Abstract
Actin, a central component of the eukaryotic cytoskeleton, plays a crucial role in determining cell shape in addition to several other functions. Recently, the structure of the archaeal actin homolog Ta0583, isolated from the archaeon Thermoplasma acidophilum, which lacks a cell wall, was reported by Roeben et al. (J. Mol. Biol. 358:145-156, 2006). Here we show that Ta0583 assembles into bundles of filaments similar to those formed by eukaryotic actin. Specifically, Ta0583 forms a helix with a filament width of 5.5 nm and an axial repeating unit of 5.5 nm, both of which are comparable to those of eukaryotic actin. Eukaryotic actin shows a greater resemblance to Ta0583 than to bacterial MreB and ParM in terms of polymerization characteristics, such as the requirement for Mg2+, critical concentration, and repeating unit size. Furthermore, phylogenetic analysis also showed a closer relationship between Ta0583 and eukaryotic actin than between MreB or ParM and actin. However, the low specificity of Ta0583 for nucleotide triphosphates indicates that Ta0583 is more primitive than eukaryotic actin. Taken together, our results suggest that Ta0583 retains the ancient characteristics of eukaryotic actin.
The domain Archaea consists of two groups: Euryarchaeota and Crenarchaeota. Euryarchaeota include methanogens, extreme halophiles, and some thermophilic archaea, such as Archaeoglobus and Thermoplasma spp. Thermoplasma acidophilum is a thermophilic archaeon that lacks a cell wall surrounding the cell membrane (4). Thermoplasma species are unique in this respect among archaeal species, which usually possess a proteinaceous or pseudomurein cell wall (11). The presence of a cytoskeleton in T. acidophilum has been postulated by Hixon and Searcy (12). Superprecipitation of the cell extract of T. acidophilum has been observed, and the cells become spherical at low temperatures, suggesting depolymerization of the internal structure (31).
Eukaryotic cells contain complex intracellular membranous structures and are usually larger and more complex than prokaryotic cells. Several models have been proposed to explain the evolution of the complex eukaryotic cell, including fusion, intrusion, or symbiosis of two or more prokaryotic cells, i.e., archaea and bacteria (5, 8, 10, 19, 20, 21, 22, 24, 39).
In addition to the evolutionary origin of eukaryotic cellular structures, the origins of eukaryote-specific components, including the cytoskeleton, have generated a great deal of interest. Although the cytoskeleton is considered to be eukaryote specific, homology between tubulin, a component of the microtubule, and the bacterial counterpart FtsZ, which participates in division ring formation, has been reported (2).
Actin is a principal element of the eukaryotic cytoskeleton. Monomeric actin has a molecular mass of about 43 kDa and can polymerize to form F-actin, which comprises two protofilaments that form a right-handed double helix (6, 13, 23, 29). Actin can form various types of polymer depending on the conditions (1, 9). Actin homologs, encoded by mreB genes, are conserved among rod-shaped, filamentous, and helical bacteria, suggesting that MreB protein is important for generating a nonspherical shape in bacteria (15). van den Ent et al. have reported the X-ray crystallographic structure of the actin homolog MreB from the thermophilic bacterium Thermotoga maritima (35). Despite a very weak sequence similarity between actin and MreB (∼15%), the structural similarity of these two proteins is striking. MreB and its close relative Mbl are important in regulating the cell shape of Bacillus subtilis (15). The possible involvement of MreB in the cytoskeletal structure of another bacterium, Spiroplasma melliferum, has also been postulated (17).
Recently, the structure of the archaeal actin homolog Ta0583, from T. acidophilum, was reported by Roeben et al. (25). Ta0583 displays significant structural homology to actin, MreB, and ParM. Interestingly, Ta0583 is able to hydrolyze GTP, UTP, and CTP as well as ATP. In this report, we analyzed the polymerization characteristics of Ta0583 from T. acidophilum. Our results suggest that eukaryotic actin displays a greater resemblance to Ta0583 than to bacterial MreB and ParM.
MATERIALS AND METHODS
Phylogenetic analysis of actin and MreB homologs.
Initially, we performed a BLAST search against the protein database (GenBank) using T. maritima MreB as a key sequence under the default conditions and obtained six actin like archaeal sequences (Table 1). We also selected 21 homologs of bacterial MreB, ParM, actin and hsp70 as listed in Table 1. The 27 amino acid sequences were aligned with CLUSTAL X (34). The well-aligned regions (159 sites in total) were selected for further phylogenetic analyses. The 27 sequences were then used for construction of a phylogenetic tree using the MrBayes 3.1.1 program with the WAG model and the 4-class discrete gamma distribution model (26). From this analysis, we were able to classify actin, MreB, and their related proteins into seven classes: hsp70 proteins; ParM proteins; Thermotoga MreB proteins; NP_276159 and NP_613455; NP_07845; Ta0583, NP_111160, and EAM93814; and eukaryotic actin proteins. To perform more-credible phylogenetic analysis, we selected 14 proteins (see Table 1) as the representatives of the seven groups and performed maximum-likelihood (ML) analysis. The relationship of three proteins, Ta0583, NP_111160, and EAM93814, was fixed to be the following: Ta0583 and NP_111160 are monophyletic and EAM93814 is the sister group. Under these conditions, there are 945 possible topologies. The log likelihoods and other related statistical values of the 945 trees were estimated with CODEML in PAML 3.13 (36). Further statistical tests were performed with CONSEL (32).
