septicemia, arthritis, and abortion (10, 34, 41, 42). Here, we report the genome sequence of H. somnus 129Pt, a commensal preputial isolate. We compared H. somnus 129Pt with the finished genomes of Haemophilus influenzae Rd, an avirulent laboratory strain that can invade certain human epithelial cell lines in culture (12), and Haemophilus ducreyi 35000HP, which causes the human sexually transmitted disease chancroid. H. somnus 129Pt and H. influenzae Rd are moderately related (14, 25), do not cause disease, and colonize mucosal niches in different hosts. Phylogenetic analyses have shown that H. ducreyi is not closely related to members of the genus Haemophilus (1, 14, 30) but is a member of the Pasteurellaceae that colonizes genital mucosal surfaces. Its surface lipooligosaccharides (LOS) are similar in structure to those from Haemophilus and Neisseria strains (8), and it occupies a (mucosal) niche similar to that of H. somnus but in a different host. Therefore, the identification of common genes that these organisms share as well as genes unique to each organism may provide some clues about their host specificities and niche preferences.
Through a genome-wide comparative analysis of H. somnus 129Pt, H. influenzae Rd, and H. ducreyi 35000HP, we found similarities and differences among these organisms in terms of the numbers and compositions of genes involved in metabolism, host colonization, and persistence. Our results provide a foundation of information about the commensal 129Pt strain of H. somnus, revealing clues about its preference for the bovine urogenital tract environment. Our work will be useful in future genomic comparisons with pathogenic isolates to gain insight into virulence mechanisms and potentially lead to new strategies for the prevention and control of diseases caused by H. somnus.
MATERIALS AND METHODS
Sequencing of the H. somnus 129Pt genome.
The random shotgun method was used for sequencing of the genome of H. somnus 129Pt. Large-insert (40-kb), medium-insert (8-kb), and small-insert (3-kb) random sequencing libraries were sequenced for this genome project, with an average success rate of 96% and average high-quality read lengths of 685 nucleotides. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible misassemblies were corrected with Dupfinisher (C. Han, unpublished data) or a transposon bomb of bridging clones (EZ-Tn5 <P6Kyori/KAN-2> Tnp transposome kit; Epicenter Biotechnologies). Gaps between contigs were closed by editing, custom primer walks, or PCR amplification. The completed genome sequence of H. somnus 129Pt contains 59,147 reads, achieving an average of 18-fold sequence coverage per base with an error rate of less than 1 in 100,000.
Annotation.
Gene predictions were obtained using Glimmer (13, 60), and tRNAs were identified using tRNAScan-SE (46). Basic analysis of the gene predictions was performed by comparing coding sequences against the Pfam, BLOCKS, COGS, and ProDom databases. Gene definitions and functional classes were added manually by a team of annotators by using BLAST results in addition to information from the basic analysis.
Sequence analysis.
To obtain lists of common and unique genes in the H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd genomes, we used blastp with a cutoff of 1e−5 to obtain best hits for the proteins of each genome blasted against each other. We used the blast2gi program from the Seals package (69) to format the BLAST results in tabular form.
Nucleotide sequence accession numbers.
The sequences of the H. somnus 129Pt chromosome and plasmid pHS129 have been deposited in the GenBank database under accession numbers CP000436 and CP000019.
RESULTS AND DISCUSSION
General genome features.
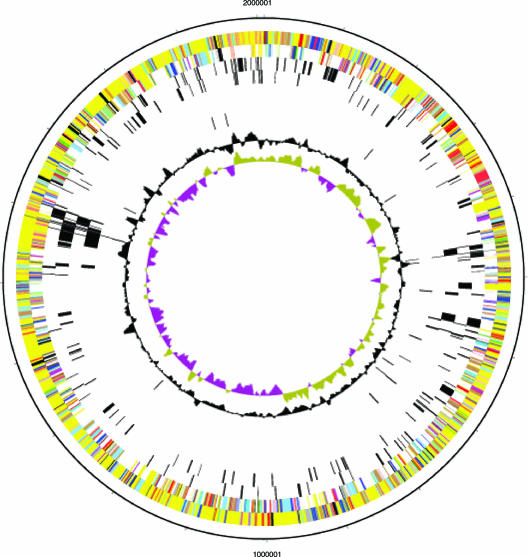
The 2.0-Mb H. somnus 129Pt chromosome (Fig. 1) contained 1,844 predicted coding sequences (CDSs), 5 rRNA operons, and 50 tRNAs. The average open reading frame (ORF) length was 980 bp, and the overall coding density was 89.5%. The overall GC content was 37% (Table 1). H. somnus 129Pt had five candidate prophage regions (see Table S1 in the supplemental material): HS_0423 to HS_0433 spanned 9,814 bases, with a GC content of 31%; HS_0519 to HS_0539 spanned 11,509 bases, with a GC content of 35%; HS_0554 to HS_0558 spanned 3,289 bases, with a GC content of 33%; HS_1331 to HS_1337 spanned 4,076 bases, with a GC content of 36%; and HS_1365 to HS_1425 spanned 52,616 bases, with a GC content of 40%. H. somnus 129Pt had approximately 15 pseudogenes (see Table S2 in the supplemental material); this number may not include all genes with frameshifts or early stop codons. In H. somnus 129Pt, the origin of replication (oriC) was located at approximately base 1926721 (within HS_1702 atpB at positions 1927397 to 1926603); the replication terminus region (terC) was located at approximately base 953326 (in IGR0751, positions 952932 to 953616 between HS_0872 and HS_0873). The dnaA gene (HS_0136) was translocated relative to oriC in H. somnus 129Pt. IS1016 elements are novel insertion elements present in H. influenzae (43). We found five complete and three partial copies of IS1016 in H. somnus 129Pt and one in H. ducreyi 35000HP (see Table S3 in the supplemental material).
FIG. 1.
Circular representation of the H. somnus 129Pt genome. The outermost two circles indicate start sites of genes and assigned functional categories. Circle 1 consists of forward-strand gene products. Circle 2 consists of reverse-strand gene products. Colors represent the following functional categories: amino acid biosynthesis (magenta); biosynthesis of cofactors, prosthetic groups, and carriers (red); cell envelope (dark salmon); cellular processes (light gray); central intermediary metabolism (orange); conserved hypothetical and hypothetical (yellow); DNA metabolism (light blue); energy metabolism (cyan); fatty acid and phospholipid metabolism (pale salmon); Haemophilus-specific protein (salmon); other categories (dark gray); protein synthesis and protein fate (blue); purines, pyrimidines, nucleosides, and nucleotides (green); regulatory functions (black); transcription and translation (light green); transport and binding proteins (brown); and unassigned (palest salmon). Circles 3 and 4, unique genes; circles 5 and 6, prophage genes; circles 7 and 8, insertion elements; circle 9, G+C content; circle 10, GC bias (G − C/G + C). Khaki indicates values >1, and purple indicates values <1.
