Abstract
The impressive disease spectrum of Streptococcus pyogenes (the group A streptococcus [GAS]) is believed to be determined by its ability to modify gene expression in response to environmental stimuli. Virulence gene expression is controlled tightly by several different transcriptional regulators in this organism. In addition, expression of most, if not all, GAS genes is determined by a global mechanism dependent on growth phase. To begin an analysis of growth-phase regulation, we compared the transcriptome 2 h into stationary phase to that in late exponential phase of a serotype M3 GAS strain. We identified the arc transcript as more abundant in stationary phase in addition to the sag and sda transcripts that had been previously identified. We found that in stationary phase, the stability of sagA, sda, and arcT transcripts increased dramatically. We found that polynucleotide phosphorylase (PNPase [encoded by pnpA]) is rate limiting for decay of sagA and sda transcripts in late exponential phase, since the stability of these mRNAs was greater in a pnpA mutant, while stability of control mRNAs was unaffected by this mutation. Complementation restored the wild-type decay rate. Furthermore, in a pnpA mutant, the sagA mRNA appeared to be full length, as determined by Northern hybridization. It seems likely that mRNAs abundant in stationary phase are insensitive to the normal decay enzyme(s) and instead require PNPase for this process. It is possible that PNPase activity is limited in stationary phase, allowing persistence of these important virulence factor transcripts at this phase of growth.
Streptococcus pyogenes (the group A streptococcus [GAS]) is a common gram-positive human pathogen able to cause varied diseases with very different outcomes. GAS causes syndromes ranging from mild, superficial, self-limiting infections (“strep throat,” pyoderma) to severe invasive and often life-threatening illnesses (myositis, fasciitis, streptococcal toxic shock syndrome) (10). Since the GAS can grow in many different niches in its human host, it must be able to adapt to a variety of environmental conditions. This requires differential expression of genes, including those encoding virulence factors, in response to the local surroundings. Therefore, attempts to understand GAS pathogenesis have focused on identification of virulence factors and their regulation in response to local growth conditions in the host.
Virulence factors of GAS that have been characterized include surface-exposed adhesins, proteins that aid in evasion of the host immune response, toxins (including those that lyse host cells and therefore produce further changes in the microenvironment), and enzymes (proteases and DNases) that enable the GAS to spread through host tissues (10). Expression of these virulence factors in response to environmental stimuli is controlled by several transcriptional regulators, including “stand-alone” regulators such as Mga and RofA/Nra, as well as two-component signal transduction systems (26). In the GAS genomes whose sequences are available, 13 different two-component regulatory systems have been identified. Several of these are global regulators that affect transcription of a large number of different promoters, both directly and indirectly.
However, for all pathways of transcriptional control of gene expression examined in the GAS, growth-phase regulation is epistatic to the specific regulatory mechanism being studied (4, 31). For all genes studied in this organism, any specific transcript is only present at a restricted phase of growth, even under the most favorable regulatory conditions (presence of an activator and absence of a repressor). Thus, it seems important to understand the mechanism for what has been called “growth-phase regulation.”
As bacteria grow, they produce changes in their immediate environment that may trigger modifications in gene expression and entry into stationary phase. For example, when GAS are cultivated in the laboratory in Todd-Hewitt yeast extract broth, the pH drops as acid is produced from sugars, metabolic by-products accumulate in the medium, and specific nutrients are depleted, and it is also possible that specific “quorum-sensing” signals may be secreted (29, 30). Any of these environmental conditions may trigger the molecular alterations in the bacteria that occur on entry into stationary phase. Loughman and Caparon (28) found a correlation between genes expressed on entry into stationary phase in the laboratory with those expressed following subcutaneous infection in a murine model, which implies that events that occur during growth-phase transition are similar to those that occur during development of soft tissue infection. This suggests that an understanding of the mechanisms that control growth-phase regulation would be very helpful in understanding GAS pathogenesis.
It has long been recognized that most GAS mRNAs are not easily detectable in stationary phase when the cells are grown under standard laboratory conditions (21). A change in the transcriptome in stationary phase in most bacteria results from production of an accessory sigma factor (19). Its association with RNA polymerase leads to efficient transcription of some promoters and decreased transcription of others. However, because GAS lacks such a stationary-phase sigma factor, there must be a different explanation for phase-specific mRNA abundance in this organism. The amount of mRNA in a cell is determined by a combination of its rate of synthesis and its rate of decay. Although many studies have focused on the former, there are no reports yet on regulation of mRNA decay in the GAS. In this work, we found that, for the subset of transcripts studied, growth-phase-specific stability is the major contributor to growth-phase regulation.
MATERIALS AND METHODS
Bacterial growth conditions.
GAS strain MGAS315, a serotype M3 strain whose genome sequence has been determined (3), was used. GAS strains were grown in Todd-Hewitt medium with 0.2% yeast extract at 37°C in closed sidearm flasks, and Escherichia coli strains were grown in LB medium (36). Antibiotics were used at the following concentrations: spectinomycin, 50 μg/ml; tetracycline, 10 μg/ml; and kanamycin, 50 μg/ml.
RNA manipulations.
