Abstract
The asbABCDEF gene cluster from Bacillus anthracis is responsible for biosynthesis of petrobactin, a catecholate siderophore that functions in both iron acquisition and virulence in a murine model of anthrax. We initiated studies to determine the biosynthetic details of petrobactin assembly based on mutational analysis of the asb operon, identification of accumulated intermediates, and addition of exogenous siderophores to asb mutant strains. As a starting point, in-frame deletions of each of the genes in the asb locus (asbABCDEF) were constructed. The individual mutations resulted in complete abrogation of petrobactin biosynthesis when strains were grown on iron-depleted medium. However, in vitro analysis showed that each asb mutant grew to a very limited extent as vegetative cells in iron-depleted medium. In contrast, none of the B. anthracis asb mutant strains were able to outgrow from spores under the same culture conditions. Provision of exogenous petrobactin was able to rescue the growth defect in each asb mutant strain. Taken together, these data provide compelling evidence that AsbA performs the penultimate step in the biosynthesis of petrobactin, involving condensation of 3,4-dihydroxybenzoyl spermidine with citrate to form 3,4-dihydroxybenzoyl spermidinyl citrate. As a final step, the data reveal that AsbB catalyzes condensation of a second molecule of 3,4-dihydroxybenzoyl spermidine with 3,4-dihydroxybenzoyl spermidinyl citrate to form the mature siderophore. This work sets the stage for detailed biochemical studies with this unique acyl carrier protein-dependent, nonribosomal peptide synthetase-independent biosynthetic system.
In pathogenic bacteria, iron acquisition is a significant enabling factor in establishing access to and maintaining growth within the host and is often required for disease manifestation (29). Typically, bacteria that are unable to obtain physiological iron fail to grow and are subsequently eliminated by the host due to nutrient starvation or direct attack by host defense mechanisms (29). Free iron in body fluids of animals is maintained at very low levels by proteins such as transferrin or lactoferrin. This serves as a protective mechanism in animals to reduce the availability of a nutrient vital to bacterial growth (4).
As a primary mechanism for iron acquisition, pathogenic bacteria synthesize and employ high-affinity chelating molecules (known as siderophores) in order to obtain iron from host sources. Siderophores are low-molecular-weight compounds that bind ferric iron [Fe(III)] with an extremely high affinity (3, 32). Once the siderophore has scavenged ferric iron from the environment or host, the resulting holo complex binds to high-affinity receptors and is transported into the bacterial cells by a membrane-associated ATP-dependent transport system (24).
Most siderophores can be chemically categorized as catecholates, hydroxamates, or α-hydroxy carboxylates based on their ferric iron ligand-binding functional groups. Many of these molecules are polypeptides that are biosynthetically derived from the nonribosomal peptide synthetases (NRPS), a family of multifunctional enzymes that also mediate the biosynthesis of microbe-derived peptide antibiotics (7). The enzymology of NRPS-dependent siderophore biosynthesis has been studied extensively over the past decade, and details of their assembly are well understood (7). Interestingly, several bacterial siderophores have been structurally characterized that are NRPS independent, derived from alternating dicarboxylic acid and diamine or amino alcohol building blocks linked by amide or ester bonds (6).
Bacillus anthracis is a gram-positive, spore-forming bacterium that can cause serious disease and fatalities in humans and animals. Anthrax can occur after spores enter the host following inhalation or ingestion or through abrasions in the skin (10). Among the known biological-warfare agents, B. anthracis is widely accepted as one of the most serious threats because of the resilience of its spores and potential use as a bioweapon (19, 20). Recently, Cendrowski and colleagues (5) revealed that six genes in an apparent operon (asb) specify synthesis of the virulence-associated siderophore in B. anthracis. This conclusion was based on the phenotype of a B. anthracis Sterne asb mutant strain that exhibited severely limited growth in cultured macrophages and was attenuated for virulence in mice (5).
Over the past several years, a number of studies have revealed the structural details of unique NRPS-independent bacterial siderophores. Garner et al. demonstrated that B. anthracis Sterne produces a catecholate siderophore based on a novel 3,4-dihydroxybenzoate biosynthetic precursor (16). Subsequently, Koppisch and colleagues reported (23) that the primary siderophore produced by B. anthracis Sterne is identical to petrobactin, originally isolated from Marinobacter hydrocarbonoclasticus, a gram-negative bacterium that is capable of degrading hydrocarbon compounds (1). The initially reported structure of petrobactin indicated that the compound was comprised of a citrate bis-spermidine backbone, with the two 2,3-dihydroxybenzoyl moieties providing four of the requisite six donor groups for Fe(III) (1). Subsequently, total synthesis of both 2,3- and 3,4-dihydroxybenzoyl compounds unequivocally demonstrated that petrobactin utilizes a unique 3,4-dihydroxybenzoyl fragment, rather than a 2,3-dihydroxybenzoyl donor (2, 15). Wilson et al. then demonstrated (33) that the B. anthracis asb locus is necessary and sufficient for the production of petrobactin by B. anthracis (henceforth referred to as petrobactinBa, to differentiate it from the chemically identical siderophore isolated from M. hydrocarbonoclasticus, petrobactinMh).
Based on these results, we were motivated to establish experimentally the basis for assembly of petrobactin from its biosynthetic precursors, including 3,4-dihydroxybenzoate, spermidine, and citrate. In this report, we describe the first details on the biosynthesis of petrobactin by identification and structure elucidation of two petrobactinBa intermediates isolated from engineered B. anthracis asb mutant strains. In addition, we show the ability of individual asb mutants to be complemented following exogenous supplementation by petrobactinBa, petrobactinMh, and a series of heterologous siderophores. Taken together, these data provide the basis for assigning the order of assembly and presumed role for five of the six gene products encoded by the asb operon.
MATERIALS AND METHODS
Chemical materials.
Authentic siderophore standards were obtained as iron-free compounds. The hydroxamate-type siderophore aerobactin (see Fig. 1) was purchased from EMC Microcollections GmbH (Tübingen, Germany). The catecholate-type siderophore salmochelin S4 (see Fig. 1) was kindly provided by Klaus Hantke at Universität Tübingen (Tübingen, Germany). All siderophores were dissolved in pure water as stock solutions in this study.
FIG. 1.
Chemical structures of petrobactin, aerobactin, and salmochelin S4.
Bacterial strains.
