Abstract
The outer membrane of Borrelia burgdorferi, the causative agent of Lyme disease, contains very few integral membrane proteins, in contrast to other gram-negative bacteria. BBA74, a Borrelia burgdorferi plasmid-encoded protein, was proposed to be an integral outer membrane protein with putative porin function and designated as a 28-kDa outer membrane-spanning porin (Oms28). In this study, the biophysical properties of BBA74 and its subcellular localization were investigated. BBA74 is posttranslationally modified by signal peptidase I cleavage to a mature 25-kDa protein. The secondary structure of BBA74 as determined by circular dichroism spectroscopy consists of at least 78% α-helix with little β-sheet structure. BBA74 in intact B. burgdorferi cells was insensitive to proteinase K digestion, and indirect immunofluorescence microscopy showed that BBA74 was not exposed on the cell surface. Triton X-114 extraction of outer membrane vesicle preparations indicated that BBA74 is not an integral membrane protein. Taken together, the data indicate that BBA74 is a periplasmic, outer membrane-associated protein that lacks properties typically associated with porins.
Lyme disease, the most commonly reported arthropod-borne disease in the United States, is a multisystem disorder with possible neurologic, cardiac, and arthritic manifestations (21, 26). It is caused by infection with the spirochete Borrelia burgdorferi (1, 41). B. burgdorferi has a complex life cycle involving Ixodes sp. ticks and mammalian reservoir hosts, usually rodents (11, 35). To successfully complete the enzootic life cycle, the spirochete must adapt to diverse host environments and nutrient availability (15, 40). Multiple levels of gene regulation may be involved in the adaptation of B. burgdorferi to different hosts (30, 36).
Porins are members of an extended family of outer membrane-spanning proteins in gram-negative bacteria that are involved in nutrient uptake (18). Porins typically form homotrimers, allowing the diffusion of nonpolar molecules across the permeability barrier created by the outer membrane (2, 22). In addition to their essential physiological roles in nutrient acquisition, porins can potentially act as adhesins, mediate bacterial attachment to mammalian cells, and activate the complement system (12, 14, 17). These findings suggest that porins play an important role in bacterial survival and pathogenesis. Four B. burgdorferi proteins have been described as having porin activity (P66, BBA74, P13, and BBA01) (28, 29, 37, 38). Of these, BBA74 is the only protein annotated as a porin in the B. burgdorferi genome database (16). bba74 is predicted to encode a 257-amino-acid polypeptide with a conserved peptidase I signal sequence. Previous studies indicated that transcription of bba74 is regulated in response to various stimuli, including temperature, mammalian host environment, and blood (6, 24, 31, 42). In addition, bba74 may also be subject to transcriptional regulation by trans-acting proteins (27). Furthermore, bba74 transcript levels are 8- to 30-fold higher in B. burgdorferi clinical isolates attenuated for hematogenous dissemination (25). For these reasons, BBA74 was selected for more careful analysis.
BBA74 was first identified as a virulent strain-associated outer membrane-spanning (Oms) protein based on its presence in isolated outer membrane fractions, hence, its designation as Oms28 (39). Subsequent black lipid membrane assays conducted with the purified recombinant protein suggested that it is a porin (37). However, its physiological role has not yet been elucidated, nor have studies confirming its surface location in B. burgdorferi been reported. Bioinformatic analysis of the BBA74 sequence does not predict membrane-spanning regions or membrane localization. In the present study, the biophysical properties of BBA74 were characterized, and its cellular localization was determined.
MATERIALS AND METHODS
Strains and growth conditions.
B. burgdorferi strain B356, a previously described isolate from a human erythema migrans lesion (46), was employed for these studies. This strain was chosen because it produces higher levels of BBA74 than many other isolates (25). The sequence of the strain B356 bba74 coding region is identical to that of the sequenced isolate, B31 MI (25). Spirochetes were grown in modified Barbour-Stoenner-Kelly medium (45) at 34°C until cultures reached the mid-log phase of growth.
N-terminal amino acid sequence analysis.
Total cellular protein (180 μg) was treated with RNase A and DNase I for 30 min at room temperature. Protein samples were subjected to reduction and alkylation and precipitated using a modified Flugge's protocol (47). The pellets were rehydrated in sample buffer containing 8 M urea, 1 M thiourea, 2% Triton X-100, and 2 mM tributylphosphine and sonicated at low intensity for 5 seconds. Duplicate protein samples were subjected to isoelectric focusing using ReadyStrip IPG linear gradient pH 5 to 8 strips (Bio-Rad, Hercules, CA) according to the manufacturer's protocol. The IPG strips were electrophoresed in the second dimension on a 12.5% sodium dodecyl sulfate (SDS)-polyacrylamide gel. Proteins were transferred to duplicate polyvinylidene difluoride (PVDF) membranes; one membrane was subjected to immunoblot analysis with anti-BBA74 rat serum, and the second membrane was stained with Coomassie blue (Bio-Rad). The protein spots corresponding to BBA74 were excised from the Coomassie-stained membrane, destained, washed extensively, and subjected to N-terminal amino acid analysis at the W. M. Keck Biotechnology resource facility at Yale University.