RNase protection assays (RPAs) were performed as described previously (11). The sequences of primers used for probe amplification are shown in Table S1 in the supplemental material. For mRNA decay experiments, rifampin was added to give 1 mg/ml and RNA was isolated and purified through a CsCl gradient as described previously (31). Primer extension was performed as described in reference 32, using primers sagA-PE2 (sagA) and sda-PE (sda). Primers are listed in Table S1 in the supplemental material. For Northern analysis, 5 μg total RNA was fractionated in 5% polyacrylamide-8 M urea gels and electroblotted to nylon membranes (Roche). An antisense 32P-labeled RNA probe was generated by in vitro transcription using T7 RNA polymerase (MaxiScript kit; Ambion) and a DNA template generated by PCR using MGAS315 DNA and primers sagA+1-S and T7sagA-R. Membranes were hybridized overnight in Ultrahyb buffer (Ambion) at 68°C and then washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) and twice in 0.1× SSC-0.1% SDS at 68°C.
Construction of RNase mutant strains.
The strategy for creation of nonpolar deletions/substitutions in pnpA, rnr and yhaM is described in the supplemental material (see Fig. S1 and primer sequences listed in Table S1 in the supplemental material). The final result is an internal deletion of the RNase gene and replacement of the deleted portion with a promoterless aad9 gene (from pUCSpec) (23) followed by a ribosome binding site and ATG in frame with the 3′ coding sequence.
Construction of pnpA-complementing plasmid pJRS1290.
Plasmid pREG696 is a theta replicon-type plasmid carrying aad9 into which the postsegregation killing system encoded by axe-txe has been cloned (16). This is a single-copy plasmid, and, in GAS, less than 1 in 200 colonies lose the plasmid following overnight culture (C. Bates and J. Scott, unpublished data). The vector pJRS9508 was derived from pREG696 by cloning the P23 promoter from plasmid pOri23 (35) in front of a tyrA reporter gene (D. Zahner and J. Scott, unpublished data). To create a tetracycline-resistant derivative of pJRS9508, the aad9 gene was first deleted by inverse PCR using primers pREGaad9-F2-Bgl and pREGaad9-R2-Bgl. The 7-kb inverse PCR product was digested with BglII and ligated to a 3-kb BamHI ΩtetM fragment (34) from pJRS303 to create pJRS1289.
Plasmid pJRS1290 contains a copy of pnpA from MGAS315 cloned into pJRS1289 under the P23 promoter. pnpA (coding for polynucleotide phosphorylase [PNPase]) was amplified by PCR from MGAS315 DNA using primers pnpA-F4-Bam and pnpA-R4-Xho. The resulting PCR product was digested with BamHI and XhoI and ligated to BamHI-XhoI-digested pJRS1289, to create pJRS1290.
Construction of JRS1288.
Construction of JRS1288, a derivative of MGAS315 that contains gene replacement mutations in sagA and covR, is described in the supplemental material.
Construction of Psag and Phas fusion plasmids pJRS1286, pJRS1287, and pJRS1293.
The Phas-sag (pJRS1286), Psag-sag (pJRS1287), and Psag-hasABC (pJRS1293) fusions were constructed by PCR using MGAS315 genomic DNA as a template and the high-fidelity enzyme PlatinumTaq-HF (Invitrogen). For pJRS1286, the Phas-sag fusion was constructed by a two-step overlapping PCR protocol as described previously (2). First round DNA fragments were amplified using primers PhasM3-NotI-F and PhasM3-sag-R (Phas) and PhasM3-sag-F and sagA-DS-Bam-R (sagA). Primers PhasM3-sag-F and PhasM3-sag-R contain a complementary 27-nucleotide sequence which facilitated the fusion of first round products in a second round PCR using primers PhasM3-NotI-F and sagA-DS-XhoI-R2. For pJRS1287, Psag-sagA region was amplified directly from MGAS315 DNA using primers Psag-NotI-F2 and sagA-DS-XhoI-R2. The resulting DNA fragments from each of these PCRs were digested with NotI and XhoI and cloned into NotI+XhoI-digested pJRS9508, to create pJRS1286 (Phas-sag) and pJRS1287 (Psag-sag).
Similarly, for pJRS1293, the Psag-hasABC fusion was constructed by overlapping PCR using primers Psag-NotI-F2 and Psag-has-R (Psag) and Psag-has-F and hasC-DS-XhoI-R (hasABC) for first round reactions and Psag-NotI-F2 and hasC-DS-XhoI-R for a second round reaction. Primers Psag-has-F and Psag-has-R contain a complementary 30-nucleotide sequence, which allowed the fusion of first round PCR products in the second round reaction. The resulting DNA fragment was digested with NotI and XhoI and cloned into NotI-XhoI-digested pJRS1289, to create pJRS1293.
RESULTS
Identification of mRNAs that are more abundant in stationary phase in MGAS315.
To gain insight into the mechanism of stationary-phase gene regulation, differential expression profiles were determined for a wild-type M3 strain (MGAS315) (3) grown to late exponential phase and 2 h following entry into stationary phase (see Fig. S2 in the supplemental material), times at which all of our previous growth-phase experiments were performed (e.g., reference 13). Data were acquired, normalized, and analyzed as described in Materials and Methods and the supplemental material.
To compare two transcriptomes, some normalization process is required to adjust for variation between different experiments. Because the amount of most transcripts was very small or undetectable in stationary phase in our study, there were no transcripts expressed equally well in both growth phases that would have been suitable for normalization. Therefore, we performed a “phase-specific” global normalization: i.e., the amount in stationary phase was normalized to the total in stationary phase and similarly for exponential phase (see supplemental material). From this analysis, the ratios of relative mRNA abundance in stationary phase versus exponential phase for individual genes were comparable to those measured by RNase protection and other assays (13; data not shown), confirming the validity of the approach. The cutoff we employed to identify significant quantitative differences in transcripts between the two growth phases was sufficiently stringent to result in 0.028% false positives (see Fig. S3 in the supplemental material). However, this stringency may have resulted in lack of identification of some transcripts that are only slightly more abundant in stationary phase.