Bacterial strains used for this study are summarized in Table 1. All B. anthracis strains were derived from the wild-type strain Sterne 34F2 (31) and were propagated at 37°C in LB or LB plus kanamycin (40 μg/ml) as necessary. Marinobacter hydrocarbonoclasticus DSM 8798 (ATCC 49840) was obtained from the DSMZ GmbH (Braunschweig, Germany) and was propagated at 28°C in Bacto marine broth (Difco). XL10-Gold Kan cells were used in DNA cloning/plasmid construction experiments. All plasmid DNA was passaged through the dam dcm INV110 strain prior to introduction into B. anthracis.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| Bacillus anthracis | ||
| Sterne 34F2 | Wild type (pXO1+, pXO2−) | 31 |
| SC107 | 34F2, Δasb::kmr | 5 |
| BA850 | 34F2, ΔasbABCDEF | This work |
| BA851 | 34F2, ΔasbA | This work |
| BA852 | 34F2, ΔasbB | This work |
| BA853 | 34F2, ΔasbC | This work |
| BA854 | 34F2, ΔasbD | This work |
| BA855 | 34F2, ΔasbE | This work |
| BA866 | 34F2, ΔasbF | This work |
| BA867 | 34F2, ΔasbAB | This work |
| BA868 | 34F2/pHP13 | This work |
| BA869 | BA851/pHP13 | This work |
| BA870 | BA851/pBKJ358 | This work |
| BA871 | 34F2/pAD123 | This work |
| BA872 | BA852/pAD123 | This work |
| BA873 | BA852/pBKJ359 | This work |
| Escherichia coli | ||
| XL10-Gold Kan | Tetr Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′ proAB lacIqZΔM15 Tn10 (Tetr) Tn5 (Kanr) Amy] | Stratagene |
| INV110 | F′ {traΔ36 proAB lacIqlacZΔM15} rpsL (Strr) thr leu endA thi-1 lacY galKgalT ara tonAtsx dam dcm supE4Δ(lac-proAB) Δ(mcrC-mrr)102::Tn10(Tetr) | Invitrogen |
| Marinobacter hydrocarbonoclasticus | ||
| DSM 8798 | Wild type | 1 |
| Plasmids | ||
| pBKJ258 | Allelic exchange vector, Emr | This work |
| pBKJ351 | ΔasbA construct cloned in pBKJ258 | This work |
| pBKJ352 | ΔasbB construct cloned in pBKJ258 | This work |
| pBKJ353 | ΔasbC construct cloned in pBKJ258 | This work |
| pBKJ354 | ΔasbD construct cloned in pBKJ258 | This work |
| pBKJ355 | ΔasbE construct cloned in pBKJ258 | This work |
| pBKJ356 | ΔasbF construct cloned in pBKJ258 | This work |
| pBKJ357 | ΔasbAB construct cloned in pBKJ258 | This work |
| pHP13 | Gram-positive, E. coli shuttle vector; Cmr Emr | 17 |
| pAD123 | Gram-positive, E. coli shuttle vector; Apr Emr | 11 |
| pBKJ358 | asbA cloned into pHP13 | This work |
| pBKJ359 | asbB cloned into pAD123 | This work |
Construction of mutants in the asb operon.
The mutant strains isolated for this work were all constructed via allelic exchange. Each mutant allele was constructed via PCR (oligonucleotide primer sequences are presented in Table 2) using Phusion High Fidelity DNA polymerase (New England Biolabs) according to the manufacturer's instructions. The PCR product was cloned into the allelic exchange vector pBKJ258 and the DNA sequence verified. Each construct contained approximately 500 bp both upstream and downstream of the target gene, with the DNA sequence CCCGGG (the recognition sequence for the restriction endonuclease SmaI) introduced between these two flanking regions. For the ΔasbA mutation, the first 18 nucleotides were retained, as well as the last 18 (this number includes the predicted stop codon). This resulted in a deletion of 98% (1,773 out of 1,809 nucleotides) of asbA. A similar strategy was followed for each mutant. For the ΔasbB mutation, the initial 21 nucleotides were fused with the final 105 nucleotides of the gene, resulting in a deletion of 93% (1,713 out of 1,839 nucleotides) of asbB. For the ΔasbC mutation, the initial 21 nucleotides were fused with the final 186 nucleotides of the gene, resulting in a deletion of 83% (1,032 out of 1,239 nucleotides) of asbC. For the ΔasbD mutation, the initial 15 nucleotides were fused with the final 30 nucleotides of the gene, resulting in a deletion of 84% (231 out of 276 nucleotides) of asbD. For the ΔasbE mutation, the initial 18 nucleotides were fused with the final 21 nucleotides of the gene, resulting in a deletion of 96% (945 out of 984 nucleotides) of asbE. For the ΔasbF mutation, the initial 15 nucleotides were fused with the final 18 nucleotides of the gene, resulting in a deletion of 96% (810 out of 843 nucleotides) of asbF. For the entire operon deletion, ΔasbABCDEF, the initial 18 nucleotides of asbA were fused with the final 18 nucleotides of asbF, resulting in a deletion of 99% (7,054 out of 7,090 nucleotides) of the entire operon. Finally, for the ΔasbAB mutation, the initial 18 nucleotides of asbA were fused with the final 105 nucleotides of asbB, resulting in a deletion of 97% (3,583 out of 3,706 nucleotides) of the region containing asbA and asbB. Care was taken to avoid deleting sequences important for expression of the downstream gene in these mutants, especially for the ΔasbB mutant (asbB and asbC are predicted to overlap by 14 bp) and the ΔasbC mutant (asbC and asbD are predicted to overlap by 4 bp). When the gene directly upstream was deleted, the following number of base pairs was retained for each mutant: for asbB (in the ΔasbA mutant), 76 bp; for asbC, 91 bp; for asbD, 182 bp; for asbE, 44 bp; and for asbF, 55 bp. To isolate the various mutations described above, allelic exchange was performed as described previously (21) with the following modification: the allelic exchange vector pBKJ258, which has a larger polylinker region, was used instead of the otherwise identical pBKJ236. Allelic exchange was verified by PCR, and the resulting mutants are otherwise isogenic to the parental strain. Spores were generated as described previously, except that growth of the cultures was performed at 37°C instead of 30°C (28). All subsequent experiments were performed from these stocks.
TABLE 2.
Oligonucleotide primers used in this study
| Targeta | Nucleotide sequence (5′ to 3′) |
|---|---|
| ΔasbA | CGCGCGGCCGCCAAGAGTATCAAGGAATGAAAATAGGAC |
| CGCCCCGGGTTGTTTCGCATGCTTCATACTTGCCCTC | |
| CGCCCCGGGGTGGCTGTTCGTTCATAAATAAGGTTAAAATTTC | |
| CGCGGATCCCTTATTACATCTTTCATATTGTGCAATCGTATC | |
| ΔasbB | CGCGCGGCCGCCAGCTAAAGGATGGTTATCCGGTC |
| CGCCCCGGGATGATACATATCCATTCTCATACCCCCT | |
| CGCCCCGGGATAGATGTGGAATCGCTAGTTCATGAAGTTCCA | |
| CGCGGATCCCCATAACAATCGGTACTTCATCTATGTCAC | |
| ΔasbC | CGCGCGGCCGCCATAAAGAAGGATTACCTGTCCGTATTGC |
| CGCCCCGGGTTCTCTATTAACAATTAGCATAATTTCCCCCT | |
| CGCCCCGGGATGGGGGAAATTGTTAAAGCGAAAGTAATC | |
| CGCGGATCCCTGTTATATCAAAGTCTCCATCCCATAGTCC | |
| ΔasbD | CGCGCGGCCGCGATGCAACAATACGGTTGTTCTGAAGC |
| CGCCCCGGGCGCTTCCCGTCTCATGTTGTAACC | |
| CGCCCCGGGCCGTTACAGGACGTAAATGTGAATAACTAAC | |
| CGCGGATCCCCCTTCCCAGCGAAAATCAGGATC | |
| ΔasbE | CGCGCGGCCGCGCAAGAGGCGATTGTATATCGAGGG |
| CGCCCCGGGCACTTTAATTGAAGTCATACTATCATCCACTTTC | |
| CGCCCCGGGGAGTCTGTCTTAGTATTTTAAAAACTTTACTG | |
| CGCGGATCCGAAGGCAACGTATCCGTTAACGTATTTG | |
| ΔasbF | CGCGCGGCCGCCAGGAGCCAACAGCAGAAATGGTAG |
| CGCCCCGGGTAGTGAATATTTCATAGGTTTGAACTCCC | |
| CGCCCCGGGGAAGTAGTAACTTCTTAATATAAAATGATGAAAGAG | |
| CGCGGATCCGGCTATTACATCTTTACTACTTCCCATTC | |
| ΔasbAB | AACTAGCGATTCCACATCCCCGGGTTGTTTCGCATGCTTCATACTTG |
| ATGAAGCATGCGAAACAACCCGGGGATGTGGAATCGCTAGTTCATG | |
| Diagnosticb | TAACATATGAGCAACGAGAAAAACAATGGG |
| CAAACTCGGAACCATTACCATATTTCTCCC | |
| TTCCCCTCGTAATTGCAGATAACGTACCAC | |
| GGGATCGCACTCGAATCACA | |
| asbA complementation | CGCGGTACCCAAGAGTATCAAGGAATGAAAATAGGACAAAAG |
| CGCGGTACCTTATGAACGAACAGCCACTTCTCTAACG | |
| asbB complementation | CGCGGTACCCAAGAGTATCAAGGAATGAAAATAGGACAAAAG |
| ATACATATCCATTCTCATACTTGCCCTCCTCTTTCTATAAACAC | |
| AAGAGGAGGGCAAGTATGAGAATGGATATGTATCATACGAAAATATTG | |
| CGCGGTACCTTAACAATTAGCATAATTTCCCCCTG |
Four oligonucleotide primers (i.e., two pairs) were used to generate each allelic exchange construct. One pair (the first two listed) generated a fragment with homology upstream to the specific gene, and the other pair (the last two listed) generated a fragment with homology downstream to the gene. The entire operon deletion (ΔasbABCDEF) was constructed using the upstream primer pair from the ΔasbA set and the downstream primer pair from the ΔasbF set. The upstream homology for the ΔasbAB mutant was generated using the first primer from the ΔasbAB set and the first primer from the ΔasbA set; the downstream homology was generated using the second primer from the ΔasbAB set and the fourth primer from the ΔasbB set.