Recombinant BBA74 expression and purification.
B. burgdorferi BBA74 was cloned and expressed in Escherichia coli as follows. bba74, from B. burgdorferi strain 297 minus the sequence encoding the leader peptide, was cloned into the pProEx Htb expression vector (Invitrogen, Carlsbad, CA) using primers bba75 5′BamHI (5′-TGGGATCCTTAAATGTGTTTGCAGATTCTAAC-3′) and bba74 3′-XhoI (5′-ATCTCGAGCTATCTCATGTATAAAGAAATAGCC-3′) according to the manufacturer's instructions. The vector was used to transform E. coli TOP10 cells and screened by negative selection on LB broth containing 50 μg/ml ampicillin. The clones were sequenced, which confirmed that the cloned bba74 sequence was identical to that of strain B31MI with the exception of a single amino acid difference at position 177 (T→I). The selected clone was used to transform E. coli Rosetta2 cells (Novagen) for expression of the recombinant protein. An overnight starter culture from a single colony was diluted 1:20 in prewarmed LB broth containing chloramphenicol (25 μg/ml) and ampicillin (50 μg/ml) and grown with shaking at 37°C until the optical density at 600 nm reached 0.4. Expression of BBA74 was induced by addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.2 mM, and cultures were grown for an additional 3 h.
Cells were harvested by centrifugation at 7,000 × g for 15 min, washed once in 1× phosphate-buffered saline (PBS), resuspended in 4 ml lysis buffer 1 (50 mM Tris [pH 8.2], lysozyme [0.4 mg/ml], 50 μl of protease inhibitor cocktail [Sigma-Aldrich, St. Louis, MO]) and frozen overnight at −20°C. The suspension was thawed by the addition of 14 ml of lysis buffer 2 (20 mM Tris [pH 8.8], 10% glycerol, 0.1% Elugent [Calbiochem], 0.2 M NaCl), and the suspension was centrifuged at 17,000 × g for 20 min at 4°C. The resulting pellet was resuspended in 100 mM NaH2PO4, 6 M guanidine-HCl, and 10 mM Tris-HCl, pH 8.0, incubated with end-over-end mixing for 30 min at room temperature, and centrifuged at 15,000 rpm for 30 min at 4°C. The supernatant was removed and filtered through a 5-mm prefilter (Millipore).
Ni-nitrilotriacetic acid resin and filtered supernatant were mixed for 30 min at room temperature with rocking and applied to a column, and flowthrough was collected. The column was washed with 25 ml of denaturing buffer B (100 mM NaH2PO4, 8 M urea, and 10 mM Tris-HCl, pH 8.0) with 20 mM imidazole, and the recombinant protein was eluted with 25 ml of denaturing buffer B containing 250 mM imidazole. Elution fractions of 1 ml were collected from the column, and 5 μl of each fraction was assessed for the presence of BBA74 by 12.5% SDS-polyacrylamide gel electrophoresis (PAGE). Fractions containing BBA74 were pooled and dialyzed overnight at 4°C against 2 liters of buffer containing 4 M urea and 50 mM Tris-HCl (pH 7.6). The fractions were then sequentially dialyzed overnight against 50 mM Tris-HCl buffer containing 2 M and 1 M urea, respectively, followed by final dialysis in 1× PBS. The renatured, dialyzed BBA74 was assayed for protein concentration and stored at −20°C.
Generation of antiserum directed against BBA74 and OppA IV.
Recombinant poly-histidine OppA IV protein was cloned into pProEX Htb using primers oppA IV 5′-BamHI (5′-CACCGGATCCTGTGTTAATGAAAGTAATA GAAACAAA-3′) and oppA IV 3′-XhoI (5′-CTCGAGTTTAATTGGTTTTATTTCAG ATAAATT-3′), and the fusion protein was purified as described above for BBA74. Monospecific polyclonal antisera against BBA74 and OppA IV were generated by hyperimmunizing female Sprague-Dawley rats with a 1:1 mixture of Freund's complete adjuvant (Sigma-Aldrich, St. Louis, MO) and 40 μg of either BBA74 or OppA IV in PBS (pH 7.4) administered intraperitoneally. After 3 and 5 weeks, the animals were boosted intraperitoneally with BBA74 or OppA IV in a 1:1 mixture of Freund's incomplete adjuvant (Sigma-Aldrich). Two weeks following the second boost, animals were exsanguinated and serum was collected. Chemiluminescent immunoblot analysis revealed that the resulting antisera reacted with single polypeptides of the expected molecular masses in B. burgdorferi whole-cell lysates.