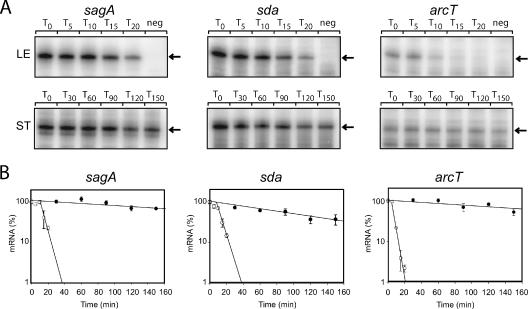
Using the above approach, we identified six genes whose transcripts constituted a greater fraction of the total mRNA 2 h into stationary phase than in late exponential phase (hereafter referred to as stationary-phase transcripts [SPTs]; Table 1), while the transcripts for 735 genes were less abundant in stationary phase than in exponential phase. The SPTs included two that had been previously identified, sagA and sda (13), thus validating the microarray results. In addition to these previously studied genes, we identified another member of the sag operon (sagI), genes encoded in a putative arginine deiminase operon (arcA and arcT), and a putative transposase (SPyM3_0221). These results were confirmed in independent experiments for three SPTs (sagA, sda, and arcT; Fig. 1 and Table 2) and three control transcripts (hasA, gyrA, and slo). The lack of identification of all members of the sag or arc operons may result either from processing of the transcript to fragments with different stabilities or from technical difficulties. We did not pursue this further.
TABLE 1.
Genes displaying increased transcript abundance in stationary phase
| M3 locusa | M1 locusb | Genea | Predicted functiona | Increase (fold) in stationary-phase transcript abundancec |
|---|---|---|---|---|
| SpyM3_0221 | NAd | NA | Putative IS1548-like transposase | 2.4 |
| SpyM3_0480 | Spy0738 | sagA | Streptolysin S precursor | 12 |
| SpyM3_0488 | Spy0746 | sagI | Streptolysin S biogenesis | 2.2 |
| SpyM3_1192 | Spy1542 | arcT | Peptidase | 4.8 |
| SpyM3_1196 | Spy1547 | arcA | Arginine deiminase | 2.3 |
| SpyM3_1745 | Spy2043 | sda | Streptodornase | 6.7 |
Genomic loci, gene names, and predicted function are based on the current annotation of the genome of S. pyogenes M3 strain MGAS315 (3) in The Institute of Genomic Research Complete Microbial Resource database (http://cmr.tigr.org).
M1 locus refers to strain SF370 (14).
Increase (fold) in transcript abundance of cultures grown 2 h into stationary phase versus that of cultures grown to late exponential phase as determined by microarray analysis.
NA, not applicable because there is no M1 homologue.
FIG. 1.
Decay rate of sagA, sda, and arcT transcripts in late exponential (LE) and stationary (ST) phases determined by RPAs. (A) RNA was extracted from MGAS315 at the times (minutes) indicated above each lane following the addition of rifampin to halt transcription. RPAs were performed as described previously (11) using 10 μg total RNA from exponential phase and 5 μg from stationary phase. A negative control for each probe (neg) consists of labeled probe without RNA. The arrow to the right of each gel indicates the expected size of the protected RNA probe following digestion with RNase T1. (B) The amount of each transcript in Fig. 1A was quantified using ImageQuant 5.0 software (Molecular Dynamics). The average intensity of each band was calculated for each time point. The y axes of the graphs represent the fraction (%) of each message remaining at the indicated time after addition of rifampin. Error bars represent the standard deviation of values obtained for each time point from two independent reactions. Regression analysis was performed using SigmaPlot 9.0 software (SPSS Inc.) and used to calculate the slope of each line. Open symbols, late exponential phase; closed symbols, stationary phase.
TABLE 2.
Approximate half-lives of individual transcripts in exponential and stationary phases
| Gene | Half-life (min) in:
|
|
|---|---|---|
| Exponential phase | Stationary phase | |
| sagA | 3.0a | 220a |
| sda | 5.1a | 100a |
| arcT | 2.7a | 170a |
| gyrA | 2.4b | NDc |
| hasA | 2.7b | ND |
| slo | 3.5b | ND |
Half-life calculated from RPAs in Fig. 2.
Half-life calculated from RPAs (data not shown).
ND, transcript not detected.
The sagA, sda, and arcT transcripts are more stable in stationary phase than in exponential phase.
The steady-state level of an mRNA is determined by its rate of synthesis (transcription) and its rate of decay. Because no SPTs encode transcriptional regulators, we thought that message stability might be an important determinant of stationary-phase mRNA abundance. Therefore, we compared decay rates of three SPTs (sagA, sda, and arcT) in late exponential phase and 2 h into stationary phase. For these experiments, transcription was arrested by the addition of 1 mg/ml rifampin, the concentration that we found to reproducibly inhibit an increase in cell density of exponential-phase cultures. Gene-specific antisense RNA probes were used in RPAs to quantitate transcripts at successive times following addition of rifampin (Fig. 1A and B). We found that the stability of each SPT increased at least 20-fold in stationary phase (Fig. 1). In contrast, three different transcripts, hasA, gyrA, and slo, whose half-lives in exponential phase were similar to those of the SPTs (Table 2), were undetectable in stationary phase (data not shown).