TABLE 1.
General features of the H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd genomes
| Parameter | Value for organism
|
||
|---|---|---|---|
| H. somnus 129Pt | H. ducreyi 35000HP | H. influenzae Rda | |
| Size (Mb) | 2.0 | 1.7 | 1.8 |
| Plasmid | pHS129 (5.2 kb) | None | None |
| Coding sequence (Mb) | 1.8 | 1.5 | 1.6 |
| Coding sequence (%) | 89.5 | 86.6 | 89 |
| GC% | 37 | 38 | 38 |
| Average ORF size (bp) | 980 | 828 | 915 |
| Total no. of CDSs | 1,844 | 1,781 | 1,743 |
| No. of CDSs with assigned function | 1,313 | 1,092 | 1,007 |
| No. of conserved hypothetical CDSs | 440 | 495 | 347 |
| No. of hypothetical CDSs | 61 | 130 | 389 |
| No. of tRNAs | 50 | 45 | 54 |
| No. of rRNA operons | 5 | 6 | 6 |
| No. of possible pseudogenes | 15b | 19b | 62c |
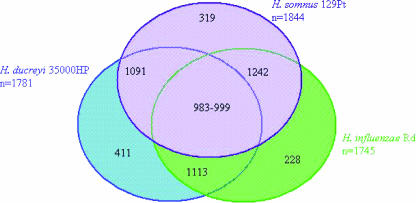
Comparisons of the H. somnus 129Pt, H. influenzae Rd, and H. ducreyi 35000HP chromosomes using ACT (http://www.sanger.ac.uk/Software/ACT/) revealed no large regions of synteny. Pairwise BLAST comparisons of the protein sequences from H. somnus 129Pt, H. influenzae Rd, and H. ducreyi 35000HP revealed 319 CDSs unique to H. somnus 129Pt, 411 CDSs unique to H. ducreyi 35000HP, and 228 CDSs unique to H. influenzae Rd (Fig. 2). H. somnus 129Pt and H. ducreyi 35000HP had 1,091 sequences in common, H. somnus 129Pt and H. influenzae Rd had 1,242 sequences in common, and H. ducreyi 35000HP and H. influenzae Rd had 1,113 sequences in common. The three organisms had approximately 983 to 999 genes in common. Genes unique to H. somnus 129Pt (see Table S4 in the supplemental material) included genes involved in LOS biosynthesis, carbohydrate uptake and metabolism, cation transport, amino acid metabolism, ubiquinone and menaquinone biosynthesis, cell surface adhesion, biosynthesis of cofactors, energy metabolism, and electron transport. A genome-wide comparative analysis of H. somnus 129Pt, H. influenzae Rd, and H. ducreyi 35000HP revealed similarities and differences in the numbers and compositions of genes involved in various aspects of metabolism and host colonization and persistence.
FIG. 2.
Common and unique genes in H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd.
Carbohydrate and amino acid utilization.
H. somnus 129Pt had approximately 94 genes involved in carbohydrate utilization, encoding metabolic enzymes or transporters, while H. ducreyi 35000HP had approximately 27 genes and H. influenzae Rd had about 58 genes (see Table S5 in the supplemental material). The approximate numbers of genes involved in amino acid utilization in H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd were 81, 59, and 103, respectively (see Table S6 in the supplemental material). Our results show that H. somnus 129Pt had more genes for carbohydrate utilization than did H. ducreyi 35000HP and H. influenzae Rd, while both H. somnus 129Pt and H. influenzae Rd had more genes for amino acid utilization than did H. ducreyi. These results suggest that the bovine urogenital tract, in which H. somnus 129Pt lives, and the human respiratory tract, where H. influenzae lives, contain more abundant nutrient sources than human genitalia, where H. ducreyi resides.
Nucleotide biosynthesis.
H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd all had genes encoding components of the purine biosynthesis pathway. However, the H. ducreyi 35000HP genes purC, purD, purF, purH, purK, and purL were fragmented; these genes were each represented by two or three separate adjacent ORFs (see Table S7 in the supplemental material), indicating that they may not be functional. In terms of pyrimidine biosynthesis genes, H. somnus 129Pt and H. ducreyi 35000HP were missing carA, carB, pyrB, pyrC, and pyrI, as is H. influenzae Rd (65). In H. influenzae Rd, added citrulline can salvage both arginine and pyrimidine biosynthesis via the argI gene product (65). H. ducreyi also had argI, so it should be able to use citrulline. Since H. somnus 129Pt did not have argI, the lack of carA, carB, pyrB, pyrC, and pyrI explains the need for uracil in the growth medium (35).
TCA cycle, Embden-Meyerhof, pentose phosphate, and Entner-Doudoroff pathways.
Like H. influenzae (65), both H. somnus 129Pt and H. ducreyi 35000HP lacked genes encoding aerobic respiratory chain enzymes and were missing key enzymes that are part of the oxidative branch of the tricarboxylic acid (TCA) cycle. However, in terms of the missing enzymes, H. ducreyi 35000HP and H. influenzae Rd were more similar to each other than to H. somnus 129Pt, as they were missing the genes encoding citrate synthase, aconitase, and isocitrate dehydrogenase, while H. somnus 129Pt was missing only the gene encoding isocitrate dehydrogenase. In terms of the rest of the TCA cycle, all three organisms were missing succinate dehydrogenase as well as the entire glyoxylate bypass. H. ducreyi 35000HP was also missing the gene encoding succinyl coenzyme A synthetase, which both H. somnus 129Pt and H. influenzae Rd had. H. ducreyi 35000HP was also missing succinyl coenzyme A synthetase (sucCD), while the other two organisms had these genes (HS_0956 to HS_0957 and HI1196 to HI1197). H. somnus 129Pt, H. influenzae Rd, and H. ducreyi 35000HP all had the genes aspC (HS_0838, HD1419, and HI1617) and aspA (HS_0466, HD0564, and HI0534), whose products convert oxaloacetate to aspartate and alpha-ketoglutarate (aspC) and aspartate to fumarate (aspA), thus bypassing the steps catalyzed by the missing GltA, AcnB, and Icd.