CD spectroscopy.
Circular dichroism (CD) spectroscopy was performed on purified BBA74 using a Jasco J-715 spectropolarimeter (Jasco, Easton, MD). Three scans were averaged for each spectrum. The background signal (buffer) was subtracted prior to calculating mean residue molar ellipticity (MRE [θ]) values. The percentages of helix, sheet, and random coil structure were calculated by using the K2d algorithm available at http://www.embl-heidelberg.de.
Proteinase K digestion and immunoblot analysis.
Spirochetes (1 × 109 cells) were harvested by centrifugation, washed with 1× PBS, resuspended in 950 μl of 1× PBS containing 5 mM MgCl2, and treated with either 200 μg of proteinase K or distilled water for 40 min at room temperature. Proteinase K digestion was terminated by addition of 500 μg of phenylmethylsulfonyl fluoride to each reaction mixture. Cells were harvested by centrifugation, washed with 1× PBS, and boiled for 5 min in 1× SDS-PAGE sample buffer. Whole-cell lysates obtained in this manner were separated by 10 to 20% gradient SDS-PAGE, and proteins were transferred to a PVDF membrane for immunoblot analysis. Membranes were probed with either rat anti-BBA74 (1:1,000), rat anti-OppAIV (1:500), rabbit anti-P66 (1:1,500) (kindly provided by J. Coburn) or rabbit anti-FlaB (1:10,000) (13) sera. Alkaline phosphatase-conjugated goat anti-rat and anti-rabbit immunoglobulin G (IgG; Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD) were used appropriately as the secondary antibodies. The blots were washed three times, and visualization was achieved by treating membranes with 5-bromo-4-chloro-3-indolyl-phosphate-nitroblue tetrazolium solution (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD) for 1 to 3 min at room temperature. The color reaction was stopped by washing the membranes thoroughly with deionized water.
Indirect immunofluorescence microscopy.
Indirect immunofluorescence staining was performed essentially as described by Cox et al. (13). B. burgdorferi B356 cells were grown to mid-log phase, and 1 × 107 intact cells were resuspended in 0.1 ml 1× PBS supplemented with 10% fetal calf serum (FCS). The cells were incubated in a solution consisting of rabbit anti-OspA (1:100; kindly provided by D. Bucher) or anti-FlaB (1:100) and rat anti-BBA74 (1:50) antibodies. Prior to use, the anti-BBA74 antibodies were preadsorbed with intact B. burgdorferi B31-A3 cells lacking BBA74 to minimize cross-reactivity. After incubation with the primary antibodies for 2 h at 34°C with intermittent gentle mixing, cells were washed three times with 0.5 ml of 1× PBS, 1% FCS and resuspended in 0.1 ml 1× PBS with 10% FCS, and 10 μl was placed on glass chamber slides and air dried. Slides were incubated with Alexa Fluor 488-labeled goat anti-rabbit IgG (1:200) and Alexa Fluor 594-labeled goat anti-rat IgG (1:200) antibodies (Molecular Probes, Invitrogen) in 1× PBS, 10% FCS for 1 h at 34°C with intermittent gentle mixing. After incubation, slides were gently washed three times with 1× PBS, 1% FCS and once with 1× PBS and were fixed with 4% PBS-buffered formaldehyde (methanol free; Ted Pella Inc.) for an additional 15 min at room temperature. The labeled cells were mounted in VectaShield medium containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA) and were visualized using a Zeiss inverted Axiovert 200 motorized microscope with the 100× objective and Zeiss filter sets 31, 34, and 38 for Alexa Fluor 594 and 488 and DAPI, respectively. Images were acquired with a Zeiss Axiocam MRM cooled charge-coupled device camera and were analyzed using Axiovision 4.3 software.
Similar indirect immunofluorescence assays were performed with cells initially fixed with methanol to identify subsurface antigens. A 10-μl aliquot of cell suspension in 1× PBS, 10% FCS was air dried, and cells were fixed by immersing the slides in 60% methanol at room temperature for 10 min, followed by washing with 1× PBS. Slides were air dried and incubated with anti-OspA (1:100) or anti-FlaB (1:100) and anti-BBA74 (1:50) antibodies in 1× PBS, 10% FCS for 1 h at 34°C. Cells were washed three times with 1× PBS, 1% FCS and incubated with secondary antibodies as above. After incubation, cells were washed three times with 1× PBS, 1% FCS and once with 1× PBS and visualized as described above. To rule out the possibility of nonspecific cross-reactivity for the immunofluorescence labeling, methanol-fixed B356 cells were labeled with preimmune rabbit and rat sera. The cells were washed, incubated with the appropriate secondary antibodies, and visualized as described above.