TABLE 1.
Protein sequences used for phylogenetic analysis
| Protein and organisma | Domain | GenBank accession no. | ||
|---|---|---|---|---|
| MreB
|
||||
| Bacillus subtilis | Bacteria | Q01465 | ||
| B. subtilis | Bacteria | P39751 | ||
| B. subtilis | Bacteria | P39763 | ||
| Escherichia coli | Bacteria | P0A9X4 | ||
| Thermotoga maritima* | Bacteria | AAD35673 | ||
| T. maritima* | Bacteria | AAD36611 | ||
| ParM
|
||||
| E. coli* | Bacteria | A24920 | ||
| E. coli | Bacteria | NP_053130 | ||
| E. coli | Bacteria | NP_288357 | ||
| E. coli | Bacteria | P11904 | ||
| Salmonella enterica serovar Typhimurium | Bacteria | NP_058227 | ||
| Serratia marcescens | Bacteria | AAB37120 | ||
| Shigella flexneri* | Bacteria | NP_085361 | ||
| Shigella sonnei | Bacteria | NP_052472 | ||
| Actin
|
||||
| Homo sapiens* | Eukarya | P12814 | ||
| Saccharomyces cerevisiae* | Eukarya | P60010 | ||
| hsp70
|
||||
| S. cerevisiae | Eukarya | P09435 | ||
| Thermoplasma acidophilum* | Archaea | P50023 | ||
| B. subtilis* | Bacteria | P17820 | ||
| E. coli | Bacteria | AAN78519 | ||
| T. maritima | Bacteria | AAC79725 | ||
| Hypothetical
|
||||
| Archaeoglobus fulgidus* | Archaea | NP_07845 | ||
| Ferroplasma acidarmanus* | Archaea | EAM93814 | ||
| Methanobacterium thermoautotrophicum* | Archaea | NP_276159 | ||
| Methanopyrus kandleri* | Archaea | NP_613455 | ||
| T. acidophilum* | Archaea | NP_394057 (Ta0583) | ||
| Thermoplasma volcanium* | Archaea | NP_111160 | ||
Asterisked proteins appear in Fig. 1.
Protein expression and purification.
Genomic DNA of T. acidophilum 122-1B2 (type strain) (37) was prepared by phenol-chloroform extraction.
The gene encoding MreB homolog Ta0583 (27) (GenBank accession number NP_394057) in T. acidophilum was amplified from genomic DNA by PCR with primers ATGGTAGTTGTAGGATTGGATG and TTATCTACATCGATCTTTCCGC. The amplified DNA fragment was cloned into the TA expression vector pCRT7/NT-TOPO, in which 35 residues (MRGSHHHHHHGMASMTGGQQMGRDLYDDDDKDPTL), including a hexahistidine tag, originating from the vector were added to the N terminus. Mutation A (Trp37Glu and Lys42Glu) and mutation AB (Trp37Glu, Lys42Glu, Arg271Glu, and Met326Asp) were introduced into Ta0583 using mutagenic primers TTGATAAGGATCCAACCCTTATG and CTGTGCTGAGGACGGGTATTTCTCCGCCTATACCTTCGCTCTCGGTCTCCGTCAC for mutation A and mutagenic primers GAGAACATAAGGCTGAACCTCGAAGGAGAGGTTGACAGGGTTACTTCTCTGATACCAGTAGGGGGAGGGTCCAACCTGATAGGAGACCG and TCCGGATCAAGCTTCGAATTCTCGCCCTTTTATTAGTCCGATCTTTCCGCCGCATCC for mutation AB (mutation sites are underlined). The protein was expressed in Escherichia coli BL21RIL(DE3) cells, grown at 37°C in LB medium. The induced cells were harvested after 16 h of cultivation and lysed by sonication in a buffer containing 100 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 5 mM imidazole. The lysate was then incubated at 60°C for 10 min. After centrifugation (5,000 × g, 10 min), the supernatant was applied to a Ni2+-nitrilotriacetic acid column (Amersham Biosciences, Piscataway, NJ). The column was washed with a buffer containing 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 60 mM imidazole, and bound protein was then eluted with a buffer containing 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 1 M imidazole. The eluate was dialyzed against 20 mM Tris-HCl (pH 8.0) and stored at 4°C. The purity of the preparation was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The protein concentration was estimated by the bicinchoninic acid method.
Light-scattering measurements.