These results indicate that H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd (65) have an incomplete, reductive TCA cycle. Many other bacteria have an incomplete TCA cycle, and very few have a complete one (33). This incomplete reductive type of cycle probably functions primarily in carbon assimilation and the generation of precursor metabolites for biosynthesis; phylogenetic evidence indicates that the original state of the TCA cycle was a reductive biosynthetic pathway (59).
Since H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd were all lacking the complete energy-producing branch of the TCA cycle, they must have other means for energy generation. Both H. somnus 129Pt and H. influenzae Rd had complete Embden-Meyerhof (glycolysis) pathways. H. somnus 129Pt had a homolog of the Escherichia coli glucokinase (glk) (HS_0379). H influenzae Rd did not have a homolog of the E. coli glucokinase but did have two genes (HI0144 and HI0182) encoding proteins that are weakly homologous to two glucokinases from Streptomyces coelicolor A3 (20 to 30% amino acid identity) (65). Based on BLAST sequence comparisons to E. coli, S. coelicolor, H. somnus 129Pt, and H. influenzae Rd genes, H. ducreyi 35000HP did not appear to have glucokinase at all (see Table S8 in the supplemental material). However, it did have the rest of the glycolysis pathway, so it should be able to convert glucose-6-phosphate to pyruvate. All three organisms had the complete pentose phosphate pathway, but only H. somnus 129Pt and H. influenzae Rd had the Entner-Doudoroff pathway. In terms of fermentation, all three organisms had the genes necessary for the conversion of pyruvate to lactate, formate, and acetate but not to ethanol (see Table S8 in the supplemental material).
Ubiquinone and menaquinone biosynthesis.
Chorismate is the starting point for ubiquinone and menaquinone synthesis (49). H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd all had the shikimate pathway for chorismate biosynthesis. In addition, H. somnus 129Pt had all of the genes encoding components of both the ubiquinone (ubiABCDEFGHX) and menaquinone (menABCDEFG) synthesis pathways, and H. ducreyi 35000HP had genes encoding all of the components of the ubiquinone synthesis pathway except chorismate lyase (ubiC), which catalyzes the first step in the pathway, the conversion of chorismate to p-hydroxybenzoate (see Table S8 in the supplemental material). Without this enzyme, H. ducreyi 35000HP cannot synthesize ubiquinone; we were unable to find genes comprising a complete pathway to bypass this step, such as the pathway utilizing chrorismate mutase, prephenate dehydrogenase, and phenylalanine ammonia lyase commonly found in plants and studied in Escherichia coli mutants (5). H. influenzae Rd had none of the genes for ubiquinone biosynthesis (see Table S8 in the supplemental material). Like H. somnus 129Pt, both H. ducreyi 35000HP and H. influenzae Rd had the complete menaquinone biosynthesis pathway.
NAD biosynthesis.
In culture, H. somnus (3) and H. ducreyi (48, 53) do not require NAD (V factor) for growth, but H. influenzae Rd does (73). This indicates that H. somnus may be able to use nicotinamide as a precursor in NAD biosynthesis, as H. ducreyi does (48). The H. ducreyi 35000HP genome contained two full copies and a fragment of the gene nadV (48, 53), which encodes nicotinamide phosphoribosyl transferase (HD1445 fragment, HD1447, and HD1455), which allows strains that possess it to be grown in the presence of nicotinamide (48). H. somnus 129Pt had a nadV homolog (HS_0002) with 54% amino acid identity to HD1447 and HD1455. This explains the requirement for nicotinamide in the defined medium for growth of H. somnus (35). In H. ducreyi 35000HP, the nadV gene is part of the pNAD1 plasmid sequence, which is integrated into the chromosome of H. ducreyi 35000HP as two tandem copies (HD1440 to HD1455) (53). H. somnus 129Pt had one chromosomal copy of nadV (HS_0002) and did not contain integrated copies of the pNAD1 plasmid, as it had none of the other genes encoded by pNAD1.
H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd did not have the nadA, nadB, nadC, nadD, and nadE genes, which encode the de novo pathway for the synthesis of NAD from l-aspartate (see Table S9 in the supplemental material). In eukaryotes, the de novo pathway starts with tryptophan, and some bacteria may utilize that pathway rather than the aspartate pathway (44). However, none of these organisms had the eukaryotic tryptophan pathway. The salvage pathway in H. influenzae involves a bifunctional protein encoded by the nadR gene. H. somnus 129Pt had a homolog (HS_0087) of the H. influenzae nadR (HI0763) gene. In H. ducreyi 35000HP, nadR was split into two ORFS (HD0274 and HD0275), which may not be functional.
Host colonization and persistence.
Although H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd do not cause systemic infection (51, 62), they do colonize and persist in their hosts. We investigated whether these three organisms shared any genes involved in host colonization and persistence and whether they had any genes in common with invasive strains of H. influenzae. Colonization of mucosal surfaces involves the circumvention of mucociliary function so that the clearance of the bacteria is impeded and adherence of bacterial cells to host epithelium (reviewed in references 58 and 72). Interference with mucosal ciliary activity is accomplished by LOS (38) (see Table S10 in the supplemental material).
Successful colonization of mucosal surfaces by bacteria depends on their adherence to host mucosal epithelial cells (58, 72). Evidence indicates that adherence of Haemophilus spp. to host cells involves multiple mechanisms including pili and a wide variety of adhesins (reviewed in reference 36). The H. influenzae pilus gene cluster (hifA, hifY3, hifY2, hifY1, hifB, hifC, hifD, and hifE) is present in pathogenic isolates but not in strains like Rd (21, 68). We also did not find similar genes in H. somnus 129Pt (see Table S11 in the supplemental material). H. ducreyi 35000HP had genes with low similarity (∼30% protein sequence identity) to hifA and hifB (HD0284), hifC (HD0283), and hifD (HD0281). The protein translations of these genes were also ∼30% identical to the type 1 pilus components encoded by fimA (HD0281), fimC (HD0284), and fimD (HD0283) of E. coli (39). If not involved in the formation of type 1 pili, these H. ducreyi 35000HP genes might be part of a secretion system. The H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd genomes did contain candidate pilA, pilB, pilC, and pilD genes (see Tables S11 and S13 in the supplemental material), which encode the components necessary to process and export the PilA subunit of type IV pili (15). However, the H. somnus 129Pt pil genes were not clustered as in H. ducreyi 35000HP and H. influenzae Rd.