The first and third diagnostic primers were used to identify the ΔasbA, ΔasbB, and ΔasbAB mutations; the second and fourth diagnostic primers were used to identify the ΔasbC, ΔasbD, ΔasbE, and ΔasbF mutation; the first and second diagnostic primers were used to identify the ΔasbABCDEF mutation.
Bacterial growth conditions.
For growth in iron-depleted conditions, vegetative cells from rich media were washed and inoculated into iron-depleted medium (IDM) (5). Overnight cultures started from 1 μl of purified spore preparation were grown in LB, brain heart infusion (BHI), or LB plus kanamycin (40 μg/ml) and were diluted 1:10 in fresh medium and allowed to recover for 1 h at 37°C. Cells were collected by centrifugation at 3,000 rpm, and the pellets were washed twice in phosphate-buffered saline followed by a final wash in IDM; the resulting pellet was resuspended in 5 ml of IDM. The optical density at 600 nm (OD600) was determined, and cultures were diluted to an OD600 of 0.01 in a 60- or 70-ml volume of IDM; cultures were shaken at 250 rpm at 37°C, and the OD600 was read every hour for 8 to 10 h. For feeding experiments with exogenous siderophores, each purified molecule was prepared and added to a final concentration of 2 μM. Outgrowth from B. anthracis spores was performed in a microplate reader (SpectraMax M2; Molecular Devices) essentially as described previously (25). Spores (5 × 104) were inoculated into 200 μl of medium in a 96-well microplate and incubated at 37°C with near-constant agitation. Growth was followed by measurement of optical density at 600 nm for 10 h.
Disc diffusion assays.
Vegetative cells of the wild type and the various asb mutants, grown to mid- to late exponential phase in LB medium, were diluted 1:1,000 in sterile water plus 0.7% agar cooled to 45°C. Spores were treated in a similar manner; purified spores were added to sterile water plus 0.7% agar cooled to 45°C to a final concentration of approximately 105 spores/ml. Three milliliters of these suspensions was overlaid onto LB -plus-0.5 mM 2,2′-dipyridyl (an iron chelator) agar plates and allowed to solidify for at least 30 min. Sterile paper discs (6 mm in diameter) infused with water and purified petrobactinBa (2 μg and 10 μg in 10 μl water) were placed on the agar overlays and incubated at 37°C for 48 h, and zones of growth around the paper discs were measured. Note that support of growth on these plates is useful only for those compounds that have an iron affinity comparable to or greater than that of 2,2′-dipyridyl.
Production and isolation of petrobactin.
To isolate petrobactinBa, the wild-type B. anthracis strain Sterne 34F2 was grown in IDM (described above) to induce siderophore production. Five milliliters of vegetative cells grown to mid-exponential phase in LB were used to inoculate 500 ml of IDM. This culture was then incubated at 250 rpm at 37°C for 12 to 17 h. Cells were removed from the culture medium using filtration through 0.20-μm-filter flask units (Corning). These filtrates (from 5 liters) were adjusted to pH 7.0 and subsequently applied to a column packed with Amberlite XAD-16 (Supelco, PA) (500 ml). The column was washed with pure water several times and then eluted with 100% methanol. All fractions with siderophore activity (see below) were pooled and were subjected to further purification by preparative high-performance liquid chromatography (HPLC). Highly purified petrobactinBa was obtained by preparative HPLC using a C18 reverse-phase semiprep column (SymmetryPrep C18; 7 μm, 7.8 by 300 mm; Waters). The HPLC was performed with a Beckman Coulter system (Fullerton, CA) with a diode-array detector using a linear stepwise gradient from 5% to 50% aqueous acetonitrile in 0.1% (vol/vol) trifluoroacetic acid at a flow rate of 1.5 ml/min over 40 min. The HPLC peaks with siderophore activity were collected and lyophilized, resulting in purified petrobactinBa. The chemical structure of petrobactinBa was verified by Fourier transform ion cyclotron resonance (FTICR) mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectral analysis. To isolate petrobactinMh, cultures of M. hydrocarbonoclasticus DSM 8798 precultured in Bacto marine broth (Difco) were grown in a synthetic seawater medium at 28°C for 7 days, as has been described previously (1, 18). After fermentation broths were harvested by centrifugation, XAD-16 resin was added to each supernatant and the mixtures were agitated by shaking for 6 h at 150 rpm in the dark. Each filtered XAD-16 resin was used for further isolation of petrobactinMh.
Complementation of ΔasbA and ΔasbB mutations.
To provide asbA for in trans complementation, pBKJ358 was constructed. PCR was used to amplify a 2,330-bp fragment that contained the entire asbA gene (1,809 bp) and 512 bp of upstream sequence presumably containing the entire native asb promoter. This DNA fragment was cloned into the pCR8/GW/TOPO cloning vector (Invitrogen) according to the manufacturer's instructions, and the DNA sequence was verified. This construct was then transferred into the gram-positive, Escherichia coli shuttle vector pHP13 as an EcoRI fragment. The resulting plasmid (pBKJ358) was introduced into the ΔasbA strain using electroporation. To provide asbB for a similar in trans complementation experiment, pBKJ359 was constructed. PCR was used to fuse the 512 bp of upstream sequence used in pBKJ358 to the asbB gene (1,839 bp). This PCR product was cloned and sequenced in a manner identical to that for the asbA product, transferred into the gram-positive, E. coli shuttle vector pAD123 as a KpnI fragment and introduced into the ΔasbB strain using electroporation. The following control strains were also constructed: the wild type containing each vector (pHP13 or pAD123), the ΔasbA strain containing pHP13, and the ΔasbB strain containing pAD123. Each strain was grown in IDM as described above, and complementation was shown by the restoration of petrobactinBa production in each mutant by liquid chromatography-mass spectrometry (LCMS) analysis using a Shimadzu LCMS-2010EV system. (A detailed description of LCMS analysis can be found below). Further complementation experiments for ΔasbC, ΔasbD, ΔasbE, ΔasbF, ΔasbAB, or ΔasbABCDEF were not conducted.
Isolation of biosynthetic intermediates from Bacillus anthracis asb mutants.