Polymerization of Ta0583 was monitored by right-angle light scattering in a Shimadzu (Kyoto, Japan) RF-5300PC fluorometer at 340 nm. The standard polymerization buffer contained 50 mM sodium acetate (pH 5.5), 4 mM MgCl2, and 4 mM ATP. Ta0583 was added to the buffer (final concentration, 0.1 mg ml−1) in a 1-cm-path-length cell maintained at 56°C. The mixture was stirred rapidly with a plastic stick, and light scattering was monitored. To test the pH dependence, 50 mM sodium acetate (pH 4.0 to 5.0) or 50 mM morpholineethanesulfonic acid (MES; pH 6.0) was used in place of sodium acetate (pH 5.5). To test the effect of monovalent cations, 0.1 to 200 mM NaCl or KCl was added. To test the effect of divalent cations, 0 to 1 mM MgCl2/CaCl2 plus 1 mM EGTA was added. Alternatively, 1 to 50 mM MgCl2/CaCl2 was added in place of 4 mM MgCl2. To test the effect of nucleotides, 4 mM nucleotide (GTP, CTP, UTP, dATP, dGTP, dCTP, dUTP, dTTP, ADP, GDP, CDP, or UDP) was added in place of 4 mM ATP. To test the effect of adenosine 5′-(β,γ-imido)triphosphate (AMP-PNP), 4 mM AMP-PNP was added in place of 4 mM ATP. Alternatively, 4 or 8 mM AMP-PNP was added in addition to 4 mM ATP. To test repolymerization, Ta0583 filament (polymerized in standard polymerization buffer at 56°C for 10 min) was placed at 0 or 56°C for 1 h. Polymerization was then measured using the light-scattering assay at 56°C. To determine the critical concentration of Ta0583 for polymerization, the light-scattering intensity was plotted as a function of the total concentration of Ta0583 in solution, and the plot was extrapolated to zero intensity.
Precipitation assay.
Ta0583 (0.5 mg ml−1) was polymerized in the standard buffer with or without ATP at 56°C for 5 min. Ta0583 fiber was centrifuged (at 10,000 × g for 10 min at 25°C), and the supernatant and pellet were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. To test the effect of nucleotides, 4 mM nucleoside triphosphate (NTP) (ATP, UTP, CTP, or GTP) was added in place of 4 mM ATP.
Electron microscopy.
One milligram of Ta0583 per milliliter in the standard polymerization buffer was incubated at 25°C for 5 min (see Fig. 3A and B), or 0.1 mg ml−1 Ta0583 in buffer (50 mM sodium acetate [pH 5.0], 4 mM MgCl2, 0.1 mM CaCl2, and 4 mM ATP) was incubated at 56°C for 5 min (see Fig. 3C). Then 5 μl of the Ta0583 solution was placed on a Formvar-coated copper electron microscopic grid. Excess liquid was blotted, and the grid was washed with a drop of water. The grids were stained with 1.5% uranyl acetate solution, dried, and observed with a JEM-1010 electron microscope (JEOL, Tokyo, Japan). The images were digitized using a line scanner (Leafscan45; Scitex, Herzliya, Israel) at a pixel size of 5 μm, whose contrast transfer function was compensated. Fourier diffraction patterns were computed from the electron micrographs. All images were analyzed with the Eos package as reported previously (38).
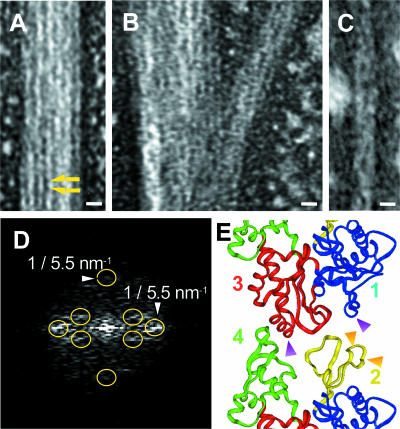
FIG. 3.
(A to C) Electron micrographs of the negatively stained Ta0583 bundle structure. (A) Typical view of bundle of Ta0583 fibers; (B) branched bundle; (C) filament with shortest width (0.1 mg ml−1). Bars, 10 nm. (D) Computed Fourier transform of Ta0583 fiber. Yellow circles indicate diffraction spots due to layered lines. (E) Polymerization model of T. maritima MreB. Shown is a contact site of MreB fiber. Orange arrows, A mutation points; purple arrows, B mutation points. Cyan, yellow, magenta, and green, domains 1, 2, 3, and 4, respectively.
RESULTS
Phylogenetic tree of actin and MreB homologs.
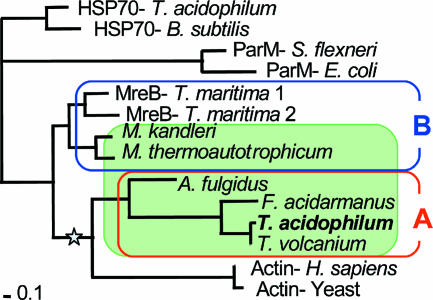
To investigate archaeal homologs of actin, we retrieved archaeal homologs of MreB from the databases. We found MreB and actin homologs only in Euryarchaeota (Table 1). Actin and MreB share low sequence similarity with the heat shock protein hsp70. The log likelihoods and related statistical values of 945 possible trees of 14 selected sequences were compared as described in Materials and Methods. Figure 1 shows the ML tree of MreB homologs and eukaryotic actins, rooted by hsp70s. Archaeal actin homologs are divided into two groups. The homologs of thermophilic archaea, including T. acidophilum (group A), form a group with actin. However, our analysis shows that MreB homologs of methanogens (Methanobacterium thermoautotrophicum and Methanopyrus kandleri) comprise a separate group (group B) with bacterial MreB. In the ML tree, the group A MreB homologs and eukaryotic actin form a clade with a bootstrap value of 0.766. In contrast, the bootstrap value of the possible cluster made of Ta0583 and ParM, proposed by Roeben et al. (25), is only 0.086. Therefore, the group A actin clade is supported by a much higher bootstrap value than the possible Ta0583-ParM cluster, although the monophyletic status of Ta0583 and ParM cannot be statistically rejected.
FIG. 1.