The H. somnus 129Pt and H. ducreyi 35000HP pil genes were similar in sequence to the H. influenzae genes encoding the components of the DNA uptake machinery (see Table S13 in the supplemental material). Therefore, based on sequence similarity alone, it is impossible to know which function is provided by these pil genes in H. somnus 129Pt. Evidence indicates that the type IV pilin-like proteins encoded by the H. influenzae Rd pilABCD genes are necessary for transformation (15). However, it has recently been demonstrated that H. influenzae strain 86-028NP expresses type IV pilus-like structures and twitching motility when grown under certain conditions (4). Twitching motility and the presence of type IV pili have not yet been investigated in H. somnus 129Pt. In the absence of demonstrated twitching motility, it is likely that the H. somnus 129Pt genes function in DNA uptake, as H. somnus 129Pt does have genes involved in DNA uptake and transformation. If twitching motility by H. somnus is demonstrated in the future, then these components may function in both motility and DNA uptake, as has been described previously for Neisseria gonorrhoeae (24).
H. ducreyi expresses fine, tangled pili, and the major pilin protein is encoded by the gene ftpA (7) (HD0068), which is not present in H. influenzae Rd or H. somnus 129Pt. In addition, H. ducreyi has the tad genes, which are responsible for tight adherence to surfaces. The tad loci are part of a 15-gene cluster in H. ducreyi that includes the rcpABC and flp genes, which are necessary for H. ducreyi microcolony formation in vitro (55) and may be important for colonization in a variety of environmental niches (40).
As expected, H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd lacked the adhesin genes found in pathogenic strains of H. influenzae (see Table S11 in the supplemental material). However, they did have other adhesin genes, which may be involved in colonization and persistence. Most notably, H. somnus 129Pt had 12 genes encoding large adhesins, which had low sequence similarity to two filamentous hemagglutinins from H. ducreyi 35000HP (HD1156 and HD1505) and three genes from H. influenzae Rd (HI1718, HI1685, and HI0383) (see Table S11 in the supplemental material). The protein sequences of these adhesins had C-terminal DsrA-like and Hia-like domains, indicating that they are probably autotransporters, which would use the Sec secretion system to move across the inner membrane (reviewed in reference 11). All three organisms had the sec secretion system genes.
Spinola's group previously identified several genes required for H. ducreyi infection and colonization of the skin. These genes include dsrA (autotransporter adhesin) (6), lspA1 and lspA2 (filamentous hemagglutinins) (37), dltA (lectin A), hgbA (hemoglobin receptor) (2), peptidoglycan-associated lipoprotein (P6) (22), ncaA (collagen-binding outer membrane protein) (23), and tadA (tight adherence protein A) (63). H. somnus 129Pt had genes with significant sequence similarity to dsrA (HS_1543) and P6 (HS_0263) (see Table S11 in the supplemental material) and also had two ORFS (the result of a frameshift) representing the N-terminal and C-terminal regions of hgbA (HS_0720 and HS_0721) (see Table S12 in the supplemental material). H. influenzae Rd had hgbA (HI0661) and P6 (HI0381) genes. H. somnus 2336 has a trimeric autotransporter adhesin (11), which had 47% amino acid identity to two genes (HS_1085 and HS_1234) in strain 129Pt. In addition, Both H. influenzae Rd and H. ducreyi 35000HP had oapA (HI0330/HD0651) and oapB (HI0331/HD0652) genes. H. somnus 129Pt had only oapA (HS_1287). OapA is required for H. influenzae colonization of the rat nasopharynx (70). OapB may be a lipoprotein (70), but its function is currently unknown. Therefore, it is likely that many of these genes in H. somnus 129Pt and H. influenzae Rd play a role in colonization.
The persistence of bacteria at the original site of colonization depends on the ability to evade the immune system and the ability to acquire iron from the host (58, 72). Immune evasion is accomplished in part by phase variation in the structures of surface LOS. The LOS biosynthesis genes in H. ducreyi, H. influenzae, and H. somnus have been studied in detail (28, 50, 62). Table S10 in the supplemental material shows all LOS biosynthetic genes identified in H. somnus 129Pt and H. ducreyi 35000HP, compared to H. influenzae Rd (21).
H. somnus 129Pt genes that were not found in H. influenzae or H. ducreyi included HS_0116 and HS_0275, encoding glycosyltransferases. H. somnus also has the lob locus, consisting of lob1, lob2A, lob2B, lob2C, and lob2D; the products of these lob genes are glycosyltransferases that may be homologs of the Neisseria lgt genes responsible for building the lacto-N-neotetraose chain (62). As previously reported, strain 129Pt has lob1 (HS_0638) and lob2D (HS_0636) but has only the 5′ ends of lob2A (HS_0637) and lob2C (HS_0636a). The lob2B gene was not present at all. H. ducreyi 35000HP had genes annotated as lgtA and lgtB (similar to lob2A/lob2B and lob2C, respectively) as well as potential homologs of lob1 and lob2D. H. influenzae Rd had lic2A (lob2A homolog) and genes with similarity to lob2C and lob2D (see Table S10 in the supplemental material).
The persistence of bacteria in the host may also be facilitated by antigenic variation, such as that which occurs in the outer membrane protein P2 of H. influenzae Rd (HI0139), which is immunogenic (58). H. ducreyi 35000HP had two copies of the gene encoding P2 (HD1433 and HD1435), but H. somnus 129Pt did not have any. H. ducreyi 35000HP and H. influenzae Rd also had genes encoding P4, which H. somnus 129Pt did not have. All three organisms had the genes encoding P1, P5, and P6 (see Table S11 in the supplemental material); this finding is consistent with the observed reactivity of H. somnus with a monoclonal antibody (kindly provided by Tim Murphy) to P6 (unpublished data).
In addition to evading the immune system, bacteria must be able to synthesize heme or acquire it and iron from the host in order to survive and persist (reviewed in references 58 and 72). All of these organisms contained genes that should enable them to satisfy their iron requirement (see Table S12 in the supplemental material). Both H. ducreyi 35000HP and H. influenzae Rd lack the complete pathway for heme biosynthesis (20, 71), while H. somnus 129Pt had genes encoding all components of the heme biosynthesis pathway from l-glutamate to protoheme (see Table S12 in the supplemental material). Although H. influenzae Rd does not encode the genes required to make heme from 5-amino-levulinate, it does have ferrochetalase (hemH), which adds an iron molecule to protoporphyrin IX to make protoheme (71).