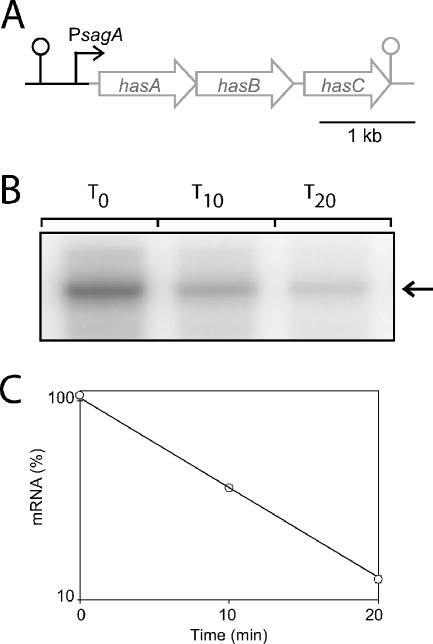
As a control to demonstrate that rifampin is active in stationary phase, we needed a transcript that was detectable but unstable at that stage of growth. Because we thought the hasA transcript would be unstable in stationary phase, we sought a strong promoter from which it could be expressed at that growth phase. We chose Psag, since we observed a significant amount of sagA transcript in stationary phase. For this experiment, we constructed plasmid pJRS1293 in which Psag was placed in front of hasABC (Fig. 2A; see Materials and Methods). To further improve expression from this promoter, a covR mutant strain (JRS1288) was used because CovR represses Psag (13). We found that in JRS1288/pJRS1293, hasA mRNA was detectable in stationary phase and was unstable. Following the addition of rifampin 2 h after entry into stationary phase, the hasA transcript decayed with about the same kinetics as in the exponential phase (Fig. 2B and C and Table 2) (data not shown) Note that the half-life was about 6.6 min in stationary phase and 2.7 min in exponential phase.) This demonstrates that rifampin is active in stationary-phase cells and that mRNA decay can be measured at that stage of growth. Therefore, this control validates the conclusion that the transcripts of the SPTs sagA, sda, and arcT are more stable in stationary phase than in exponential phase.
FIG. 2.
Decay of the hasABC transcript expressed from the Psag promoter in stationary-phase cells. (A) Diagram showing pJRS1293. The fusion consists of the entire hasABC transcript (postitions +1 to 3837; gray) fused to the Psag promoter region (black). The lollipop symbols represent predicted Rho-independent transcriptional terminators. RNase protection assays (B) and the decay curve calculations (C) were performed as described for Fig. 1.
The sagA and sda transcripts have the same 5′ ends in stationary and exponential phases.
In Escherichia coli and Bacillus subtilis, decay of most mRNAs is initiated by an endonuclease that binds at or near the 5′ end of the message. Consequently, the 5′ end of an mRNA is the primary determinant of stability (9). It seemed possible, therefore, that SPTs might have different 5′ ends in stationary and exponential phases. Results of primer extension experiments (see Fig. S4 in the supplemental material) demonstrated that sagA and sda mRNAs have identical 5′ ends in exponential phase and stationary phase, indicating that the 5′ end of SPTs is not the major determinant of their increased stationary-phase stability.
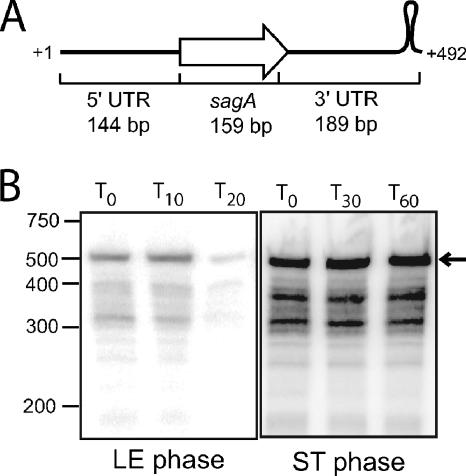
The sizes of the sagA transcript are the same in exponential and stationary phases.
Although the 5′ ends of the sagA transcript are the same in exponential and stationary phases, it was possible that stationary-phase mRNA was cleaved downstream and did not include the entire sagA open reading frame. To test this, we performed a high-resolution Northern blot using a probe for sagA that includes the entire open reading frame (ORF) as well as the 5′ untranslated region (UTR) (144 bases upstream of the ORF) and the 3′ UTR (189 bases), including the presumed RNA polymerase pause site stem-loop structure (Fig. 3A). The hybridization experiment (Fig. 3B) showed that the sagA transcript is the same length in exponential and stationary phases and that there is little accumulation of mRNA of an intermediate size in either phase.
FIG. 3.
Northern blot analysis of sagA mRNA decay in late exponential (LE) and stationary (ST) phases. (A) Diagram showing the region complementary to the RNA probe used to detect sagA messages. (B) RNA was extracted from MGAS315 at the times (minutes) indicated above each lane following the addition of rifampin to halt transcription. The size of RNA markers (in nucleotides) run on the same gel is indicated to the left. The expected size of the full-length sagA transcript is indicated with an arrow to the right.
PNPase is rate-limiting for decay for SPTs in exponential phase, but not for control transcripts.