Maximum-likelihood tree of actin and MreB homologs rooted by the heat shock protein hsp70. The internal mode indicated by a star has a bootstrap value of 0.766 for monophyly of group A MreB homologs and eukaryotic actin. Archaeal homologs are shown against a green background. Group A and B homologs are surrounded by red and blue lines, respectively. Standard bar represents 0.1 nucleotide change per site. See Table 1 for accession numbers of the sequences used.
Polymerization assays.
To test the resemblance between group A MreB and eukaryotic actin, we analyzed T. acidophilum Ta0583. The time course for the polymerization of Ta0583 under various buffer conditions was monitored by light scattering (Fig. 2). Because fibrous structures were detected by electron microscopy (Fig. 3), we refer to the observed increase in scattering as polymerization below.
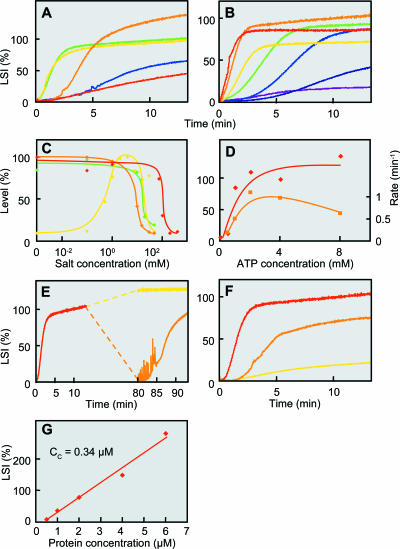
FIG. 2.
Polymerization of Ta0583 monitored by light scattering. (A) pH dependence. The light-scattering intensity (LSI) is expressed as a percentage of that attained after 10 min under standard conditions. Colors represent pH values as follows: red, pH 4.0; orange, pH 4.5; yellow, pH 5.0; green, pH 5.5; blue, pH 6.0. (B) Temperature dependence. Red, 65°C; orange, 56°C; yellow, 45°C; green, 35°C; sky blue, 25°C; blue, 15°C; purple, 5°C. (C) Effects of cations on the level of polymerization. Yellow, MgCl2; green, CaCl2; red, NaCl; orange, KCl. (D) ATP concentration dependence. Red diamonds, levels (expressed as percentages) attained at 10 min; orange rectangles, increasing rate (per minute). (E) Depolymerization and repolymerization experiment. Initial polymerization (red) was done under standard conditions. After attainment of the plateau, one sample was chilled on ice for 1 h, while another was kept at 56°C; both samples were then incubated again at 56°C. Orange and yellow, second measurements after incubation at 0 and 56°C, respectively. (F) Polymerization activities of Ta0583 mutants. Red, wild type; orange, mutant A; yellow, mutant AB. (G) Protein concentration dependence of LSI. LSI at 10 min after polymerization is plotted against Ta0583 concentration.
After a lag period, the light-scattering signal increased and reached a plateau. A lag period has been noted during actin polymerization and is considered to be a nucleation phase (23). The slope and the plateau level for Ta0583 polymerization were found to depend on pH (Fig. 2A). The plateau level of light scattering was greatest at pH 4.5, although the polymerization rate was low under these conditions. A fast and similarly high level of scattering was attained at pH 5.5, which is close to the previously estimated internal pH of T. acidophilum (30). The pH 5.5 buffer was used in subsequent experiments.
Figure 2B shows the temperature dependence of polymerization. The plateau level increased with increasing temperature up to 56°C. This finding is compatible with the fact that the optimal growth temperature of T. acidophilum is around 60°C.
Mg2+ was required for polymerization (Fig. 2C), and 4 mM MgCl2 was sufficient to enhance the polymerization. However, higher concentrations of MgCl2 suppressed polymerization, with half suppression by about 18 mM. Polymerization was also suppressed by addition of NaCl, KCl, or CaCl2, with half suppression by about 100 mM, 10 mM, and 18 mM, respectively.
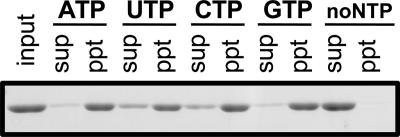
Actin polymerization depends specifically on ATP, whereas polymerization of T. maritima MreB was supported by both ATP and GTP. We found that the polymerization of Ta0583 requires NTP and is optimal at a concentration of about 4 mM ATP (Fig. 2D). The ATP-dependent polymerization of Ta0583 was also confirmed by precipitation analysis (Fig. 4). Ta0583 polymerization was found to be supported not only by ATP and GTP but also by UTP and CTP (Fig. 4; Table 2). The low specificity of Ta0583 for NTPs is consistent with a previous study by Roeben et al. (25), which reported the hydrolysis of GTP, CTP, and UTP as well as ATP. However, even the deoxy form of NTP could support the polymerization reaction (Table 2). This low specificity for NTP species suggests that Ta0583 retains primitive characteristics of ancestral actin. Low specificity is often associated with antiquity of enzymes (14).
FIG. 4.
Precipitation analysis of Ta0583. Shown are the Ta0583 bands in the supernatant (sup) and the precipitate (ppt) after centrifugation (at 10,000 × g for 10 min at 25°C) and the input sample (no centrifugation) after polymerization of 0.5 mg ml−1 Ta0583 under standard conditions with ATP, UTP, CTP, or GTP or without NTPs for 5 min. The Ta0583 band corresponds to a molecular mass of 39 kDa, which is consistent with the gene sequence.