Preparation of B. burgdorferi OMVs.
Spirochetes were fractionated into outer membrane vesicle (OMV) and protoplasmic cylinder (PC) fractions essentially as described by Skare et al. (39). Spirochetes (2.5 × 1010) were agitated in 25 mM citrate buffer (pH 3.2) for 2 h at room temperature. The resulting OMV and PC fractions were separated on a discontinuous sucrose gradient composed of 56%, 42%, and 25% (wt/wt) sucrose. The OMV fraction was further purified by centrifugation on a continuous 10 to 42% (wt/wt) sucrose gradient. The OMV fraction was removed by needle aspiration, diluted in 1× PBS, and collected by centrifugation at 141,000 × g for 5 h at 5°C. The final OMV pellet was resuspended in 100 μl 1× PBS containing 1 mM phenylmethylsulfonyl fluoride and was stored in aliquots at −80°C until use. The PC material was recovered by centrifugation at 10,000 × g for 20 min at 5°C, resuspended in 1 ml 1× PBS, and stored at −80°C until use.
For high-molar salt treatment of B. burgdorferi OMV fractions, 200 μg of OMV protein was suspended in 150 μl of 1 M NaCl and incubated for 15 min on ice. Samples were diluted to 1 ml with 1× PBS and centrifuged at 43,000 × g for 2 h at 4°C. The obtained membrane pellets were suspended in 30 μl of 1× SDS-PAGE loading buffer and boiled for 5 min. The proteins in the supernatant were precipitated by addition of 7 volumes of ice-cold acetone, boiled in 30 μl of 1× SDS-PAGE loading buffer for 5 min, and used for immunoblot analysis as above, except that SDS-PAGE was performed with 12.5% gels and horseradish peroxidase-conjugated secondary antibodies were employed.
TX-114 phase partitioning.
OMV and PC fractions (150 μg) were solubilized overnight in 1 ml of PBS containing 2% Trion X-114 (TX-114) at 4°C as described elsewhere (5, 8, 39). After removing TX-114-insoluble material, the supernatant was incubated at 37°C for 15 min followed by separation of the two phases by centrifugation at 15,000 × g for 15 min at room temperature. The aqueous phase was removed and reextracted five times with 2% TX-114. The detergent phases were combined, and proteins in both the detergent and aqueous phases were precipitated by addition of 7 volumes of ice-cold acetone. Precipitated proteins were recovered by centrifugation at 15,000 × g for 30 min at 4°C and employed for immunoblot analysis.
RESULTS
BBA74 is processed at its N terminus.
The BBA74 amino acid sequence contains a predicted signal peptidase I cleavage site between A24 and D25. To investigate whether BBA74 is processed by signal peptidase I, total B. burgdorferi cellular proteins were separated by two-dimensional PAGE. BBA74 focused at a pH of ∼5.7 and migrated as a 25- to 26-kDa protein. These characteristics are consistent with the predicted pI and molecular mass of a peptidase I-cleaved BBA74 and suggested posttranslational processing of the protein by signal peptidase I cleavage.
Protein spots corresponding to BBA74 were excised and subjected to N-terminal sequence analysis. The results identified the first seven N-terminal amino acids of BBA74 as DSNNANI. This peptide sequence is identical to the predicted BBA74 amino acid sequence from residues 25 to 31. This demonstrates that the mature BBA74 sequence lacks the first 24 amino acid residues, as predicted for signal peptidase I cleavage.
BBA74 lacks an extensive β-sheet structure.
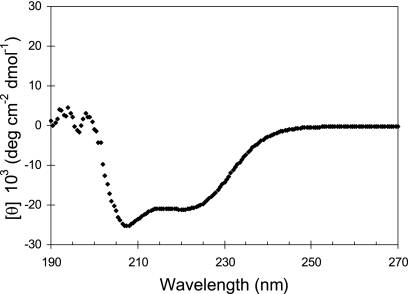
CD spectral analysis was performed to determine whether the BBA74 protein contains the extensive β-sheet secondary structure typical of integral outer membrane proteins in gram-negative bacteria (43). The MRE values were calculated from the obtained data and plotted as a function of the far-UV spectrum (Fig. 1). The obtained CD spectrum indicated the presence of double minima at 208 and 220 nm as observed in α-helical proteins. This suggests the presence of a pronounced α-helical secondary structure in BBA74. The MRE values were analyzed using the K2d algorithm, which predicted that the mature BBA74 protein contains 78% α-helical, 21% random coil, and 1% β-sheet secondary structures.