One possible explanation for the increased stability of SPTs in stationary phase is that they are insensitive to the RNase that initiates decay of most messages and that the enzyme required for their decay is less active in stationary than in exponential phase. Because the major endonuclease required for mRNA decay in gram-positive bacteria has not been identified, and because there is no difference in the 5′ ends of SPTs isolated from the late exponential phase and 2 h following entry into stationary phase, we focused on 3′-5′ exonucleases that could be identified by homology. We identified three candidates: SPyM3_0195 (referred to as yhaM) encodes a protein which has 45.3% identity and 60.4% similarity to YhaM from B. subtilis, SPyM3_0352 (referred to as rnr) encodes a protein with 29.1% identity and 44.3% similarity to Rnr from E. coli and 40.4% identity and 54.7% similarity to Rnr from B. subtilis, and SPyM3_1676 (referred to as pnpA) encodes a protein with 45.1% identity and 58.4% similarity to Pnp from E. coli and 60.0% identity and 73.0% similarity to PnpA from B. subtilis. A nonpolar deletion mutation in each of these potential RNase genes was constructed (see Materials and Methods and see Fig. S1 in the the supplemental material).
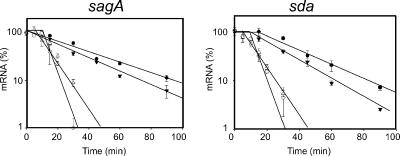
We were unable to detect any major difference in the decay rate of sda mRNA in the yhaM or the rnr mutant strains (data not shown), and these were not analyzed further. However, in the pnpA mutant (JRS1392), both SPT transcripts investigated (sagA and sda) decayed more slowly than they did in the parental strain (Fig. 4 and Table 3). However, it should be noted that these transcripts are more stable in stationary phase (Fig. 1) than in a pnpA mutant in exponential phase. As controls, hasA, slo, and gyrA were used. Little difference was observed in stability of these transcripts between the pnpA mutant and the wild-type parental strain (Table 3).
FIG. 4.
Exponential-phase decay of sagA and sda transcripts in MGAS315 (wild type), JRS1392 (pnpA), and complemented strains. Ten micrograms of total RNA was used for each RPA, and regression analyses were performed as described for Fig. 1. ○, MGAS315 (wild type); •, JRS1392 (ΔpnpA); ▾, JRS1392/pJRS1289 (vector control); ▿, JRS1392/pJRS1293 (pnpA complementation).
TABLE 3.
Approximate half-lives of individual transcripts in exponential phase
| Gene | Half-life (min) ina:
|
|||
|---|---|---|---|---|
| Wild type (MGAS315) | ΔpnpA (JRS1392) | pnpA complementation (JRS1392/pJRS1293) | Vector control (JRS1392/pJRS1289) | |
| sagA | 3.0 | 25 | 6.5 | 21 |
| sda | 5.1 | 19 | 7.1 | 17 |
| arcT | 2.7 | NTb | NT | NT |
| hasA | 2.7 | 5.5 | NT | NT |
| slo | 3.5 | 6.4 | NT | NT |
| gyrA | 2.4 | 3.2 | NT | NT |
Half-lives calculated from RPAs (data not shown).
NT, not tested.
To ensure that the phenotype observed in JRS1392 was caused by the pnpA mutation and not an unknown mutation elsewhere in the genome, complementation analysis was performed. The pnpA gene was amplified by PCR and cloned into the stably inherited single-copy vector pJRS1289 under the P23 promoter to create pJRS1290 (see Materials and Methods). We found that the pnpA mutation in JRS1392 was complemented by pnpA on plasmid pJRS1290, but not by the vector alone, since the rates of sagA and of sda mRNA decay returned to that of the wild type in the complemented strain (Fig. 4).
The stable sagA transcript in the pnpA mutant strain JRS1392 is full length.
The sagA transcript detected by RPA in the pnpA mutant strain could be either the full-length transcript or a stabilized decay intermediate resulting from endonuclease cleavage. To differentiate between these two possibilities, we analyzed total RNA isolated from MGAS315 and JRS1392 at various times following rifampin addition by Northern blot analysis using the probe described above (Fig. 3A). There was no observable accumulation of stable decay intermediates in either strain in late exponential phase, and the stable decay intermediates detected in stationary phase were present in the wild type as well as the mutant and represent a minor fraction of the sagA mRNA (see Fig. S5 in the supplemental material). We conclude from these results that the sagA transcript measured in the RPA experiments is full length and that the absence of PNPase does not result in accumulation of a stable decay intermediate.
Initiation of transcription also plays a role in determining the amount of sagA mRNA in stationary phase.
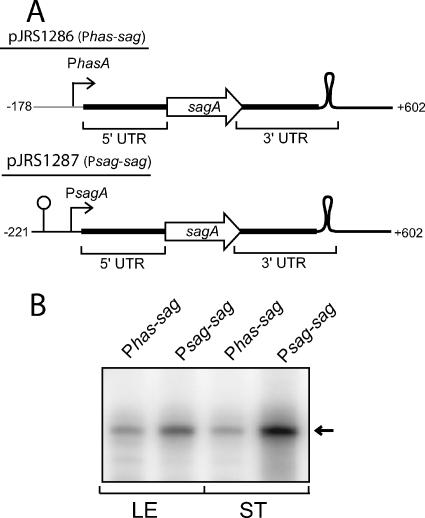
To gain insight into the contribution of transcription initiation to the abundance of sagA mRNA in stationary phase, we used the sagA transcript as a reporter of promoter activity since we have shown it to be stable in stationary phase. We investigated one promoter producing a message that is undetectable in stationary phase (Phas) and one producing an SPT (Psag). The Phas promoter fragment used includes the −10 element and 6 nucleotides downstream and is fused in front of the untranslated sagA leader (Fig. 5A). The sag DNA includes the sagA ORF and the region downstream, including 110 bases downstream of the transcriptional pause site that is present between sagA and sagB (Fig. 5A). For these experiments, the reporter fusion constructs were cloned into pJRS1289, a stable single-copy vector (see Materials and Methods). This allowed direct comparison of promoter strength based on the quantity of reporter sagA mRNA present. To ensure that the only sagA transcript produced would be that of the reporter, the chromosomal copy of the sagA gene was deleted from MGAS315. In addition, because both Phas and Psag are repressed by CovR, the covR gene was deleted to produce the strain used (JRS1288).