TABLE 2.
Effects of NTPs on the polymerization of Ta0583
| NTPa | Level (%)b | Rate (1/min)c |
|---|---|---|
| No nucleotide | 9.0 | — |
| ATP | 100.0 | 0.71 |
| UTP | 77.9 | 0.48 |
| GTP | 98.8 | 0.37 |
| CTP | 101.4 | 0.33 |
| dATP | 88.1 | 0.87 |
| dTTP | 68.0 | 0.51 |
| dUTP | 62.9 | 0.38 |
| dCTP | 78.8 | 0.79 |
| dGTP | 91.6 | 0.32 |
| ADP | 10.3 | — |
| UDP | 9.9 | — |
| CDP | 7.7 | — |
| GDP | 10.3 | — |
| AMP-PNP | 11.7 | — |
| ATP + AMP-PNP | 92.1 | 0.41 |
| ATP + 8 mM AMP-PNP | 58.2 | 0.37 |
Except where otherwise indicated, the concentration of NTP, dNTP, NDP, and AMP-PNP was 4 mM.
Level represents steady state light scattering intensity attained at 10 min after addition of Ta0583. The level attained in the presence of ATP was assumed 100.0 %.
The rate indicates 1/t1/2, where t1/2 represents the time needed for scattering to reach half of the level attained at 10 min. —, the rate was not reliable under conditions where the level attained was less than 15%.
Nucleoside diphosphate or AMP-PNP did not support polymerization (Table 2), suggesting that the hydrolysis at the third phosphate linkage in NTP is required for the polymerization reaction. The effect of ATP was competitively suppressed by AMP-PNP (Table 2).
The polymerization reaction was accelerated at higher temperatures up to a maximum of 65°C. Not only was the polymerization reaction suppressed by low temperatures, but the polymerized Ta0583 was depolymerized when the polymer was placed at 0°C (Fig. 2E). In this experiment, Ta0583 polymerized under standard conditions was placed at 0°C for 1 h. Then the solution was placed in the cell folder maintained at 56°C. The initial level of the light scattering was as low as the level before polymerization and was followed by an increase in scattering. The depolymerization at 0°C cannot be attributed to the consumption of ATP, because the high level of scattering was observed when the mixture was maintained at 56°C for 1 h after the polymerization reaction under standard conditions.
A 3-dimensional model of polymerized MreB has recently been postulated (35). Based on the model (Fig. 3E), we designed mutations that were anticipated to inhibit the polymerization reaction. We introduced negative charges into domain 2, in Ta0583 mutant A (Fig. 3E). Additional negative charges were introduced into domains 1 and 3, which were expected to interact with domains 2 and 4 of the neighboring subunit, respectively (Fig. 3E), in Ta0583 mutant AB. Polymerization was half inhibited in mutant A and totally abolished in mutant AB (Fig. 2F). These results suggest that Ta0583 has subunit interactions similar to those of actin and T. maritima MreB in the fibrous structure and that domains 2 and 1 or 3 in Ta0583 are also responsible for the polymerization. Our results are consistent with the intersubunit interactions found in the crystal structure of Ta0583 (25), as described in the Discussion.
Polymerization of eukaryotic actin is accelerated by phalloidin and inhibited by cytochalasin D (3). However, the polymerization of Ta0583 was unaffected by both phalloidin and cytochalasin D, at least up to 10 μM (Table 3).
TABLE 3.
Effects of compounds on the polymerization of Ta0583
| Compound(s) | Polymerizationa |
|---|---|
| KCl | − (10 mM) |
| NaCl | − (100 mM) |
| MgCl2 | + (0.5 mM)/− (18 mM) |
| CaCl2 | − (18 mM) |
| ATP, GTP, CTP, UTP | + |
| dATP, dGTP, dCTP, dTTP, dUTP | + |
| ADP, GDP, CDP, UDP | ± |
| Phalloidin | ± |
| Cytochalasin D | ± |
+, enhancement; −, suppression; ±, no effect. The concentration having the half effect is given in parentheses.
The protein concentration dependence of Ta0583 light scattering was measured (Fig. 2G). The critical concentration of Ta0583 was estimated from the plot and is 0.34 μM. This value is much higher than that of MreB (0.003 μM) (7) and is similar to that of actin fiber (0.25 μM) (16).
Electron microscopy.
We used electron microscopy to examine negatively stained bundles of Ta0583 fibers. Figure 3A shows a typical bundle of fibers observed in the Ta0583 sample under standard polymerizing conditions at 25°C. The structure of the bundle resembles the paracrystalline structure of actin formed at high concentrations of KCl (1). The total width of the bundle ranged from 14 nm to more than 1.5 μm (Fig. 3A), with frequent branching (Fig. 3B).
Figure 3C shows a typical view of the narrowest fiber in which a partial helical structure could be observed. Each protofilament in the Ta0583 bundle had a width of about 5.6 nm (5.6 ± 1.0 nm). A subunit-like structure of about 5.0 nm (5.0 ± 1.0 nm) in the axial direction was also seen in some parts of the bundle (Fig. 3C). The repeating unit was analyzed by Fourier transformation of electron micrographs (Fig. 3D). The equatorial and meridian spots indicate the transversal and longitudinal repeats of monomers within Ta0583 fiber. The repeating unit of ∼5.5 nm by ∼5.5 nm was obtained by diffraction analysis (Fig. 3D). The Fourier pattern also indicated the existence of a helical structure, as indicated by some off-meridian/diagonal spots. The off-meridian/diagonal spots indicate the twisting of the fibers like an actin filament.