FIG. 1.
BBA74 does not have an extensive β-sheet secondary structure. BBA74 was purified, and the CD spectrum was obtained using a J-715 spectropolarimeter (Jasco, Easton, MD). Three scans were averaged for each spectrum, and the calculated MRE values were plotted as a function of the far-UV spectrum.
BBA74 is not surface exposed.
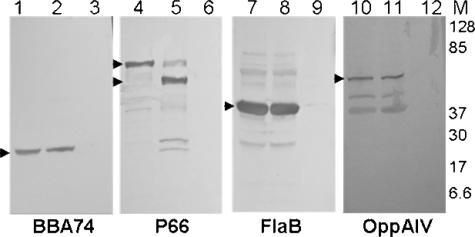
To investigate possible surface exposure of BBA74, intact B. burgdorferi was subjected to proteinase K digestion. As shown in Fig. 2 (lanes 1 and 2), no digestion of BBA74 was detected in the intact cells. The efficiency of proteolysis was assessed by evaluating cleavage of P66, a protein previously shown to contain surface-exposed regions (10). P66 was cleaved, as expected (Fig. 2, lane 5). The periplasmic marker FlaB and the inner membrane marker OppA IV were used as controls for assessing the integrity of the outer membrane (34, 44). Neither of these proteins was digested by proteinase K (Fig. 2, lanes 8 and 11). In cells permeabilized by pretreatment with 0.6% Triton X-100, all proteins, including BBA74, were completely digested on proteinase K treatment. The lack of BBA74 cleavage in intact spirochetes indicates that this protein does not have accessible surface-exposed regions.
FIG. 2.
BBA74 does not have surface-exposed epitopes. B. burgdorferi cells were incubated with proteinase K with or without permeabilization. Total lysates were separated on 10 to 20% SDS-PAGE gels and blotted to PVDF membranes. The membranes were probed with polyclonal rat sera against BBA74 or OppA IV or rabbit sera against P66 or FlaB. Lanes 1, 4, 7, and 10, intact cells without proteinase K treatment; lanes 2, 5, 8, and 11, intact cells with proteinase K treatment; lanes 3, 6, 9, and 12, permeabilized cells with proteinase K treatment. Arrowheads indicate native and digested proteins. The migration positions of molecular mass markers are indicated by numbers in the right margin.
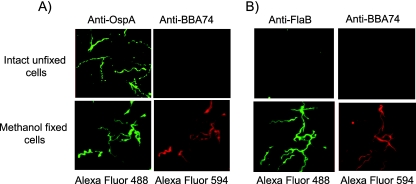
Immunofluorescence microscopy was employed as an alternative method to elucidate localization of BBA74. As shown in Fig. 3, BBA74 could not be detected on the surface of intact bacteria with anti-BBA74 antibody. As expected, OspA was detectable on the surface of these intact cells. When cells were permeabilized by methanol fixation, both BBA74 and OspA could be visualized with specific antibodies. The latter result is consistent with a previously published study indicating that, in addition to being associated with the outer membrane, a substantial amount of OspA is associated with the cytoplasmic membrane of B. burgdorferi (13). The integrity of the outer membranes of the nonpermeabilized B. burgdorferi cells was confirmed by staining of intact B. burgdorferi cells with anti-FlaB antibodies. Immunostaining with preimmune rabbit and rat sera in combination with appropriate secondary antibodies ruled out any possibility of nonspecific cross-reactivity that may have contributed to the observed localization (data not shown). The results are in full agreement with the proteinase K digestion results and indicate that BBA74 is not exposed on the bacterial surface.
FIG. 3.
Localization of BBA74 by immunofluorescence microscopy. Intact unfixed or methanol-fixed B. burgdorferi cells were incubated with either anti-OspA and anti-BBA74 (A) or anti-FlaB and anti-BBA74 (B) antibodies. Alexa Fluor 488-labeled goat anti-rabbit IgG and Alexa Fluor 594-labeled goat anti-rat IgG were employed as secondary antibodies. Immunofluorescence staining was visualized using a Zeiss inverted Axiovert 200 microscope, and the images were acquired using AxioVision software. The images were converted to JPEG format, and brightness and contrast were linearly adjusted for each panel. The top and bottom labels indicate the primary and secondary antibodies used for each image panel. All the images were acquired at 1,000× final magnification.
BBA74 is an outer membrane-associated protein.