FIG. 5.
Stationary-phase expression of sagA mRNA from different promoters. (A) Diagram of the Phas-sagA and Psag-sagA constructs. The Phas-sagA fusion consists of the entire sagA transcript (+1 to +602; thick black line) fused to the Phas promoter (gray line) by overlap-extension PCR. The loop downstream of sagA represents a transcriptional pause site at which most of the mRNA produced from Psag is terminated in vivo. (B) RPAs were performed on 100 ng RNA. The arrow to the right indicates the expected size of the protected RNA probe.
The Phas and Psag reporter plasmids pJRS1286 and pJRS1287, respectively (Fig. 5A), were transformed into strain JRS1288. Because the plasmids and chromosomal mutations are stable, no antibiotics were used in the growth medium. RNA was isolated in the late exponential phase and 2 h into stationary phase, and the amount of sagA mRNA was quantitated by RNase protection assays using the sagA probe used for Fig. 1. In the exponential phase, there was little difference (about twofold) in the amount of sag transcript from the two different promoters, while in stationary phase, there was much more transcript (about ninefold) from Psag (Fig. 5B). Since transcription from both promoters produces the same transcript, mRNA stability is not relevant to this quantitative difference. Therefore, we conclude that Psag is more active than Phas in stationary phase. These results indicate that the steady-state level of sagA mRNA in stationary phase is determined both by the strength of the promoter and by the increased stability of the message.
DISCUSSION
The stationary-phase transcriptome.
The success of GAS as a pathogen that can grow in many different environmental niches in its human host is dependent on its ability to regulate gene expression in response to its microenvironment. Although many transcriptional regulators have been characterized in GAS, all of these systems are affected by a global mechanism of growth-phase regulation. To gain insight into the mechanisms of stationary-phase gene regulation in GAS, we identified the stationary-phase transcriptome of the serotype M3 GAS strain MGAS315 using microarray analysis.
We confirmed the previous reports that the transcripts for the secreted virulence factors sagA and sda are more abundant in stationary phase. This is reminiscent of virulence factor regulation in Staphylococcus aureus, where most secreted virulence factors are predominantly expressed in stationary phase, while other virulence factors are predominantly transcribed in exponential phase. While their mechanisms of control may be completely different, it seems possible that increased stationary-phase production of secreted enzymes, with the concomitant decrease in the expression of adhesins, could favor the spread of these organisms through host tissues. This would also allow them to escape the poor local growth conditions that caused their initial entry into stationary phase to begin with.
We also identified transcripts for the arginine catabolism operon (arc) as being SPTs, in agreement with a proteomics analysis that identified the proteins encoded by these genes as more abundant in stationary phase (7). Increased expression of these genes may reflect the need to use arginine as an energy source when other carbon sources (e.g., glucose) become limited. In addition, utilization of arginine by this pathway has been shown to protect oral streptococci (5) and Lactobacillus sakei (6) against the harsh stationary-phase environment, either through the production of ammonia to neutralize the low pH or by a pH-independent mechanism.
Although we identified two transcripts in both the sag operon and the arc operon as more abundant in stationary phase, the other genes in these operons were not identified by our microarrays. This suggests that the stability of different parts of a polycistronic transcript may differ (1, 18). While differential decay rates of fragments generated by endonucleolytic cleavage of the sag and arc mRNAs could account for our observations, it should also be remembered that the stringency of the cutoff for our microarrays might lead to underreporting of transcripts more abundant in stationary phase.
Transcription initiation plays a role in stationary-phase abundance of sagA mRNA.
It appears from our results using fusions to sagA as a reporter transcript that the sag promoter, Psag, is recognized and active in stationary phase, while the has promoter, Phas, may not be active at that stage of growth (Fig. 5). In late exponential phase, the promoters appear to have similar strengths, since the amount of sagA transcribed from Psag is within about twofold of that transcribed from Phas. In addition, the strengths of these promoters appear to be similar when transcribed in vitro using GAS RNA polymerase (15, 17). However, using a sagA reporter transcript, we found more mRNA when it is transcribed from Psag 2 h following entry into stationary phase than when it is transcribed from Phas (Fig. 5). Since the same message is produced from the two different promoters, mRNA stability should not affect the relative abundance of these transcripts. In addition, we were able to detect incorporation of 32P into sagA mRNA expressed from Psag when added to the growth medium 2 h after the onset of stationary phase (data not shown). Therefore, we conclude that Psag is active in stationary phase, while Phas may not be recognized well by RNA polymerase in stationary phase.