DISCUSSION
The phylogenetic analysis of actin homologs showed that archaeal homologs form two groups: group A and group B. Eukaryotic actin showed a closer relationship to group A, including Ta0583, than to group B homologs, MreB and ParM.
We found that Ta0583, which shows sequence homology to eukaryotic actin, polymerizes to form bundles. The polymerization of Ta0583, monitored by light scattering, was found to be optimal at pH 4.5 to 5.5. Intriguingly, this pH range is similar to the intracellular pH of T. acidophilum (i.e., pH 5.5) (30). The polymerization reaction accelerated with increasing temperature up to a maximum at 56°C, which is consistent with the optimal growth temperature of T. acidophilum, about 60°C. Several other characteristics of Ta0583 are summarized in Table 4 together with those of its eukaryotic and bacterial counterparts. The effects of MgCl2 on Ta0583 are similar to the effects of this salt on eukaryotic actin, though the optimal concentrations are different: 4 mM for Ta0583 and 2 mM for actin (23). MgCl2 has been reported to suppress the polymerization of MreB from the bacterium T. maritima (7, 35).
TABLE 4.
Comparison of actin homologs
The electron microscopic observations of the negatively stained samples suggest the presence of a protofilament with a size and shape similar to those of actin. The size of this repeating unit (5.5 nm by 5.5 nm) is identical to that of actin protofilaments (13). The unit size is different from those of MreB (5.1 nm by 3.9 nm) and ParM (4.9 nm) (25, 35). The filamentous structure of actin is a right-handed double helix (13) and that of ParM is helical (25), whereas that of T. maritima MreB is straight (35). A helical structure was detected for Ta0583. The branched and curved flexible fibers that are observed in actin and MreB (33, 35) are also observed in Ta0583 fiber (Fig. 3B). However, the structure of the narrowest fiber of Ta0583 may be different from that of actin fiber and rather similar to that of MreB fiber. The narrowest actin fiber is made from two protofilaments with a right-handed helix (13). On the other hand, the simplest fiber of MreB is pairs of thin filaments, namely, four protofilaments (35). The width of the narrowest fiber of Ta0583 is 10 to 14 nm (Fig. 3C). This width is more than twice the size of the Ta0583 monomer; thus, the narrowest fiber of Ta0583 may consist of more than two protofilaments. Taken together, however, these findings suggest that filaments of actin are more similar to those of Ta0583 than to those of T. maritima MreB.
Roeben et al. have analyzed the 3-dimensional structure of Ta0583 (25). They also reported that Ta0583 forms sheets with spacing resembling the crystal lattice in vitro, indicating an inherent propensity to form a filamentous structure. The fold of Ta0583 contains the core structure of actin and clearly belongs to the actin/hsp70 superfamily of ATPases. Based on these findings, Ta0583 is approximately equidistant from actin and MreB at the structural level and combines features from both eubacterial actin homologs, MreB and ParM (25). However, Roeben et al. did not report the ATP- or GTP-dependent polymerization of Ta0583 (25).
Our results are similar to those of Roeben et al. in the sense that Ta0583 can form (para)crystalline structures, although there are significant discrepancies. Specifically, Roeben et al. have not reported nucleotide-dependent polymerization, while our observations show that ATP or GTP can induce Ta0583 polymerization. These discrepancies may be attributed to different conditions used for the polymerization analysis. For example, Roeben et al. used a more neutral pH (pH 6.8) and a higher MgCl2 concentration (10 mM), both of which are inhibitory to the polymerization reaction, for their experiments.
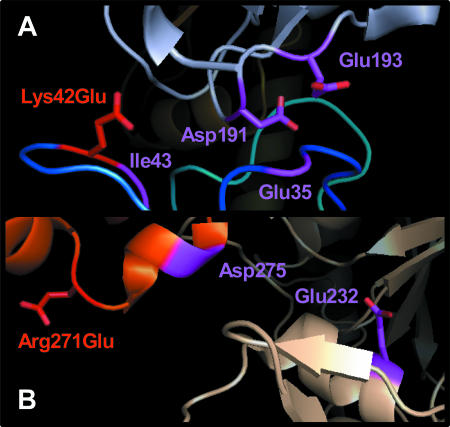
Roeben et al. reported that Glu35, Ile43, and Asp275 are protofilament contacts in the Ta0583 crystal sheet (25). Our mutation analysis is compatible with the protofilament contact reported by Roeben et al. (25). In the structure reported by Roeben et al., charged residues could be located near the introduced charged residues. The distance between the mutated residue Lys42Glu and Asp191 is 10.71 Å, and the distance between Lys42Glu and Glu193 is 12.04 Å in mutant A (Fig. 5A). The distance between mutated residue Arg271Glu and Glu232 is 22.61 Å in mutant AB (Fig. 5B). These negatively charged residues are anticipated to cause electrostatic repulsion in the known structure of Ta0583.
FIG. 5.