We found that H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd had approximately 15, 9, and 17 iron and heme transporter genes, respectively (see Table S12 in the supplemental material). H. influenzae can use heme, heme-hemopexin, and iron-bound transferrin as sources of iron as well as iron salts and chelates if protoporphyrin IX is available (71). H. influenzae does not produce siderophores (52, 54), can use human transferrin as a sole source of iron (31), and has two genes encoding the transferrin-binding proteins Tbp1 and Tbp2 (HI0994 [tbpA] and HI0995 [tbpB], respectively) (26), which facilitate the uptake of iron in a TonB-dependent process (58). H. ducreyi does not produce siderophores and cannot use heme-hemopexin (64). H. ducreyi can fulfill its iron requirement by using free heme, hemoglobin, catalase, and hemoglobin complexed with haptoglobin but not human transferrin or lactoferrin (45). Since it cannot use transferrin as a source of iron, it is not surprising that H. ducreyi 35000HP lacked transferrin-binding protein genes (tbpA and tbpB). H. ducreyi 35000HP also did not have homologs of the H. influenzae hxuABC genes (HI0262 to HI0264) for heme-hemopexin utilization. H. somnus 129Pt does not make siderophores (56) and did not have homologs of the hxuABC genes, indicating that it cannot use heme-hemopexin as a heme source either. H. somnus 129Pt had tbpA (HS_0449) and tbpB (HS_0448) genes, encoding the bipartite transferrin receptor system (16) (see Table S12 in the supplemental material). Strain 129Pt had three additional, separate ORFs representing C-terminal (HS_0097), central (HS_0098), and N-terminal (HS_0102) fragments of TbpA, which may represent a frameshifted version of the gene encoding the single-component TbpA2 protein of strain 649 (16). In support of this idea, HS_0097 encodes a 482-amino-acid protein, which corresponded to the last two-thirds of TbpA2 from strain 649, as reported previously (16). We also found a poly(G) tract containing eight G's in HS_0098; a poly(G) tract present in Histophilus ovis strain 3384Y and H. somnus strain 649 tbpA2 genes controls the expression of TbpA2 through phase variation (16, 18). In strain 649 grown under iron-replete conditions, the presence of eight G's in the poly(G) tract introduces a premature stop codon into the reading frame of the gene (16), as do the eight G's in the HS_0098 fragment. When grown under iron-restricted conditions in the presence of goat transferrin, the poly(G) tract of the strain 649 tbpA2 gene contains nine G's, removing the frameshift and resulting in a full-length TbpA2 precursor (16). H. somnus 129Pt had yet another copy of tbpA (HS_0582) that had 35% amino acid identity to the H. ovis and H. somnus 649 tbpA genes.
In gram-negative bacteria, the TonB system is used for the active transport of iron and heme from host proteins, such as transferrin and hemoglobin, into the periplasmic space (57). The TonB system consists of three essential proteins, TonB, ExbB, and ExbD. H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae Rd had the tonB, exbB, and exbD genes as well as genes encoding outer membrane hemoglobin and heme receptors, many with sequence similarity to TonB-dependent receptors. For example, H. ducreyi 35000HP has hgbA/hupA (HD2025), which encodes an outer membrane hemoglobin receptor (19), and tdhA/hemR (HD0388), encoding a heme receptor, both with amino acid sequence similarity to TonB-dependent outer membrane receptors (19, 66). H. somnus 129Pt had hgbA, but due to a frameshift, it was split into two ORFS (HS_0720 and HS_0721). The hgbA genes of H. somni ovine strains 9L and 3384Y are also frameshifted, and this finding correlates with an inability of these strains to bind and utilize hemoglobin (67); in the ovine strains 9L and 3384Y, the premature stop codon in hgbA occurs much earlier in the sequence (base 642) than in the 129Pt gene (base 2958). The frameshifted H. somnus 129Pt hgbA gene was adjacent to two genes involved in heme utilization (HS_0722 and HS_0723). Only one of these genes (HS_0722) had similarity to genes in H. ducreyi 35000HP (HD0620) and H. influenzae Rd (HI0854). H. influenzae Rd had a gene encoding a hemoglobin binding protein with sequence similarity to H. ducreyi hgbA (HI0661) as well as two copies of hgbB (HI0635 and HI0712). H. somnus 129Pt and H. ducreyi 35000HP did not have hgbB. H. somnus 129Pt had a gene annotated as hemR (HS_0728), encoding an outer membrane heme receptor with 50% amino acid sequence identity to H. ducreyi tdhA/hemR; H. influenzae Rd also had hemR (HI0113). Additional TonB-dependent receptors included H. somnus 129Pt gene HS_0181 and H. influenzae Rd genes HI1565 and HI1567. The H. influenzae Rd hemoglobin receptor genes hgbA (HI0661), hgbB (HI0635 and HI0712), and HI1565 have 20 (HI0661 and HI0635), 36 (HI0712), and 18 (HI1565) CAAC repeats close to the gene start that are involved in phase variation of gene expression (32). However, the H. somnus 129Pt and H. ducreyi 35000HP hgbA genes, which were the closest hits to all of the H. influenzae Rd hemoglobin receptor genes, did not have these repeats. Therefore, these genes in H. somnus 129Pt and H. ducreyi 35000HP are unlikely to undergo phase variation. In fact, as the hgbA gene in H. somnus 129Pt was split into two ORFs (HS_0720 and HS_0721), it is probably nonfunctional.
Following the transfer of extracellular protein-bound iron into the periplasm by TonB-dependent receptors, an iron transporter moves iron from the periplasmic space to the cytoplasm (reviewed in references 27 and 58). In H. influenzae, this activity is encoded by the hitABC (fbpABC) genes (61). H. somnus 129Pt contained genes with similarity to the H. influenzae hitABC genes, but H. ducreyi 35000HP did not (see Table S12 in the supplemental material). In many bacteria, iron utilization is regulated through the fur gene product. In H. influenzae, the transferrin receptor genes as well as the genes for hemopexin and hemoglobin receptors are repressed by iron, heme, and protoporphyrin IX, probably through the actions of Fur (29). H. somnus 129Pt, H. ducreyi 35000HP, and H. influenzae all had fur homologs, and H. ducreyi has a functional fur system (9). As H. influenzae Rd, H. ducreyi 35000HP, and H. somnus 129Pt had fur homologs, we looked for a Fur binding motif consensus sequence (AATTATTATC) (26) in the genomes to see if it was present in the upstream region of genes involved in iron utilization. We found putative Fur binding motifs in the 500-bp upstream region of the H. somnus 129Pt genes encoding a TonB-dependent outer membrane receptor (HS_0181) and hugX (HS_0723), in two H. influenzae Rd genes encoding hemoglobin-binding proteins (HI0635 and HI0712), and in the H. ducreyi 35000HP genes encoding hemoglobin utilization protein A (HD2025) and heme-binding protein A (HD0215). This suggests that the transcription of these genes may be regulated by Fur.