To produce and analyze metabolic products under iron limitation, the B. anthracis asb mutant strains (ΔasbA, ΔasbB, ΔasbC, ΔasbD, ΔasbE, ΔasbF, ΔasbAB, and ΔasbABCDEF strains) were grown in IDM, and filtrates (100 ml) of the growth medium were generated as described above. To analyze intracellular accumulation of the intermediates, cells from culture filtrates were resuspended in 20 ml sterile phosphate-buffered saline buffer (pH 7.4) (without calcium chloride or magnesium chloride). Cells were subjected to a single freeze-thaw cycle at −20°C and two cycles at −80°C. Cell suspensions were sonicated six times with 30-s pulses at 4°C. Cell lysates were pelleted by centrifugation, and lysate supernatants were incubated on ice with 18 μl of DNase (RNase free) (180 Un) for 1 h. Cell lysates were then spun by centrifugation at 9,000 × g for 35 min, and supernatants (10 ml) were collected for analysis. These filtrates and cell lysates were adjusted to pH 7.0, XAD-16 resin (5 g for filtrates and 1 g for cell lysates) was added, and the mixtures were shaken for 2 h at 150 rpm in the dark. These mixtures were next filtered, and the resin was washed with pure water several times and then eluted with 100% methanol. The methanol eluates were concentrated to dryness in vacuo and dissolved in 80% methanol (1 ml for filtrates and 100 μl for cell lysates). The resin extracts from the filtrates of each mutant were analyzed using HPLC under the same conditions as for the isolation of petrobactin described above. Metabolites were identified by mass spectrometry of HPLC-purified samples and by HPLC analysis or by UV spectral analysis with purified petrobactinBa, using a synthetic sample of 3,4-dihydroxybenzoyl spermidine as a standard. Mass spectra were recorded on a Micromass LCT time-of-flight mass spectrometer equipped with an electrospray ionization mode and FTICR MS as described above. The resin extracts from the cell lysates of asb mutants were analyzed using a Shimadzu LCMS-2010EV system. (A detailed description of LCMS analysis can be found below).
LCMS analysis.
LC was performed by using a Shimadzu LC-20AD HPLC system consisting of a UV/VIS detector (SDP-20AV) and an autosampler (SIL-20AC). The HPLC was coupled to a Shimadzu LCMS-2010EV mass spectrometer with an electrospray ionization (ESI) interface. LC was carried out on an analytical column (Waters XBridge C18; 3.5 μm, 2.1 by 150 mm) utilizing a gradient elution with a flow rate of 0.1 ml/min ranging from 5% to 50% aqueous acetonitrile, including 0.1% formic acid, over 30 min. The ESI source was set at the positive mode. Selected ion monitoring was conducted to monitor ions at m/z 719.3, 282.2, and 456.2, which corresponded to the protonated molecular ions of petrobactinBa, 3,4-dihydroxybenzoyl spermidine, and 3,4-dihydroxybenzoyl spermidinyl citrate, respectively. The MS operating conditions were optimized as the following: drying gas, 1.5 liters/min; CDL temperature, 250°C; heat block temperature, 200°C and detector voltage, 1.5 kV.
MS and NMR analysis.
Gas-phase singly and doubly protonated siderophores and singly protonated siderophore intermediates were generated by ESI at 70 μl/h (Apollo ion source; Bruker Daltonics, Billerica, MA) of a solution containing 10 μM siderophore and 1 μM siderophore intermediates, respectively (1:1 methanol:water with or without 0.1% formic acid). To accurately determine the masses of the siderophores, the calibration standard (G2421A; Agilent Technologies, Palo Alto, CA) was mixed with the samples (200-fold dilution of the standard) for internal calibration. All mass spectra were collected with an actively shielded 7 T FTICR mass spectrometer with a quadrupole front end (APEX-Q; Bruker Daltonics). Structural characterization of siderophores and siderophore intermediates was performed by sustained off-resonance irradiation collision-activated dissociation tandem mass spectrometry (SORI-CAD MS/MS). NMR spectra were recorded on a Bruker AMX 500-MHz NMR spectrometer (Billerica, MA). The 1H NMR spectrum of siderophores was measured in deuterium oxide at room temperature. The 13C NMR spectrum of siderophores was recorded in deuterium oxide using broad-band proton decoupling.
Chemical synthesis of authentic 3,4-dihydroxybenzoyl spermidine.
The synthesis of 3,4-dihydroxybenzoyl spermidine as a biosynthetic intermediate standard from 3,4-dihydroxybenzoic acid was initiated by benzylation of 3,4-dihydroxybenzoic acid to afford the corresponding benzyl ester. Direct displacement of benzyl ester by 3-aminopropanol catalyzed by heterocyclic carbene afforded 3,4-bis(benzyloxy)-N-(3-hydroxypropyl)benzamide under mild conditions (27). This atom-economical process, developed by Movassaghi and Schmidt (27), involves an initial transesterification to benzyl 3,4-bis(benzyloxy)benzoate followed by a rapid intramolecular N,O-acyl shift to 3,4-bis(benzyloxy)-N-(3-hydroxypropyl)benzamide and obviates the use of stoichiometric coupling agents (27). O-iodoxybenzoic acid (IBX)-mediated oxidation of 3,4-bis(benzyloxy)-N-(3-hydroxypropyl)benzamide furnished aldehyde 3,4-bis(benzyloxy)-N-(2-formylethyl)benzamide (13, 14). Next, reductive amination of aldehyde with amine promoted by sodium triacetoxyborohydride provided the spermidine adduct N1-[3, 4-bis(benzyloxy)benzoyl]-N8-(tert-butoxycarbonyl)spermidine (13). Sequential deprotection of the benzyl ethers by catalytic hydrogenation to N1-[3,4-dihydroxybenzoyl]-N8-(tert-butoxycarbonyl)spermidine followed by the Boc carbamate with 4 N HCl in dioxane afforded 3,4-dihydroxybenzoyl spermidine.
3,4-Dihydroxybenzoyl spermidine.
1H NMR (600 MHz, CD3OD) 1.70 to 1.90 (m, 4H), 1.90 to 2.21 (m, 2H), 3.00 (t, J = 7.2 Hz, 2H), 3.02 to 3.22 (m, 4H), 3.49 (t, J = 6.0 Hz, 2H), 6.81 (d, J = 7.8 Hz, 1H), 7.25 (d, J = 8.4 Hz, 1H), 7.32 (s, 1H); 13C NMR (150 MHz, CD3OD) 24.5, 25.7, 28.0, 37.3, 40.2, 46.5, 48.3, 115.9, 116.0, 120.9, 126.3, 146.5, 150.6, 171.3; HRMS (APCI+) calculated for C14H24N3O3 [M+H]+, 282.1812. Found, 282.1819 (error, 2.4 ppm).
RESULTS
Deduced roles of AsbA and AsbB in biosynthesis of petrobactinBa.
Database comparisons revealed that AsbA and AsbB are most similar in primary amino acid sequence to IucA and IucC from Escherichia coli (5), gene products that are involved in the biosynthesis of aerobactin (Fig. 1). Based on genetic studies of E. coli mutant strains, iucA was shown to mediate condensation of Nɛ-acetyl-Nɛ-hydroxy-lysine with citrate, and iucC was shown to be responsible for linkage of a second Nɛ-acetyl-Nɛ-hydroxy-lysine molecule with the intermediate generated via the iucA gene product (8, 9). Based on the NRPS-independent assembly of petrobactin, the demonstrated role of asb in its biosynthesis, and the close amino acid sequence relationships of AsbA/AsbB and IucA/IucC, we developed a biosynthetic scheme for assembly of petrobactinBa (Fig. 2). Specifically, we propose that the siderophore is formed by the condensation of 3,4-dihydroxybenzoyl spermidine (3,4-DHB-SPD) with an activated form of citrate (coenzyme A or AMP ester) to form 3,4-dihydroxybenzoyl spermidinyl citrate (3,4-DHB-SPD-CT), followed by the condensation of a second 3,4-DHB-SPD subunit with 3,4-DHB-SPD-CT (again activated as coenzyme A or AMP ester) to form the mature siderophore. This scheme is analogous to the assembly of aerobactin and a number of other siderophores, including rhizobactin 1021, achromobactin, vibrioferrin, alcaligin, and desferrioxamine E, that have similarly annotated genes, forming a family of 40 NRPS-independent siderophore biosynthetic systems (6).