Contact site of Ta0583. This figure was made by residue replacement from the Ta0583 crystal structure (25) (Protein Data Bank accession number 2fsj) with PyMOL. Original contact sites are shown in purple. (A) Lys42Glu in mutant A (red) and Asp191/Glu193 in the neighboring subunit. (B) Arg271Glu in mutant AB (red) and Glu232 in the neighboring subunit.
In the phylogenetic tree (Fig. 1), group A and group B homologs have deep branchings with each other as well as with eukaryotic actin and bacterial MreB. These results support the structural analysis of Roeben et al., which showed that Ta0583 is approximately equidistant from actin and MreB at the structural level. However, the tree suggests a closer phylogenetic relationship between group A archaeal homologs, including Ta0583, and eukaryotic actin than between actin and ParM. Ta0583 also showed a unit size similar to that of eukaryotic actin in the filamentous structure. Combining these results, Ta0583 retains the characteristics of actin more than ParM does.
Because T. acidophilum lacks a cell wall, the cytoplasmic membrane is directly exposed to the surrounding milieu. A simple liposomal structure made up of a lipid membrane is expected to form a spherical structure. However, although the overall shape of the cells can be quite variable, they rarely form simple rod or spherical structures (12). Accordingly, the presence of a cytoskeleton in T. acidophilum has been postulated (12). Hixon and Searcy reported a superprecipitation reaction of the cell extract of T. acidophilum (12). The cell extract is superprecipitated by adding calcium and ATP, and the reaction is inhibited by EGTA. These results suggested the presence of an actin-like component in the cells of T. acidophilum. Ta0583, reported here, may represent the actin-like component postulated by Searcy a few decades ago (31). However, the cation dependence of superprecipitation is different from the cation dependence of Ta0583. This observation suggests that there may be additional components responsible for the superprecipitation.
Searcy also reported that T. acidophilum cells become spherical at low temperatures, suggesting depolymerization of the internal structure (31). The depolymerization of Ta0583 detected at low temperatures in our study is compatible with these observations and suggests that Ta0583 is involved in the formation of an internal structure that alters cell morphology. However, Roeben et al. (25) have reported that the content of Ta0583 (0.04%) in T. acidophilum cells is lower than that of eukaryotic actin in eukaryotic cells (∼8%), and the amount may not be sufficient to maintain the structure. Accordingly, components other than Ta0583 may be involved in maintaining cellular structure. Alternatively, expression of Ta0583 in T. acidophilum cells may vary depending on the precise environmental conditions (25).
Polymerization of actin and bacterial MreB is specific to ATP and ATP/GTP, respectively. However, the polymerization of Ta0583 showed very little specificity to ribo- and deoxyribonucleotides (Fig. 4; Table 2). Low specificity for substrates of ancient forms of enzymes has been postulated (14). The low nucleotide specificity found in Ta0583 may represent the ancient characteristics of eukaryotic actin.
There are several models of the evolution of eukaryotic cells (5, 8, 10, 19, 20, 21, 22, 24, 39). However, the ancestor or origin of the eukaryotic nucleus is still unclear. T. acidophilum has been proposed as a host of endosymbiotic origin of eukaryotic cells (20). A phylogenetic tree constructed from small rRNA sequences suggests that different archaeal species are all equally related to eukaryotes and that none of them has a special relationship to the ancestor of eukaryotic cells (39). However, the closest relatives of particular eukaryotic components are often found in different species of Archaea, depending on the gene of interest. For example, eukaryotic RNA polymerase resembles its counterparts in Crenarchaeota and in Thermoplasma species more closely than it resembles its homologs in other species of Euryarchaeota (18). The closest relatives of eukaryotic histone have been found in Euryarchaeota, especially in methanogens (28). Our current data suggest that Thermoplasma species, and possibly also Archaeoglobus species, retain the closest relatives of eukaryotic actin (Table 4). Because the other archaeal homologs of MreB are in the same branch as the bacterial homologs (Fig. 1), which are less similar to eukaryotic actin, our data also imply that the common ancestor of eukaryotes and Archaea already possessed a primitive actin.