Growth phase-specific transcription from a promoter may result from growth-phase-specific repression and/or activation. For example, Psag may be repressed in exponential phase by a transcriptional regulator that becomes inactive later in growth. Several negative transcriptional regulators in GAS, including CovR (13), LuxS (29), Rgg (8), and FasX (25), influence the expression of one or more of the SPT genes. However, none seems to be specific for SPTs. Alternatively, or in addition, Psag may be up-regulated in stationary phase. Evidence for positive regulation of Psag comes from the demonstration of induction of this promoter during lag phase, when expression of most genes is normally very low, by the addition of spent culture medium from late exponential phase (30). However, this investigation did not examine whether this effect was specific for Psag or other SPTs. Because no transcriptional regulator was identified in our microarrays as an SPT, if a growth-phase-specific activator exists, it is probably only expressed transiently so that its mRNA is unable to be detected 2 h into stationary phase. However, although the mechanism is not yet known, our results demonstrate that differential transcription does play a role in growth phase-specific mRNA abundance, at least for the sagA SPT.
mRNA decay is of primary importance for growth phase-specific mRNA abundance.
The steady-state level of an individual mRNA is determined both by its rate of synthesis and by its rate of decay. The results of this study suggest that differential mRNA stability is the major determinant of stationary-phase message abundance. This is supported by the finding that all three SPTs tested have dramatically increased stability in stationary phase while other transcripts remain unstable. Furthermore, in stationary phase, the amount of the SPT transcript for sagA is much greater than that of the control transcript for hasABC, even when both are transcribed from the same promoter. This conclusion is based on the need to use a large amount of RNA (20 μg) to detect the hasABC transcript (which decays rapidly) compared to the amount (100 ng) needed to detect the sag transcript (an SPT). Therefore, the stability of the mRNA synthesized from Phas has a greater effect on the abundance of the transcript from this promoter than does the strength of the promoter.
The SPTs we studied are much more stable in stationary phase than most mRNAs that have been investigated in bacteria. The half-lives of about 80% of the mRNAs of E. coli and B. subtilis are less than 8 min, as determined by global mRNA stability analyses (3a). Even for “stable” mRNAs, half-lives only in the range of 15 to 45 min have been reported (20, 22, 24). In contrast, in GAS, half-lives of SPTs ranged up to about 225 min in stationary phase.
Decay of SPTs.
While there are differences in the mRNA decay pathways of E. coli and B. subtilis, message decay in both organisms is primarily initiated by endonucleolytic cleavage by an enzyme that recognizes the 5′ end of a transcript, and this is usually the rate-limiting step in decay (9). The endonuclease cleavage products are then processively digested by 3′-to-5′ exonucleases. This decay pathway was demonstrated by the accumulation of endonuclease cleavage products in exonuclease mutants (12, 33, 37).
To define the decay pathway for SPTs, we constructed mutations in each of three putative 3′-to-5′ exonuclease genes. While deletion of the gene for either of the exonucleases encoded by rnr (RNase R) or yhaM had no major effect on SPT decay, deletion of the gene encoding polynucleotide phosphorylase had a dramatic effect on decay of sagA and sda, the two SPTs tested. In late exponential phase, these transcripts are more stable in a pnpA deletion mutant than in the parental wild-type strain (8.3-fold for sagA and 3.7-fold for sda), and their stability returns to the wild-type level when pnpA is provided in trans on a plasmid. The rate of decay of control transcripts that are not SPTs is not significantly affected by the pnpA deletion. Since PNPase appears to be required for SPT decay, but some other RNase is able to substitute for PNPase for the decay of other transcripts, it appears that SPTs are not subject to digestion by the pathway used for most GAS transcripts. We also note that there must be at least one additional RNase involved in decay of SPTs in exponential phase since SPTs are more stable in stationary phase than in a PNPase mutant.
Although mRNA decay is usually initiated by a 5′ endonuclease cleavage, this does not seem to be the case for the SPT sagA. If this occurred, mutations in the exonuclease required for further decay should have resulted in accumulation of endonuclease cleavage products. However, for sagA, in the pnpA mutant in exponential phase, no decay products were detectable and the transcript remained full length. Furthermore, the 5′ end of the sagA transcript in exponential phase, in which sagA mRNA decays, is identical to that in stationary phase and that produced in vitro from this promoter. Together, these results imply that at least this SPT is degraded by a different pathway from other GAS mRNAs and that PNPase is rate limiting for its decay. We expect, therefore, that in stationary phase, SPTs become stable because they are either protected from this specific decay pathway (27, 38) or because this pathway becomes defective or inactive in stationary phase.
Why are SPTs insensitive to the major GAS mRNA decay pathway?
One reason SPTs are insensitive to the major GAS mRNA decay pathway may be that there is no internal site in SPTs that is susceptible to cleavage by the 5′ endonuclease responsible for initiating decay of most GAS mRNAs. Alternatively, or in addition, the 5′ end of SPTs may be inaccessible to the endonuclease because of RNA secondary structure or because the message is associated with a protein, a small RNA, or ribosomes. Rapid translation may also protect SPTs from decay. Further investigation is required to identify which of these factors is/are involved in insensitivity of SPTs to the major endoribonuclease of the GAS decay pathway.
Concluding remarks.
Bacteria adapt to different niches by modifying gene expression in response to environmental stimuli. In GAS, growth-phase control of gene expression is superimposed on all other levels of gene regulation and therefore must be understood to begin to comprehend the interactions of this pathogen with its host. In this study, we demonstrated that in GAS, the rate of mRNA decay is the major factor affecting growth-phase mRNA abundance for the SPTs we studied. Although SPTs are degraded in exponential phase with kinetics similar to most GAS mRNAs, the pathway for SPT decay appears to be different. As the cells enter stationary phase, this pathway becomes defective, resulting in dramatically increased mRNA stability, while the bulk of GAS mRNAs are degraded normally. Further experimentation is required to identify the features of SPTs that make them insensitive to the normal decay pathway, to elucidate the molecular details of the pathway by which they decay, and to learn why this pathway is growth-phase specific.