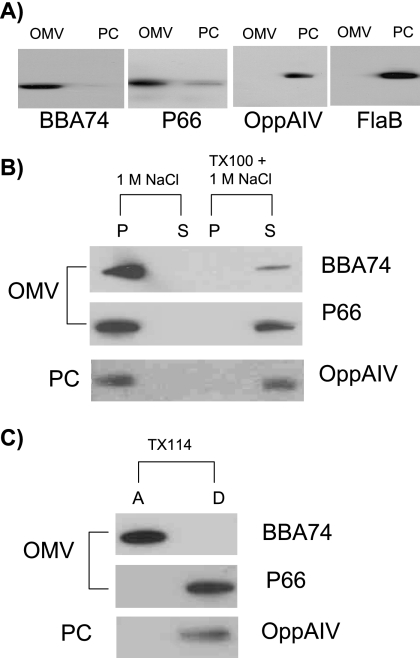
In light of the above findings and earlier reports describing BBA74 as an outer membrane-spanning protein, the cellular location of BBA74 was reevaluated using the fractionation methods originally used to localize it to the outer membrane (37, 39). B. burgdorferi cells were fractionated into OMV and PC fractions which were analyzed by immunoblot analysis. P66 and BBA74 predominantly separated into the OMV fraction, indicating that both were associated with the outer membrane (Fig. 4A). To rule out contamination of the OMV fraction with periplasmic or cytoplasmic membrane-associated proteins, the fractions were also examined for FlaB and OppA IV; both proteins separated into the PC fraction, as expected. BBA74 and P66 were closely associated with the outer membrane of the spirochete via a nonionic interaction, as both separated with the pelleted membrane fraction after 1 M NaCl treatment (Fig. 4B). Integrity of the outer membrane was important for the observed association, as pretreatment of the OMV fraction with Triton X-100 released BBA74 and P66 from the outer membrane.
FIG. 4.
Cellular localization of BBA74. OMV and PC fractions were prepared and treated as described in Materials and Methods. Protein fractions were separated by 12.5% SDS-PAGE, and the indicated proteins were visualized by immunoblotting. (A) Separation into OMV and PC fractions; (B) 1 M NaCl treatment; (C) TX-114 phase partitioning. P, pellet; S, soluble; A, aqueous; D, detergent.
BBA74 is not an integral membrane protein.
To further characterize the nature of the association of BBA74 with the outer membrane, TX-114 phase partitioning studies were performed. Such partitioning has been employed to distinguish amphiphilic proteins from hydrophilic proteins (4). The results indicated that BBA74 separated exclusively into the aqueous phase (Fig. 4C). In contrast, P66 and OppA IV were predominantly extracted into the detergent-enriched phase as expected for integral membrane or membrane-anchored proteins. When Triton X-114 phase partitioning was carried out by an alternative method (7), identical results were obtained (data not shown). These findings suggest that BBA74 is not a membrane-spanning protein.
DISCUSSION
BBA74 was initially annotated as the Oms28 (outer membrane-spanning, 28-kDa) protein based on its presence in the OMV fraction (37, 39). Subsequent studies with purified recombinant BBA74 showed that it possessed a 0.6-nS porin activity in planar membrane conductance assays. Based on these findings, it was concluded that BBA74 is an outer membrane-spanning porin (37). Some of the properties reported here for BBA74 are in agreement with this earlier investigation (37); BBA74 is associated with the OMV, partitions into the aqueous phase on Triton X-114 extraction, and is processed by signal peptidase I cleavage of the primary BBA74 translation product. However, no direct evidence existed for surface exposure or β-barrel structure, as would be expected for an outer membrane-spanning porin.
In the current study it was demonstrated that BBA74 has no surface localization, based on resistance to proteinase K treatment and indirect immunofluorescence staining. This contrasts with the behavior of three other characterized B. burgdorferi porins, P66, P13, and BBA01, all of which have surface exposure and are digested by proteinase K (10, 23, 29). The possibility that BBA74 has an innate resistance to protease digestion is unlikely, since indirect immunofluorescence staining indicated that BBA74 is not present on the cell surface. In contrast, both P66 and P13 have been shown to be present on the cell surface by immunofluorescence microscopy (9, 23).
All previously characterized gram-negative porins have been shown to assume a β-barrel topology with multiple surface-exposed loops and lack α-helical transmembrane domains (33, 48). To identify whether mature BBA74 has the substantial β-sheet secondary structure required to form the β-barrel topology, CD spectroscopy of the purified protein was performed. This analysis indicated that BBA74 has a pronounced α-helical secondary structure. Further analysis of CD data by the K2d algorithm predicted that the BBA74 secondary structure consists of 78% α-helix, 21% random coil, and 1% β-sheet. CD spectral analysis of the classical gram-negative β-barrel proteins indicated the presence of a significant β-sheet secondary structure (>40%) (43). Thus, BBA74 does not possess a secondary structure characteristic of the gram-negative β-barrel porins. Interestingly, even though mature BBA74 appears to have extensive α-helical content, none of the topology prediction algorithms used could identify any α-helical transmembrane domains. This was further substantiated by the Triton X-114 extraction experiments. A previous study noted that BBA74 partitions into the aqueous phase on Triton X-114 extraction (37). This behavior was ascribed to improper folding of the protein resulting in anomalous fractionation. The more direct explanation is that BBA74 is periplasmic and extrinsically associated with the OM.