FIG. 2.
Proposed biosynthetic scheme of petrobactinBa. Spermidine and 3,4-dihydroxybenzoic acid are condensed by a combination of AsbC, -D, -E, and -F, although other factors could be involved. The product of this reaction, 3,4-dihydroxybenzoyl spermidine, is condensed with citrate by AsbA to form a second intermediate, 3,4-dihydroxybenzoyl spermidinyl citrate. The product of the AsbA reaction is then condensed with a second molecule of 3,4-dihydroxybenzoyl spermidine by AsbB to form petrobactinBa.
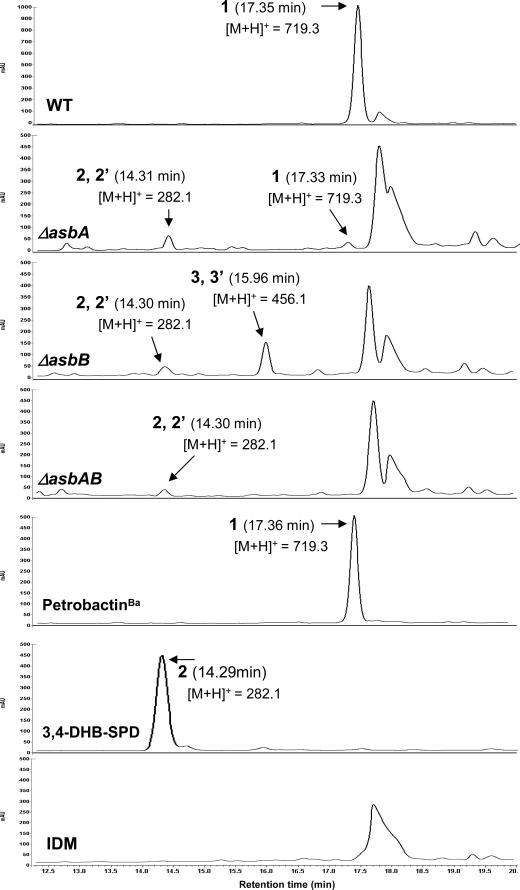
In order to test this hypothesis, in-frame deletion mutants were generated for each of the six genes of the asb operon (see Materials and Methods), as well as a mutant with a deletion that encompassed the entire operon (Fig. 3). The B. anthracis Sterne wild type and each of the mutants were grown under iron-depleted conditions, and the cells were removed by filtration. In order to identify accumulated intermediates from the petrobactinBa pathway, the filtrate and cell lysate obtained from each culture were prepared for HPLC, LCMS, and mass spectrometry analysis (see Material and Methods). As anticipated, analysis of the B. anthracis wild-type filtrate provided a single symmetrical HPLC peak corresponding to petrobactinBa (Fig. 4) with the expected molecular mass [m/z 719.3, (M+H)+]. Interestingly, inspection of the HPLC chromatograms for the ΔasbA, ΔasbB, and ΔasbAB mutants revealed that all three contained a new coincident product that was not observed in the wild type. Mass spectrometry showed that the metabolites had identical molecular masses[m/z 282.1, (M+H)+] compared to an authentic standard of 3,4-DHB-SPD. In addition to the peak corresponding to 3,4-DHB-SPD, a single asymmetric peak yielding typical UV spectra for 3,4-DHB was observed from the filtrate of the ΔasbB mutant (Fig. 4). HPLC purification of this metabolite and MS analysis indicated [molecular mass m/z 456.1, (M+H)+] that it corresponded to the more advanced biosynthetic intermediate 3,4-DHB-SPD-CT. None of the filtrates from the other mutants (ΔasbC, ΔasbD, ΔasbE, ΔasbF, and ΔasbABCDEF mutants) exhibited signals corresponding to either of these precursor metabolites.
FIG. 3.
Schematic of the various mutants of the asbABCDEF operon. More details for each mutant can be found in Materials and Methods.
FIG. 4.
HPLC profiles of filtrates from B. anthracis wild-type Sterne 34F2 and asb mutants. PetrobactinBa (1) was isolated from both the wild type and the ΔasbA mutant. The two isomers of 3,4-dihydroxybenzoyl spermidine (2 and 2′; the structures are shown in Fig. 5C and D, respectively) were isolated from the ΔasbA, ΔasbB, and ΔasbAB mutants. The two isomers of 3,4-dihydroxybenzoyl spermidinyl citrate (3 and 3′; the structures are shown in Fig. 5E and F, respectively) were isolated from the ΔasbB mutant. The mass of the [M+H]+ ion obtained by ESI MS analysis is listed below each compound.
Interestingly, close inspection of the metabolite analysis for each of the mutant strains revealed that a minor product with the same molecular mass as petrobactinBa [m/z 719.3, (M+H)+] (Fig. 4) was generated from the ΔasbA mutant. Production of petrobactinBa in a strain lacking AsbA was surprising. However, it is conceivable that AsbB (or another unknown enzyme) shares enough functional similarity to AsbA to provide partial catalysis. To test this hypothesis, a mutant lacking both AsbA and AsbB (ΔasbAB) was generated, and a siderophore metabolite analysis was performed. No apparent peak corresponding to petrobactinBa was observed for filtrates from the ΔasbAB double mutant strain (Fig. 4). Moreover, analysis of cell lysates from the wild type or the ΔasbA mutant strain by LCMS revealed that no petrobactinBa or corresponding biosynthetic intermediates accumulated (data not shown). Further studies confirmed that neither 3,4-DHB-SPD nor 3,4-DHB-SPD-CT could be detected for the cell lysates of any asb mutant. These data demonstrate that petrobactinBa and its biosynthetic intermediates 3,4-DHB-SPD and 3,4-DHB-SPD-CT are effectively excreted from B. anthracis cells.
In order to obtain higher-resolution structural information on the petrobactin biosynthetic intermediates, ESI FTICR MS of HPLC-purified molecules was conducted. Analysis of 3,4-DHB-SPD and 3,4-DHB-SPD-CT from the B. anthracis ΔasbB mutant showed abundant ion peaks at m/z 282.1811 (for 3,4-DHB-SPD) and 456.1976 (for 3,4-DHB-SPD-CT), respectively, which were then assigned as singly protonated ions, indicating the molecular formula C14H23N3O3 (for 3,4-DHB-SPD) or C20H29N3O9 (for 3,4-DHB-SPD-CT) (data not shown). A similar abundant ion peak at m/z 282.1811, corresponding to 3,4-DHB-SPD, was obtained from extracts of ΔasbA and ΔasbAB (data not shown).
To obtain more-detailed fragmentation patterns and structural information on 3,4-DHB-SPD and 3,4-DHB-SPD-CT, they were subjected to SORI-CAD MS/MS. For singly protonated HPLC-purified 3,4-DHB-SPD (Fig. 5A), major fragment ions were detected at m/z 109.03, 137.02, 194.08, 208.10, 225.12, and 265.16. The fragment ions at m/z 109.03, 137.02, 194.08, and 265.16 could be assigned to the cleavage of one terminal C—C(O) bond, one amide bond, and two amine bonds in 3,4-DHB-SPD (Fig. 5C). From this pattern, 3,4-DHB is added to the 3-carbon end of spermidine, whereas the fragment ions at m/z 208.10 and 225.12 are unlikely to result from that structure. However, the latter two ions can be produced from an isomeric structure in which 3,4-DHB is added to the 4-carbon end of spermidine (Fig. 5D). These results suggest that 3,4-DHB-SPD (Fig. 5C) and its isomeric counterpart (Fig. 5D) are synthesized in the ΔasbA, ΔasbB, and ΔasbAB mutants as the biosynthetic intermediates.