In conclusion, a comparison of H. somnus 129Pt with the genomes of H. ducreyi 35000HP and H. influenzae Rd revealed similarities and differences in gene content that belie the abilities of these organisms to utilize certain sugars and amino acids as carbon and nitrogen sources, indicating which compounds are likely to be present in their respective environments. Our finding that all three of these organisms had the components of an incomplete, reductive TCA cycle, which is probably used more for the biosynthesis of intermediates than for energy production, provides another clue about common and unique aspects of their lifestyles. Finally, we found differences in gene content among these organisms that may contribute to host specificity. These findings included differences in the numbers and types of adhesin and outer membrane protein genes as well as hemoglobin and transferrin receptor genes. In particular, the H. somnus 129Pt genome contained 12 genes that probably encode autotransporter adhesins; these genes did not have obvious counterparts in H. ducreyi 35000HP and H. influenzae Rd, so they may well represent novel functions that contribute to the preference of commensal H. somnus 129Pt for the bovine urogenital tract environment. In addition, H. somnus 129Pt had genes encoding outer membrane proteins P1, P5, and P6 but not P2 and P4; H. ducreyi 35000HP and H. influenzae Rd had genes encoding all five of these outer membrane proteins. Taken together with the finding that none of these organisms had genes with significant similarity to the adhesin genes of pathogenic H. influenzae strains, these results lead to the speculation that differences in numbers and types of adhesin and outer membrane protein genes may contribute to host specificities and niche differences among commensal and pathogenic strains.
The ability to utilize hemoglobin and transferrin may also play a role in the host specificity of H. somnus strains (17, 67). For example, bovine H. somnus isolates produce hemoglobin receptors that mediate the uptake of iron from bovine hemoglobin but not from ovine, porcine, or human hemoglobin (67). There is evidence that the ability of H. somnus isolates to cause infection in ruminants may depend on the host specificity of their transferrin receptors (74). In particular, H. somnus strains that contain single-component transferrin receptors encoded by the tbpA2 gene can acquire iron from bovine, ovine, and goat transferrins (16), but those that possess bipartite transferrin receptors encoded by the tbpA and tbpB genes can acquire iron only from bovine transferrin (16, 74). Phase variation of hemoglobin and transferrin receptor genes has been demonstrated in virulent H. somnus strains 2336 and 649 (16, 18, 67). Taken together with our finding that commensal strain 129Pt contains truncated hemoglobin (hgbA) and single-component transferrin receptor (tbpA2) genes, these results support the idea that virulence may depend in part on the ability to utilize iron from hemoglobin and transferrin.
Whole-genome comparisons of H. somnus 129Pt with pathogenic isolates will allow us to explore the potential roles of adhesins, hemoglobin and transferrin receptors, and other virulence factors, such as LOS, in host colonization, host specificity, and pathogenesis. As cell surface adhesins and outer membrane proteins are good targets for vaccine development, results of this and future studies will aid in the development of vaccines and other protective strategies against H. somnus disease.
Supplementary Material
Acknowledgments
This work was supported by the U.S. Department of Energy under contract no. W-7405-ENG-36 and by USDA-CSREES grant 2001-52100-11314 to T. J. Inzana.
We thank the anonymous reviewers of the manuscript for their insightful comments and suggestions.
Footnotes
Published ahead of print on 15 December 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Albritton, W. 1989. Biology of Haemophilus ducreyi. Microbiol. Rev. 53:377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AlTawfiq, J. A., K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 181:1049-1054. [DOI] [PubMed] [Google Scholar]
- 3.Asmussen, M. D., and C. L. Baugh. 1981. Thiamine pyrophosphate (cocarboxylase) as a growth factor for Haemophilus somnus. J. Clin. Microbiol. 14:178-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakaletz, L. O., B. D. Baker, J. A. Jurcisek, A. Harrison, L. A. Novotny, J. E. Bookwalter, R. Mungur, and R. S. Munson, Jr. 2005. Demonstration of type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect. Immun. 73:1635-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, J. L., and J. W. Frost. 2001. Microbial synthesis of p-hydroxybenzoic acid from glucose. Biotechnol. Bioeng. 76:376-390. [DOI] [PubMed] [Google Scholar]
- 6.Bong, C. T. H., R. E. Throm, K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2001. DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infect. Immun. 69:1488-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brentjens, R. J., M. Ketterer, M. A. Apicella, and S. M. Spinola. 1996. Fine tangled pili expressed by Haemophilus ducreyi are a novel class of pili. J. Bacteriol. 178:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campagnari, A. A., S. M. Spinola, A. J. Lesse, Y. A. Kwaik, R. E. Mandrell, and M. A. Apicella. 1990. Lipooligosaccharide epitopes shared among gram-negative non-enteric mucosal pathogens. Microb. Pathog. 8:353-362. [DOI] [PubMed] [Google Scholar]
- 9.Carson, S. D., C. E. Thomas, and C. Elkins. 1996. Cloning and sequencing of a Haemophilus ducreyi fur homolog. Gene 176:125-129. [DOI] [PubMed] [Google Scholar]
- 10.Corbeil, L. B., K. Blau, D. J. Prieur, and A. C. S. Ward. 1985. Serum susceptibility of Haemophilus somnus from bovine clinical cases and carriers. J. Clin. Microbiol. 22:192-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13:199-205. [DOI] [PubMed] [Google Scholar]
- 12.Daines, D. A., L. A. Cohn, H. N. Coleman, K. S. Kim, and A. L. Smith. 2003. Haemophilus influenzae Rd KW20 has virulence properties. J. Med. Microbiol. 52:277-282. [DOI] [PubMed] [Google Scholar]