To date, the sole experimental evidence that BBA74 is a porin in B. burgdorferi relies on electro-conduction activities of this protein in planar membrane assays (37). However, the reliability of this experimental procedure can be affected by several factors, including the presence of detergents like Triton X-100 (32) and contamination with other proteins (20). In the previously reported study of electro-conduction of BBA74, the protein was eluted from polyacrylamide gels in a buffer containing Triton X-100. In light of the characterized biophysical properties of BBA74, the possibility arises that the observed electro-conduction of BBA74 might be an experimental artifact due to the presence of Triton X-100.
N-terminal amino acid analysis confirmed that mature BBA74 is processed by signal peptidase I. Such processing has been associated with the Sec-dependent secretory mechanism involved in protein sorting (3, 19). Little information is available regarding sorting of membrane proteins and the role of signal peptidase I processing in B. burgdorferi. Our results demonstrate that signal peptidase I processing occurs in B. burgdorferi and suggest a possible mechanism by which BBA74 is transported into the periplasmic space. Once in the periplasmic space, BBA74 could associate with the outer leaflet of the cytoplasmic membrane or the inner leaflet of the outer membrane or remain as a soluble periplasmic protein. Fractionation of BBA74 with the OMV fraction suggests that it is associated with the inner surface of the outer membrane by interactions that are not disrupted by 1 M NaCl treatment. At present, there is no evidence for posttranslational lipid modification of BBA74 or the presence of transmembrane regions which could explain its close association with the outer membrane. Thus, the possibility that BBA74 is a soluble periplasmic protein and that its separation with OMV is an artifact of cellular fractionation cannot be ruled out at the present time.
In summary, mature BBA74 is a 25-kDa periplasmic protein with extensive α-helical content and virtually no β-sheet secondary structure. The protein may be associated with the outer membrane but does not span the membrane or have any surface exposure. It is unclear how a protein with such localization could function as a porin. Annotation of BBA74 as Oms28 porin is inconsistent with these findings. Thus, the function of BBA74 in B. burgdorferi is presently unknown.
Acknowledgments
This work was supported by NIH grants AI45801 (I.S.), AI27656 (J.R.), and AI29735 (J.R. and M.J.C.).
We thank Doris Bucher, Jenifer Coburn, and Jon Skare for antisera, Dana Mordue for guidance with immunofluorescence microscopy, and Radha Iyer for helpful advice.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Benach, J. L., E. M. Bosler, J. P. Hanrahan, J. L. Coleman, G. S. Habicht, T. F. Bast, D. J. Cameron, J. L. Ziegler, A. G. Barbour, W. Burgdorfer, R. Edelman, and R. A. Kaslow. 1983. Spirochetes isolated from the blood of two patients with Lyme disease. N. Engl. J. Med. 308:740-742. [DOI] [PubMed] [Google Scholar]
- 2.Benz, R., and K. Bauer. 1988. Permeation of hydrophilic molecules through the outer membrane of gram-negative bacteria. Review on bacterial porins. Eur. J. Biochem. 176:1-19. [DOI] [PubMed] [Google Scholar]
- 3.Black, M. T., J. G. Munn, and A. E. Allsop. 1992. On the catalytic mechanism of prokaryotic leader peptidase 1. Biochem.J. 282:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 5.Brandt, M. E., B. S. Riley, J. D. Radolf, and M. V. Norgard. 1990. Immunogenic integral membrane proteins of Borrelia burgdorferi are lipoproteins. Infect. Immun. 58:983-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks, C. S., S. R. Vuppala, A. M. Jett, and D. R. Akins. 2006. Identification of Borrelia burgdorferi outer surface proteins. Infect. Immun. 74:296-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brusca, J. S., and J. D. Radolf. 1994. Isolation of integral membrane proteins by phase partitioning with Triton X-114. Methods Enzymol. 228:182-193. [DOI] [PubMed] [Google Scholar]