FIG. 5.
FTICR MS/MS of 3,4-DHB-SPD and 3,4-DHB-SPD-CT isolated from the B. anthracis ΔasbB mutant and petrobactinBa from the wild type. A, SORI-CAD mass spectrum from 3,4-DHB-SPD; B, SORI-CAD mass spectrum of 3,4-DHB-SPD-CT; C and D, assignments of selected fragments from 3,4-DHB-SPD, suggesting the isomers 2 and 2′; E and F, assignments of selected fragments from 3,4-DHB-SPD-CT, suggesting the isomers 3 and 3′; G, SORI-CAD mass spectrum of petrobactinBa; H, assignments of selected fragments from petrobactinBa.
In SORI-CAD MS/MS evaluation of singly protonated HPLC-purified 3,4-DHB-SPD-CT, major fragment ions at m/z 109.03, 137.02, 194.08, 208.10, 211.11, 265.16, and 282.18 (Fig. 5B) were detected. The fragments at m/z 109.03, 137.02, 194.08, 211.11, 265.16, and 282.18 can be assigned to cleavage of one terminal C—C(O) bond, two amide bonds, and three amine bonds in 3,4-DHB-SPD (Fig. 5E), in which 3,4-DHB is attached to the 3-carbon end of spermidine. However, the fragment at m/z 208.10 is unlikely to result from that structure, but it can be formed directly from the isomeric structure of 3,4-DHB-SPD-CT (Fig. 5F), in which 3,4-DHB is added to the 4-carbon end of spermidine. This result indicates that the B. anthracis ΔasbB mutant strain synthesizes not only 3,4-DHB-SPD-CT (Fig. 5E) but also its chemical isomer (Fig. 5F), as unexpectedly observed in the biosynthesis of two isomeric forms of 3,4-DHB-SP in ΔasbA, ΔasbB, and ΔasbAB cultures. Significantly, both isomeric forms of 3,4-DHB-SP were observed by direct biochemical conversion to the products (28a).
Addition of selected exogenous siderophores restores growth to asb mutants in iron-depleted medium.
While the characterized asb mutant (Δasb::kmr) had been shown previously to have a growth defect in liquid IDM and failed to produce petrobactin (5, 33), it remained unclear whether exogenous petrobactin added back to IDM would restore growth. To address this question, exogenous petrobactinBa (2 μM) was provided, resulting in nearly complete restoration of the mutant strain to wild-type growth levels (Fig. 6A). These studies were extended by testing a series of heterologous siderophores against the Δasb::kmr strain in IDM to establish their ability to restore growth following addition of petrobactinBa, petrobactinMh, the hydroxamate siderophore aerobactin, and the catecholate siderophore salmochelin S4 (Fig. 6B). Of the four exogenously added siderophores, only salmochelin S4 failed to assist the growth of the Δasb::kmr mutant, with cultures showing the same poor growth kinetics as cultures with no exogenously added siderophore. As expected, petrobactinBa and petrobactinMh supplemented Δasb::kmr mutant growth in IDM to virtually equivalent levels. Interestingly, the hydroxamate siderophore aerobactin was also able to restore growth of the Δasb::kmr mutant. This suggests that B. anthracis, as shown previously with other bacteria (22, 29), maintains the ability to utilize select chemically distinct heterologous siderophores to meet its iron requirements.
FIG. 6.
A. Growth of the Δasb::kmr mutant in IDM with or without the addition of purified petrobactinBa. PetrobactinBa (2 μM) alleviated the growth defect of the Δasb::kmr mutant strain that was unable to produce this siderophore. B. Growth of the Δasb::kmr mutant in IDM supplemented with purified siderophores. PetrobactinBa, petrobactinMh, and aerobactin partially alleviated the Δasb::kmr growth defect. Salmochelin S4 failed to alleviate the Δasb::kmr growth defect. The indicated values are the averages of measurements of three independent growth curves, and each error bar corresponds to one standard deviation. All siderophore concentrations are 2 μM.
The growth curves in liquid broth described above were obtained by inoculating media with actively growing vegetative cultures of wild-type and Δasb::kmr strains. However, since the dormant spore form of B. anthracis is the actual infectious particle, we assessed the ability of purified petrobactinBa to facilitate outgrowth of newly germinated spores in extreme iron-poor conditions. Accordingly, we employed assays in which sterile paper discs infused with the test molecule (siderophore) were placed onto a solid medium limited for iron onto which either vegetative cells or spores had been spread.
Table 3 compares the results of the ability of spores and vegetative bacilli of wild-type and Δasb::kmr mutant strains to grow on solid medium containing the iron-chelating agent 2,2′-dipyridyl. For these assays, control experiments were conducted that demonstrated wild-type-level germination under all conditions used with no impact on growth due to the test medium (data not shown) containing an iron-chelating agent. Exponentially growing vegetative bacilli of the B. anthracis wild type were able to grow as a light lawn on plates containing 0.5 mM 2,2′-dipyridyl, whereas vegetative bacilli of the Δasb::kmr mutant strain were not able to propagate. Growth of Δasb::kmr vegetative cells was restored with exogenously added petrobactinBa or Fe(II)SO4 (not shown), suggesting that wild-type cells that fail to produce petrobactinBa are unable to overcome the severe iron limitation of this medium.
TABLE 3.
Growth enhancement of Bacillus anthracis by addition of petrobactinBa to 0.5 mM 2, 2′-dipyridyl medium using disc diffusion assaysa
| Strain description | Growth zone (mm) per reagent (n = 5 discs)
|
||
|---|---|---|---|
| H2O | 2 μg petrobactinBa | 10 μg petrobactinBa | |
| Wild type (spores) | No growth | 26.6 ± 2.1 | 32.8 ± 1.9 |
| Δasb::kmr (spores) | No growth | 17.0 ± 1.7 | 25.2 ± 0.5 |
| Wild type (vegetative) | Lawn | Lawna | Lawna |
| Δasb::kmr (vegetative) | No growth | 18.2 ± 0.8 | 24.4 ± 0.9 |
Student's t test comparing results for wild-type and Δasb::kmr spores, P < 0 0.01; Student's t test comparing results for Δasb::kmr spores and vegetative cells, P = 0.2.
Slightly heavier growth along edges of discs.
In contrast, spores of the B. anthracis wild type were not able to outgrow on these plates without the addition of exogenous petrobactinBa or an alternative iron source. Exogenous petrobactinBa enabled outgrowth of spores of both strains, but the wild-type strain exhibited more vigorous growth in a larger zone than the Δasb::kmr mutant. Thus, it appeared that wild-type spores, once they are able to overcome a severely iron-limited growth environment, were able to produce endogenous petrobactinBa for robust growth. In contrast, the Δasb::kmr mutant (both spores and vegetative bacilli) overcame the iron limitation to equivalent levels when exogenous petrobactinBa was provided, but then, unable to produce petrobactinBa, the cells failed to grow further.
Complementation of asbA and asbB.