- 13.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewhirst, F. E., B. J. Paster, I. Olsen, and G. J. Fraser. 1992. Phylogeny of 54 representative strains of species in the family Pasteurellaceae as determined by comparison of 16S rRNA sequences. J. Bacteriol. 174:2002-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dougherty, B. A., and H. O. Smith. 1999. Identification of Haemophilus influenzae Rd transformation genes using cassette mutagenesis. Microbiology 145:401-409. [DOI] [PubMed] [Google Scholar]
- 16.Ekins, A., F. Bahrami, A. Sijercic, D. Maret, and D. F. Niven. 2004. Haemophilus somnus possesses two systems for acquisition of transferrin-bound iron. J. Bacteriol. 186:4407-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekins, A., and D. F. Niven. 2001. Production of transferrin receptors by Histophilus ovis: three of five strains require two signals. Can. J. Microbiol. 47:417-423. [DOI] [PubMed] [Google Scholar]
- 18.Ekins, A., and D. F. Niven. 2003. Transferrin-dependent expression of TbpA by Histophilus ovis involves a poly G tract within tbpA. FEMS Microbiol. Lett. 220:95-98. [DOI] [PubMed] [Google Scholar]
- 19.Elkins, C., C. J. Chen, and C. E. Thomas. 1995. Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi. Infect. Immun. 63:2194-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkins, C., P. A. Totten, B. Olsen, and C. E. Thomas. 1998. Role of the Haemophilus ducreyi Ton system in internalization of heme from hemoglobin. Infect. Immun. 66:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 22.Fortney, K. R., R. S. Young, M. E. Bauer, B. P. Katz, A. F. Hood, R. S. Munson, Jr., and S. M. Spinola. 2000. Expression of peptidoglycan-associated lipoprotein is required for virulence in the human model of Haemophilus ducreyi infection. Infect. Immun. 68:6441-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fulcher, R. A., L. E. Cole, D. M. Janowicz, K. L. Toffer, K. R. Fortney, B. P. Katz, P. E. Orndorff, S. M. Spinola, and T. H. Kawula. 2006. Expression of Haemophilus ducreyi collagen binding outer membrane protein NcaA is required for virulence in swine and human challenge models of chancroid. Infect. Immun. 74:2651-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fussenegger, M., T. Rudel, R. Barten, R. Ryll, and T. F. Meyer. 1997. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene 192:125-134. [DOI] [PubMed] [Google Scholar]
- 25.Gonzales, H. F., and D. P. Bingham. 1983. Genetic relatedness of Haemophilus somnus to select genera of bacteria. Am. J. Vet. Res. 44:1793-1795. [PubMed] [Google Scholar]
- 26.Gray-Owen, S. D., S. Loosmore, and A. B. Schryvers. 1995. Identification and characterization of genes encoding the human transferrin-binding proteins from Haemophilus influenzae. Infect. Immun. 63:1201-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guerinot, M. L. 1994. Microbial iron transport. Annu. Rev. Microbiol. 48:743-772. [DOI] [PubMed] [Google Scholar]
- 28.Harrison, A., D. W. Dyer, A. Gillaspy, W. C. Ray, R. Mungur, M. B. Carson, H. Zhong, J. Gipson, M. Gipson, L. S. Johnson, L. Lewis, L. O. Bakaletz, and R. S. Munson, Jr. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J. Bacteriol. 187:4627-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasan, A. A., J. Holland, A. Smith, and P. Williams. 1997. Elemental iron does repress transferrin, haemopexin and haemoglobin receptor expression in Haemophilus influenzae. FEMS Microbiol. Lett. 150:19-26. [DOI] [PubMed] [Google Scholar]
- 30.Hedegaard, J., H. Okkels, B. Bruun, M. Kilian, K. K. Mortensen, and N. Norskov-Lauritsen. 2001. Phylogeny of the genus Haemophilus as determined by comparison of partial infB sequences. Microbiology 147:2599-2609. [DOI] [PubMed] [Google Scholar]
- 31.Herrington, D. A., and P. F. Sparling. 1985. Haemophilus influenzae can use human transferrin as a sole source for required iron. Infect. Immun. 48:248-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hood, D. W., M. E. Deadman, M. P. Jennings, M. Bisercic, R. D. Fleischmann, J. C. Venter, and E. R. Moxon. 1996. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 93:11121-11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huynen, M. A., T. Dandekar, and P. Bork. 1999. Variation and evolution of the citric-acid cycle: a genomic perspective. Trends Microbiol. 7:281-291. [DOI] [PubMed] [Google Scholar]
- 34.Inzana, T. J. 1999. The Haemophilus somnus complex, p. 358-361. In J. L. Howard and R. A. Smith (ed.), Current veterinary therapy 4, 4th ed. W. B. Saunders Co., Philadelphia, PA.
- 35.Inzana, T. J., and L. B. Corbeil. 1987. Development of a defined medium for Haemophilus somnus isolated from cattle. Am. J. Vet. Res. 48:366-369. [PubMed] [Google Scholar]
- 36.Jacques, M., and S. E. Paradis. 1998. Adhesin-receptor interactions in Pasteurellaceae. FEMS Microbiol. Rev. 22:45-59. [DOI] [PubMed] [Google Scholar]
- 37.Janowicz, D. M., K. R. Fortney, B. P. Katz, J. L. Latimer, K. Deng, E. J. Hansen, and S. M. Spinola. 2004. Expression of the LspA1 and LspA2 proteins by Haemophilus ducreyi is required for virulence in human volunteers. Infect. Immun. 72:4528-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson, A. P., and T. J. Inzana. 1986. Loss of ciliary activity in organ cultures of rat trachea treated with lipo-oligosaccharide from Haemophilus influenzae. J. Med. Microbiol. 22:265-268. [DOI] [PubMed] [Google Scholar]
- 39.Jones, C. H., J. S. Pinkner, R. Roth, J. Heuser, A. V. Nicholes, S. N. Abraham, and S. J. Hultgren. 1995. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 92:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, and D. H. Figurski. 2001. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 9:429-437. [DOI] [PubMed] [Google Scholar]
- 41.Kennedy, P. C., E. L. Biberstein, J. A. Howarth, L. M. Frazier, and D. L. Dungworth. 1960. Infectious meningo-encephalitis in cattle, caused by a Haemophilus-like organism. Am. J. Vet. Res. 21:403-409. [PubMed] [Google Scholar]
- 42.Kitching, J. P., and G. C. Bishop. 1994. The Haemophilus somnus disease complex in cattle, p. 1135-1142. In J. A. W. Coetzer, G. R. Thomson, R. C. Tustin, and N. P. Kriek (ed.), Infectious diseases of livestock: with special reference to Southern Africa, vol. 2. Oxford University Press, New York, NY. [Google Scholar]
- 43.Kroll, J. S., B. M. Loynds, and E. R. Moxon. 1991. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol. Microbiol. 5:1549-1560. [DOI] [PubMed] [Google Scholar]