Supplementary Material
Acknowledgments
We thank Hannah Ross-Suits for help with strain constructions.
This work was supported in part by grant RO1-AI20723 from the National Institutes of Health.
Footnotes
Published ahead of print on 22 December 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baga, M., M. Goransson, S. Normark, and B. E. Uhlin. 1988. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell 52:197-206. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, T. C., A. R. Patel, and J. R. Scott. 2004. A novel sortase, SrtC2, from Streptococcus pyogenes anchors a surface protein containing a QVPTGV motif to the cell wall. J. Bacteriol. 186:5865-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casiano-Colón, A., and R. E. Marquis. 1988. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl. Environ. Microbiol. 54:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champomier Verges, M. C., M. Zuniga, F. Morel-Deville, G. Perez-Martinez, M. Zagorec, and S. D. Ehrlich. 1999. Relationships between arginine degradation, pH and survival in Lactobacillus sakei. FEMS Microbiol. Lett. 180:297-304. [DOI] [PubMed] [Google Scholar]
- 7.Chaussee, M. A., E. A. Callegari, and M. S. Chaussee. 2004. Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J. Bacteriol. 186:7091-7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condon, C. 2003. RNA processing and degradation in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 67:157-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalton, T. L., and J. R. Scott. 2004. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J. Bacteriol. 186:3928-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donovan, W. P., and S. R. Kushner. 1986. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 83:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, J., A. A. Gusa, J. R. Scott, and G. Churchward. 2005. Binding of the global response regulator protein CovR to the sag promoter of Streptococcus pyogenes reveals a new mode of CovR-DNA interaction. J. Biol. Chem. 280:38948-38956. [DOI] [PubMed] [Google Scholar]
- 16.Grady, R., and F. Hayes. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 47:1419-1432. [DOI] [PubMed] [Google Scholar]
- 17.Gusa, A. A., J. Gao, V. Stringer, G. Churchward, and J. R. Scott. 2006. Phosphorylation of the group A streptococcal CovR response regulator causes dimerization and promoter-specific recruitment by RNA polymerase. J. Bacteriol. 188:4620-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heck, C., A. Balzer, O. Fuhrmann, and G. Klug. 2000. Initial events in the degradation of the polycistronic puf mRNA in Rhodobacter capsulatus and consequences for further processing steps. Mol. Microbiol. 35:90-100. [DOI] [PubMed] [Google Scholar]
- 19.Hengge-Aronis, R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:341-346. [PubMed] [Google Scholar]
- 20.Hienonen, E., A. Rantakari, M. Romantschuk, and S. Taira. 2004. The bacterial type III secretion system-associated pilin HrpA has an unusually long mRNA half-life. FEBS Lett. 571:217-220. [DOI] [PubMed] [Google Scholar]
- 21.Hollingshead, S. K., V. A. Fischetti, and J. R. Scott. 1987. A highly conserved region present in transcripts encoding heterologous M proteins of group A streptococci. Infect. Immun. 55:3237-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hue, K. K., S. D. Cohen, and D. H. Bechhofer. 1995. A polypurine sequence that acts as a 5′ mRNA stabilizer in Bacillus subtilis. J. Bacteriol. 177:3465-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Husmann, L. K., J. R. Scott, G. Lindahl, and L. Stenberg. 1995. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect. Immun. 63:345-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurgen, B., T. Schweder, and M. Hecker. 1998. The stability of mRNA from the gsiB gene of Bacillus subtilis is dependent on the presence of a strong ribosome binding site. Mol. Gen. Genet. 258:538-545. [DOI] [PubMed] [Google Scholar]
- 25.Kreikemeyer, B., M. D. Boyle, B. A. Buttaro, M. Heinemann, and A. Podbielski. 2001. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 39:392-406. [DOI] [PubMed] [Google Scholar]
- 26.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 27.Lee, K., X. Zhan, J. Gao, J. Qiu, Y. Feng, R. Meganathan, S. N. Cohen, and G. Georgiou. 2003. RraA. a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell 114:623-634. [PubMed] [Google Scholar]
- 28.Loughman, J. A., and M. Caparon. 2006. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J. Bacteriol. 188:399-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 30.Mangold, M., M. Siller, B. Roppenser, B. J. Vlaminckx, T. A. Penfound, R. Klein, R. Novak, R. P. Novick, and E. Charpentier. 2004. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol. Microbiol. 53:1515-1527. [DOI] [PubMed] [Google Scholar]
- 31.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opdyke, J. A., J.-G. Kang, and G. Storz. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186:6698-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oussenko, I. A., T. Abe, H. Ujiie, A. Muto, and D. H. Bechhofer. 2005. Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 187:2758-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 35.Que, Y.-A., J. A. Haefliger, P. Francioli, and P. Moreillon. 2000. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect. Immun. 68:3516-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott, J. R. 1974. A turbid plaque-forming mutant of phage P1 that cannot lysogenize Escherichia coli. Virology 62:344-349. [DOI] [PubMed] [Google Scholar]
- 37.Spickler, C., and G. A. Mackie. 2000. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J. Bacteriol. 182:2422-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, M., L. Zhou, Y. Kawarasaki, and G. Georgiou. 2006. Regulation of RraA, a protein inhibitor of RNase E-mediated RNA decay. J. Bacteriol. 188:3257-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.