To show that the mutations introduced into asbA and asbB did not disrupt downstream asb gene expression, the individual open reading frames were assessed for mutant strain complementation and restoration of petrobactinBa production. Plasmids carrying either asbA or asbB driven by the native asb promoter were introduced into the ΔasbA and ΔasbB mutants, respectively. Complementation was monitored by the restoration of petrobactinBa production from the mutant strains by LCMS analysis. As expected, ions corresponding to petrobactinBa (m/z 719.3, [M+H]+) were identified in extracts from the wild-type strains with vectors used for complementation of asbA and asbB. Providing asbA in trans restored petrobactinBa production from the ΔasbA mutant, and similarly, providing wild-type asbB also restored petrobactinBa production from the ΔasbB mutant, albeit at reduced levels compared to those for the wild type (Fig. 7).
FIG. 7.
Selected ion monitoring LCMS traces of filtrates from B. anthracis complemented ΔasbA and ΔasbB strains at the mass range of m/z 719.3 [M+H]+. The peaks at 15.960 to 16.135 min represent petrobactinBa.
Growth phenotypes of mutations in individual genes of the asb operon.
As described above, seven asb mutants were generated, including a deletion of the entire operon and an in-frame deletion for each individual gene. Two of the individual gene mutants accumulated predicted biosynthetic intermediates of petrobactin (3,4-DHB-SPD from the ΔasbA and ΔasbB mutants and 3,4-DHB-SPD-CT from the ΔasbB mutant). Each of the individual gene deletion mutants were then compared with the wild type and the entire operon deletion mutant for their ability to grow under a variety of culture conditions. The effects on growth in rich medium (BHI) and IDM were analyzed after inoculation with spores (Fig. 8A and B). No discernible growth defect was observed when the mutants were grown in rich medium; however, none of the seven asb mutants was able to outgrow under IDM conditions. Thus, outgrowth from spores in IDM for each individual gene mutant was the same as the deletion of the entire operon. The addition of Fe(II)SO4 or purified petrobactinBa (Fig. 8C and D) overcame this growth defect, leading to wild-type levels in five of the mutants (ΔasbA, ΔasbB, ΔasbC, ΔasbD, and ΔasbE mutants), with more limited growth for two others (ΔasbF and ΔasbABCDEF mutants).
FIG. 8.
Growth phenotypes of asb mutants. A to D. Growth curves of each mutant when inoculated from spores. For each curve, the growth medium was as follows: A. BHI (brain-heart infusion) broth; B. IDM; C. IDM supplemented with 100 μM ferrous sulfate; D. IDM supplemented with 1 μg/ml petrobactinBa.
DISCUSSION
Iron acquisition by bacteria is a key enabling factor in pathogenesis, and the production of petrobactin is a major route for iron acquisition in B. anthracis under iron-depleted conditions (5). In this work we have demonstrated that the first two genes of the asb operon (asbA and asbB) encode enzymes that specify the final two steps of petrobactin assembly. This involves coupling of 3,4-DHB-SPD and citrate to form 3,4-DHB-SPD-CT, followed by condensation of another molecule of 3,4-DHB-SPD to 3,4-DHB-SPD-CT, resulting in completion of the petrobactin molecule. Accumulation of 3,4-DHB-SPD-CT in the ΔasbB mutant suggests that AsbB mediates this final biosynthetic step, while the accumulation of 3,4-DHB-SPD in the ΔasbA mutant suggests that AsbA catalyzes the penultimate biosynthetic step. These findings correspond to the proposed scheme (Fig. 2) predicted from the structure of petrobactin and proposed initially due to the high sequence similarity between AsbA and AsbB with other related siderophore biosynthetic systems (6). While these results imply that AsbC, AsbD, AsbE, and AsbF are collectively responsible for production of 3,4-DHB-SPD, detailed biochemical analysis is required to assign a function for each enzyme. The close similarity between the deduced protein sequence of AsbD and thiolation domains involved in polyketide or nonribosomal peptide synthesis provides further insights into the detailed steps involved in petrobactin biosynthesis (5).
It is notable that the ΔasbA mutant can still produce low levels of petrobactinBa. This finding suggests that AsbB can also perform (albeit with low efficiency) the penultimate step in petrobactinBa biosynthesis in the absence of AsbA. The ΔasbA mutant was engineered to include a deletion of 98% of the open reading frame, ruling out a partially functional AsbA providing this activity. The amount of petrobactinBa generated by this mutant appears to be adequate to overcome the iron-limited conditions inherent in IDM, although a reproducible lag phase in the growth kinetics suggests that this requires a more extended period than that for the wild type. However, this is not the case for growth of spores from the ΔasbA mutant strain under IDM, since this organism was not able to outgrow under IDM conditions (Fig. 8B).
As expected, the addition of purified petrobactin from either B. anthracis or M. hydrocarbonoclasticus overcame the deficiency of each asb mutant for growth in IDM. Additionally, although the structurally related hydroxamate siderophore aerobactin facilitated growth, the catecholic siderophore salmochelin S4 was unable to restore growth of the B. anthracis asb mutant strains. It is likely that petrobactin and aerobactin might use a similar transport system in B. anthracis due to their structural similarity. Previous work has indicated that the B. anthracis bac operon specifies the biosynthesis of a siderophore (bacillibactin) that is structurally similar to salmochelin S4. A bac disruption mutant does not display attenuated virulence in mice and has no growth defect in IDM (5).
To our surprise, SORI-CAD MS/MS fragmentation pattern analysis revealed that each of two isomers of 3,4-DHB-SPD and 3,4-DHB-SPD-CT are formed in asb mutants (Fig. 5). The combination of the products of the remaining four biosynthetic genes, AsbCDEF, must produce these isomers where the 3,4-DHB moiety is attached to the 4-carbon end of spermidine, as opposed to the 3-carbon end that is typical for petrobactin (Fig. 2). Interestingly, the MS/MS fragmentation of petrobactinBa shows only those ions corresponding to 3,4-DHB-SPD and 3,4-DHB-SPD-CT with 3,4-DHB attached to the 3-carbon end of spermidine (Fig. 5G and H). Interestingly, there is no evidence that wild-type B. anthracis Sterne 34F2 accumulates these isomeric precursors (3,4-DHB attached to the 4-carbon end of spermidine) into an alternative form of petrobactin. Thus, these intermediates are probably degraded/recycled by an unknown mechanism. Production of the alternative isomer suggests that the enzymes responsible for the synthesis of 3,4-DHB-SPD and 3,4-DHB-SPD-CT are somewhat flexible, an attribute that has been borne out in our preliminary in vitro studies (28a).
The chemistry and biology of bacterial siderophores and their role in pathogenesis have been the subject of increasing investigation. The resulting mechanistic understanding of the metabolic systems provides numerous opportunities to design small molecule inhibitors that block siderophore biosynthesis and hence bacterial growth of pathogenic bacteria in iron-limiting environments. Recently, a bisubstrate nucleoside that inhibits domain salicylation enzymes (MbtA and YbtE) required for siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis was designed and synthesized (12, 26, 30). This compound dramatically inhibited siderophore biosynthesis and growth of M. tuberculosis and Y. pestis under iron-limiting conditions and represents a promising lead compound for the development of new antibiotics to treat tuberculosis and bubonic plague (12, 26, 30). These types of inhibitors also function as powerful tools to aid in deciphering the relevance of siderophores at specific stages of infection by probing temporal control of siderophore production for animal models of bacterial pathogenesis (12). Given the importance of the production of petrobactin in the virulence of B. anthracis, similar studies that target gene products of the asb operon could yield potential therapeutic agents for the treatment of anthrax infections.
Acknowledgments
We thank Alison Butler for an authentic sample of petrobactin.