- 44.Kurnasov, O., V. Goral, K. Colabroy, S. Gerdes, S. Anantha, A. Osterman, and T. P. Begley. 2003. NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 10:1195-1204. [DOI] [PubMed] [Google Scholar]
- 45.Lee, B. C. 1991. Iron sources for Haemophilus ducreyi. J. Med. Microbiol. 34:317-322. [DOI] [PubMed] [Google Scholar]
- 46.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mannheim, W., S. Pohl, and R. Hollander. 1980. On the taxonomy of Actinobacillus, Haemophilus, and Pasteurella: DNA base composition, respiratory quinones, and biochemical reactions of representative collection cultures. Zentralbl. Bakteriol. A 246:512-540. (In German.) [PubMed] [Google Scholar]
- 48.Martin, P. R., R. J. Shea, and M. H. Mulks. 2001. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J. Bacteriol. 183:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meganathan, R. 2001. Biosynthesis of menaquinone (vitamin K2) and ubiquinone (coenzyme Q): a perspective on enzymatic mechanisms. Vitam. Horm. 61:173-218. [DOI] [PubMed] [Google Scholar]
- 50.Melaugh, W., N. J. Phillips, A. A. Campagnari, M. V. Tullius, and B. W. Gibson. 1994. Structure of the major oligosaccharide from the lipooligosaccharide of Haemophilus ducreyi strain 35000 and evidence for additional glycoforms. Biochemistry 33:13070-13078. [DOI] [PubMed] [Google Scholar]
- 51.Morse, S. A. 1989. Chancroid and Haemophilus ducreyi. Clin. Microbiol. Rev. 2:137-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morton, D. J., and P. Williams. 1989. Utilization of transferrin-bound iron by Haemophilus species of human and porcine origins. FEMS Microbiol. Lett. 53:123-127. [DOI] [PubMed] [Google Scholar]
- 53.Munson, R. S., Jr., H. Zhong, R. Mungur, W. C. Ray, R. J. Shea, G. G. Mahairas, and M. H. Mulks. 2004. Haemophilus ducreyi strain ATCC 27722 contains a genetic element with homology to the Vibrio RS1 element that can replicate as a plasmid and confer NAD independence on Haemophilus influenzae. Infect. Immun. 72:1143-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelson, A. L., J. M. Barasch, R. M. Bunte, and J. N. Weiser. 2005. Bacterial colonization of nasal mucosa induces expression of siderocalin, an iron-sequestering component of innate immunity. Cell. Microbiol. 7:1404-1417. [DOI] [PubMed] [Google Scholar]
- 55.Nika, J. R., J. L. Latimer, C. K. Ward, R. J. Blick, N. J. Wagner, L. D. Cope, G. G. Mahairas, R. S. Munson, Jr., and E. J. Hansen. 2002. Haemophilus ducreyi requires the flp gene cluster for microcolony formation in vitro. Infect. Immun. 70:2965-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogunnariwo, J. A., C. Cheng, J. Ford, and A. B. Schryvers. 1990. Response of Haemophilus somnus to iron limitation: expression and identification of a bovine-specific transferrin receptor. Microb. Pathog. 9:397-406. [DOI] [PubMed] [Google Scholar]
- 57.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 58.Rao, V. K., G. P. Krasan, D. R. Hendrixson, S. Dawid, and J. W. St Geme III. 1999. Molecular determinants of the pathogenesis of disease due to non-typable Haemophilus influenzae. FEMS Microbiol. Rev. 23:99-129. [DOI] [PubMed] [Google Scholar]
- 59.Romano, A. H., and T. Conway. 1996. Evolution of carbohydrate metabolic pathways. Res. Microbiol. 147:448-455. [DOI] [PubMed] [Google Scholar]
- 60.Salzberg, S., A. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanders, J. D., L. D. Cope, and E. J. Hansen. 1994. Identification of a locus involved in the utilization of iron by Haemophilus influenzae. Infect. Immun. 62:4515-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siddaramppa, S., and T. J. Inzana. 2004. Haemophilus somnus virulence factors and resistance to host immunity. Anim. Health Res. Rev. 5:79-93. [DOI] [PubMed] [Google Scholar]
- 63.Spinola, S. M., K. R. Fortney, B. P. Katz, J. L. Latimer, J. R. Mock, M. Vakevainen, and E. J. Hansen. 2003. Haemophilus ducreyi requires an intact flp gene cluster for virulence in humans. Infect. Immun. 71:7178-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stevens, M. K., S. Porcella, J. Klesney-Tait, S. Lumbley, S. E. Thomas, M. V. Norgard, J. D. Radolf, and E. J. Hansen. 1996. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect. Immun. 64:1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tatusov, R. L., A. R. Mushegian, P. Bork, N. P. Brown, W. S. Hayes, M. Borodovsky, K. E. Rudd, and E. V. Koonin. 1996. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr. Biol. 6:279-291. [DOI] [PubMed] [Google Scholar]
- 66.Thomas, C. E., B. Olsen, and C. Elkins. 1998. Cloning and characterization of tdhA, a locus encoding a TonB-dependent heme receptor from Haemophilus ducreyi. Infect. Immun. 66:4254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tremblay, Y. D., F. Bahrami, and D. F. Niven. 2006. Acquisition of haemoglobin-bound iron by Histophilus somni. Vet. Microbiol. 114:104-114. [DOI] [PubMed] [Google Scholar]
- 68.VanHam, S., L. VanAlphen, F. Mooi, and J. VanPutten. 1994. The fimbrial gene-cluster of Haemophilus influenzae type-b. Mol. Microbiol. 13:673-684. [DOI] [PubMed] [Google Scholar]
- 69.Walker, D. R., and E. V. Koonin. 1997. A system for easy analysis of lots of sequences. Intell. Syst. Mol. Biol. 5:333-339. [PubMed] [Google Scholar]
- 70.Weiser, J. N., S. T. Chong, D. Greenberg, and W. Fong. 1995. Identification and characterization of a cell envelope protein of Haemophilus influenzae contributing to phase variation in colony opacity and nasopharyngeal colonization. Mol. Microbiol. 17:555-564. [DOI] [PubMed] [Google Scholar]
- 71.White, D. C., and S. Granick. 1963. Hemin biosynthesis in Haemophilus. J. Bacteriol. 85:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilson, R., R. B. Dowling, and A. D. Jackson. 1996. The biology of bacterial colonization and invasion of the respiratory mucosa. Eur. Respir. J. 9:1523-1530. [DOI] [PubMed] [Google Scholar]
- 73.Windsor, H. M., R. C. Gromkova, and H. J. Koornhof. 1993. Growth characteristics of V factor-independent transformants of Haemophilus influenzae. Int. J. Syst. Bacteriol. 43:799-804. [DOI] [PubMed] [Google Scholar]
- 74.Yu, R. H., S. D. Gray-Owen, J. Ogunnariwo, and A. B. Schryvers. 1992. Interaction of ruminant transferrins with transferrin receptors in bovine isolates of Pasteurella haemolytica and Haemophilus somnus. Infect. Immun. 60:2992-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.