Acknowledgments
We are grateful to Tetsuya Satoh for excellent technical assistance in the early stages of the experiments.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Aebi, U., W. E. Fowler, G. Isenberg, T. D. Pollard, and P. R. Smith. 1981. Crystalline actin sheets: their structure and polymorphism. J. Cell Biol. 91:340-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, E. F., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 3.Cooper, J. A. 1987. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 105:1473-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darland, G., T. D. Brock, W. Samsonoff, and S. F. Conti. 1970. A thermophilic, acidophilic mycoplasma isolated from a coal refuse pile. Science 170:1416-1418. [DOI] [PubMed] [Google Scholar]
- 5.Dyall, S. D., M. T. Brown, and P. J. Johnson. 2004. Ancient invasions: from endosymbionts to organelles. Science 304:253-257. [DOI] [PubMed] [Google Scholar]
- 6.Egelman, E. H., N. Francis, and D. J. DeRosier. 1982. F-actin is a helix with a random variable twist. Nature 298:131-135. [DOI] [PubMed] [Google Scholar]
- 7.Esue, O., M. Cordero, D. Wirtz, and Y. Tseng. 2005. The assembly of MreB, a prokaryotic homolog of actin. J. Biol. Chem. 280:2628-2635. [DOI] [PubMed] [Google Scholar]
- 8.Forterre, P. 2006. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain. Proc. Natl. Acad. Sci. USA 103:3669-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fowler, W. E., and U. Aebi. 1982. Polymorphism of actin paracrystals induced by polylysine. J. Cell Biol. 93:452-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta, R. S., and G. B. Golding. 1996. The origin of the eukaryotic cell. Trends Biochem. Sci. 21:166-171. [PubMed] [Google Scholar]
- 11.Hartmann, E., and H. Konig. 1994. A novel pathway of peptide biosynthesis found in methanogenic Archaea. Arch. Microbiol. 162:430-432. [DOI] [PubMed] [Google Scholar]
- 12.Hixon, W. G., and D. G. Searcy. 1993. Cytoskeleton in the archaebacterium Thermoplasma acidophilum? Viscosity increase in soluble extracts. Biosystems 29:151-160. [DOI] [PubMed] [Google Scholar]
- 13.Holmes, K. C., D. Popp, W. Gebhard, and W. Kabsch. 1990. Atomic model of the actin filament. Nature 347:44-49. [DOI] [PubMed] [Google Scholar]
- 14.Jensen, R. A. 1976. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 30:409-425. [DOI] [PubMed] [Google Scholar]
- 15.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 16.Korn, E. D., M. F. Carlier, and D. Pantaloni. 1987. Actin polymerization and ATP hydrolysis. Science 238:638-644. [DOI] [PubMed] [Google Scholar]
- 17.Kurner, J., A. S. Frangakis, and W. Baumeister. 2005. Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science 307:436-438. [DOI] [PubMed] [Google Scholar]
- 18.Langer, D., J. Hain, P. Thuriaux, and W. Zillig. 1995. Transcription in archaea: similarity to that in eucarya. Proc. Natl. Acad. Sci. USA 92:5768-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Garcia, P., and D. Moreira. 1999. Metabolic symbiosis at the origin of eukaryotes. Trends Biochem. Sci. 24:88-93. [DOI] [PubMed] [Google Scholar]
- 20.Margulis, L., and D. Bermudes. 1985. Symbiosis as a mechanism of evolution: status of cell symbiosis theory. Symbiosis 1:101-124. [PubMed] [Google Scholar]
- 21.Martin, W. 2005. Archaebacteria (Archaea) and the origin of the eukaryotic nucleus. Curr. Opin. Microbiol. 8:630-637. [DOI] [PubMed] [Google Scholar]
- 22.Martin, W., and M. Muller. 1998. The hydrogen hypothesis for the first eukaryote. Nature 392:37-41. [DOI] [PubMed] [Google Scholar]
- 23.Pollard, T. D., and J. A. Cooper. 1986. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 55:987-1035. [DOI] [PubMed] [Google Scholar]
- 24.Rivera, M. C., and J. A. Lake. 2004. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431:152-155. [DOI] [PubMed] [Google Scholar]
- 25.Roeben, A., C. Kofler, I. Nagy, S. Nickell, F. U. Hartl, and A. Bracher. 2006. Crystal structure of an archaeal actin homolog. J. Mol. Biol. 358:145-156. [DOI] [PubMed] [Google Scholar]
- 26.Ronquist, F., and J. P. Huelsenbeck. 2001. MyBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 27.Ruepp, A., W. Graml, M. L. Santos-Martinez, K. K. Koretke, C. Volker, H. W. Mewes, D. Frishman, S. Stocker, A. N. Lupas, and W. Baumeister. 2000. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407:508-513. [DOI] [PubMed] [Google Scholar]
- 28.Sandman, K., F. B. Perler, and J. N. Reeve. 1994. Histone-encoding genes from Pyrococcus: evidence for members of the HMf family of archaeal histones in a non-methanogenic Archaeon. Gene 150:207-208. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt, A., and M. N. Hall. 1998. Signaling to the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 14:305-338. [DOI] [PubMed] [Google Scholar]
- 30.Searcy, D. G. 1976. Thermoplasma acidophilum: intracellular pH and potassium concentration. Biochim. Biophys. Acta 451:278-286. [DOI] [PubMed] [Google Scholar]
- 31.Searcy, D. G., D. B. Stein, and K. B. Searcy. 1981. A mycoplasma-like archaebacterium possibly related to the nucleus and cytoplasm of eukaryotic cells. Ann. N. Y. Acad. Sci. 361:312-324. [DOI] [PubMed] [Google Scholar]
- 32.Shimodaira, H., and M. Hasegawa. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246-1247. [DOI] [PubMed] [Google Scholar]
- 33.Steinmetz, M. O., K. N. Goldie, and U. Aebi. 1997. A correlative analysis of actin filament assembly, structure, and dynamics. J. Cell Biol. 138:559-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Ent, F., L. A. Amos, and J. Lowe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 36.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 37.Yasuda, M., H. Oyaizu, A. Yamagishi, and T. Oshima. 1995. Morphological variation of new Thermoplasma acidophilum isolates from Japanese hot springs. Appl. Environ. Microbiol. 61:3482-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yasunaga, T., and T. Wakabayashi. 1996. Extensible and object-oriented system Eos supplies a new environment for image analysis of electron micrographs of macromolecules. J. Struct. Biol. 116:155-160. [DOI] [PubMed] [Google Scholar]
- 39.Zillig, W. 1991. Comparative biochemistry of Archaea and Bacteria. Curr. Opin. Genet. Dev. 1:544-551. [DOI] [PubMed] [Google Scholar]