This research was supported by grants RP1 and DP18 from the NIH Great Lakes Center of Excellence for Biodefense & Emerging Infectious Diseases Research and the J. G. Searle Professorship to D.H.S. J.Y.L. was supported by the Korea Research Foundation Grant funded by the government of Korea (MOEHRD; Basic Research Promotion Fund) (KRF-2003-214-C00118). H.L. is supported through an award to K.H. from the Searle Scholars Program.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Barbeau, K., G. Zhang, D. H. Live, and A. Butler. 2002. Petrobactin, a photoreactive siderophore produced by the oil-degrading marine bacterium Marinobacter hydrocarbonoclasticus. J. Am. Chem. Soc. 124:378-379. [DOI] [PubMed] [Google Scholar]
- 2.Bergeron, R. J., G. Huang, R. E. Smith, N. Bharti, J. S. McManis, and A. Butler. 2003. Total synthesis and structure revision of petrobactin. Tetrahedron 59:2007-2014. [Google Scholar]
- 3.Braun, V. 2001. Iron uptake mechanisms and their regulation in pathogenic bacteria. Int. J. Med. Microbiol. 291:67-79. [DOI] [PubMed] [Google Scholar]
- 4.Bullen, J. J., H. J. Rogers, P. B. Spalding, and C. G. Ward. 2006. Natural resistance, iron and infection: a challenge for clinical medicine. J. Med. Microbiol. 55:251-258. [DOI] [PubMed] [Google Scholar]
- 5.Cendrowski, S., W. MacArthur, and P. Hanna. 2004. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol. Microbiol. 51:407-417. [DOI] [PubMed] [Google Scholar]
- 6.Challis, G. L. 2005. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. ChemBioChem 6:601-611. [DOI] [PubMed] [Google Scholar]
- 7.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lorenzo, V., A. Bindereif, B. H. Paw, and J. B. Neilands. 1986. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J. Bacteriol. 165:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., and J. B. Neilands. 1986. Characterization of iucA and iucC genes of the aerobactin system of plasmid ColV-K30 in Escherichia coli. J. Bacteriol. 167:350-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon, T. C., M. Meselson, J. Guillemin, and P. C. Hanna. 1999. Anthrax. N. Engl. J. Med. 341:815-826. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, A. K., and J. Handelsman. 1999. A vector for promoter trapping in Bacillus cereus. Gene 226:297-305. [DOI] [PubMed] [Google Scholar]
- 12.Ferreras, J. A., J. S. Ryu, F. Di Lello, D. S. Tan, and L. E. Quadri. 2005. Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nat. Chem. Biol. 1:29-32. [DOI] [PubMed] [Google Scholar]
- 13.Frigerio, M., and M. Santagostinoand. 1994. A mild oxidizing reagent for alcohols and 1,2-diols: o-iodoxybenzoic acid (IBX) in DMSO. Tetrahedron Lett. 35:8019-8022. [Google Scholar]
- 14.Frigerio, M., M. Santagostino, and S. Sputore. 1999. A user-friendly entry to 2-iodoxybenzoic acid (IBX). J. Org. Chem. 64:4537-4538. [Google Scholar]
- 15.Gardner, R. A., R. Kinkade, C. Wang, and O. Phanstiel IV. 2004. Total synthesis of petrobactin and its homologues as potential growth stimuli for Marinobacter hydrocarbonoclasticus, an oil-degrading bacteria. J. Org. Chem. 60:3530-3537. [DOI] [PubMed] [Google Scholar]
- 16.Garner, B. L., J. E. L. Arceneaux, and B. R. Byers. 2004. Temperature control of a 3,4-dihydroxybenzoate (protocatechuate)-based siderophore in Bacillus anthracis. Curr. Microbiol. 49:89-94. [DOI] [PubMed] [Google Scholar]
- 17.Haima, P., S. Bron, and G. Venema. 1987. The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol. Gen. Genet. 209:335-342. [DOI] [PubMed] [Google Scholar]
- 18.Hickford, S. J., F. C. Küpper, G. Zhang, C. J. Carrano, J. W. Blunt, and A. Butler. 2004. Petrobactin sulfonate, a new siderophore produced by the marine bacterium Marinobacter hydrocarbonoclasticus. J. Nat. Prod. 67:1897-1899. [DOI] [PubMed] [Google Scholar]
- 19.Inglesby, T. V., T. O'Toole, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. M. Friedlander, J. Gerberding, J. Hauer, J. Hughes, J. McDade, M. T. Osterholm, G. Parker, T. M. Perl, P. K. Russell, and K. Tonat. 2002. Anthrax as a biological weapon, 2002, updated recommendations for management. JAMA 287:2236-2252. [DOI] [PubMed] [Google Scholar]
- 20.Ireland, J. A. W., and P. C. Hanna. 2002. Macrophage-enhanced germination of Bacillus anthracis endospores requires gerS. Infect. Immunol. 70:5870-5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janes, B. K., and S. Stibitz. 2006. Routine markerless gene replacement in Bacillus anthracis. Infect. Immun. 74:1949-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi, F., G. Archana, and A. Desai. 2006. Siderophore cross-utilization amongst rhizospheric bacteria and the role of their differential affinities for Fe(3+) on growth stimulation under iron-limited conditions. Curr. Microbiol. 53:141-147. [DOI] [PubMed] [Google Scholar]
- 23.Koppisch, A. T., C. C. Browder, A. L. Moe, J. T. Shelley, B. A. Kinkel, L. E. Hersman, S. Iyer, and C. E. Ruggiero. 2005. Petrobactin is the primary siderophore synthesized by Bacillus anthracis str. Sterne under conditions of iron starvation. Biometals 18:577-585. [DOI] [PubMed] [Google Scholar]
- 24.Köster, W. 2001. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res. Microbiol. 152:291-301. [DOI] [PubMed] [Google Scholar]
- 25.Martin, V. J. J., D. J. Pitera, S. T. Withers, J. D. Newman, and J. D. Keasling. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21:796-802. [DOI] [PubMed] [Google Scholar]
- 26.Miethke, M., P. Bisseret, C. L. Beckering, D. Vignard, J. Eustache, and M. A. Marahiel. 2006. Inhibition of aryl acid adenylation domains involved in bacterial siderophore synthesis. FEBS J. 273:409-413. [DOI] [PubMed] [Google Scholar]
- 27.Movassaghi, M., and M. A. Schmidt. 2005. N-heterocyclic carbene-catalyzed amidation of unactivated esters with amino alcohols. Org. Lett. 7:2453-2456. [DOI] [PubMed] [Google Scholar]
- 28.Passalacqua, K. D., N. H. Bergman, A. Herring-Palmer, and P. Hanna. 2006. The superoxide dismutases of Bacillus anthracis do not cooperatively protect against endogenous superoxide stress. J. Bacteriol. 188:3837-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Pfleger, B. F., J. Y. Lee, R. V. Somu, C. C. Aldrich, P. C. Hanna, and D. H. Sherman. 2007. Characterization and analysis of early enzymes for petrobactin biosynthesis in Bacillus anthracis. Biochemistry, in press. [DOI] [PubMed]
- 29.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 30.Somu, R. V., H. Boshoff, C. Qiao, E. M. Bennett, C. E. Barry III, and C. C. Aldrich. 2006. Rationally designed nucleoside antibiotics that inhibit siderophore biosynthesis of Mycobacterium tuberculosis. J. Med. Chem. 49:31-34. [DOI] [PubMed] [Google Scholar]
- 31.Sterne, M. 1939. The immunization of laboratory animals against anthrax. Onderstepoort J. Vet. Sci. Anim. Ind. 13:313-317. [Google Scholar]
- 32.Weinberg, E. D. 1978. Iron and infection. Microbiol. Rev. 42:45-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, M. K., R. J. Abergel, K. N. Raymond, J. E. L. Arceneaux, and B. R. Byers. 2006. Siderophores of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Biochem. Biophys. Res. Commun. 348:320-325. [DOI] [PubMed] [Google Scholar]