- 9.Bunikis, J., and A. G. Barbour. 1999. Access of antibody or trypsin to an integral outer membrane protein (P66) of Borrelia burgdorferi is hindered by Osp lipoproteins. Infect. Immun. 67:2874-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunikis, J., L. Noppa, Y. Ostberg, A. G. Barbour, and S. Bergstrom. 1996. Surface exposure and species specificity of an immunoreactive domain of a 66-kilodalton outer membrane protein (P66) of the Borrelia spp. that cause Lyme disease. Infect. Immun. 64:5111-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgdorfer, W. 1984. Discovery of the Lyme disease spirochete and its relation to tick vectors. Yale J. Biol. Med. 57:515-520. [PMC free article] [PubMed] [Google Scholar]
- 12.Coburn, J., J. R. Fischer, and J. M. Leong. 2005. Solving a sticky problem: new genetic approaches to host cell adhesion by the Lyme disease spirochete. Mol. Microbiol. 57:1182-1195. [DOI] [PubMed] [Google Scholar]
- 13.Cox, D. L., D. R. Akins, K. W. Bourell, P. Lahdenne, M. V. Norgard, and J. D. Radolf. 1996. Limited surface exposure of Borrelia burgdorferi outer surface lipoproteins. Proc. Natl. Acad. Sci. USA 93:7973-7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen, P. A., D. A. Haake, and B. Adler. 2004. Outer membrane proteins of pathogenic spirochetes. FEMS Microbiol. Rev. 28:291-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Silva, A. M., and E. Fikrig. 1997. Arthropod- and host-specific gene expression by Borrelia burgdorferi. J. Clin. Investig. 99:377-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fuji, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 17.Galdiero, F., M. A. Tufano, L. Sommese, A. Folgore, and F. Tedesco. 1984. Activation of complement system by porins extracted from Salmonella typhimurium. Infect. Immun. 46:559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock, R. E. 1987. Role of porins in outer membrane permeability. J. Bacteriol. 169:929-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacob-Dubuisson, F., R. Fernandez, and L. Coutte. 2004. Protein secretion through autotransporter and two-partner pathways. Biochim. Biophys. Acta 1694:235-257. [DOI] [PubMed] [Google Scholar]
- 20.Labarca, P., and R. Latorre. 1992. Insertion of ion channels into planar lipid bilayers by vesicle fusion. Methods Enzymol. 207:447-463. [DOI] [PubMed] [Google Scholar]
- 21.Nadelman, R. B., and G. P. Wormser. 1998. Lyme borreliosis. Lancet 352:557-565. [DOI] [PubMed] [Google Scholar]
- 22.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noppa, L., Y. Ostberg, M. Lavrinovicha, and S. Bergstrom. 2001. P13, an integral membrane protein of Borrelia burgdorferi, is C-terminally processed and contains surface-exposed domains. Infect. Immun. 69:3323-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ojaimi, C., V. Mulay, D. Liveris, R. Iyer, and I. Schwartz. 2005. Comparative transcriptional profiling of Borrelia burgdorferi clinical isolates differing in capacities for hematogenous dissemination. Infect. Immun. 73:6791-6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orloski, K. A., E. B. Hayes, G. L. Campbell, and D. T. Dennis. 2000. Surveillance for Lyme disease—United States, 1992-1998. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 49:1-11. [PubMed] [Google Scholar]
- 27.Ostberg, Y., J. A. Carroll, M. Pinne, J. G. Krum, P. Rosa, and S. Bergstrom. 2004. Pleiotropic effects of inactivating a carboxyl-terminal protease, CtpA, in Borrelia burgdorferi. J. Bacteriol. 186:2074-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostberg, Y., M. Pinne, R. Benz, P. Rosa, and S. Bergstrom. 2002. Elimination of channel-forming activity by insertional inactivation of the p13 gene in Borrelia burgdorferi. J. Bacteriol. 184:6811-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinne, M., K. Denker, E. Nilsson, R. Benz, and S. Bergstrom. 2006. The BBA01 protein, a member of paralog family 48 from Borrelia burgdorferi, is potentially interchangeable with the channel-forming protein P13. J. Bacteriol. 188:4207-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porcella, S. F., and T. G. Schwan. 2001. Borrelia burgdorferi and Treponema pallidum: a comparison of functional genomics, environmental adaptations, and pathogenic mechanisms. J. Clin. Investig. 107:651-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rostovtseva, T. K., C. L. Bashford, A. A. Lev, and C. A. Pasternak. 1994. Triton channels are sensitive to divalent cations and protons. J. Membr. Biol. 141:83-90. [DOI] [PubMed] [Google Scholar]
- 33.Schulz, G. E. 2002. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta 1565:308-317. [DOI] [PubMed] [Google Scholar]
- 34.Schulze, R. J., and W. R. Zuckert. 2006. Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol. Microbiol. 59:1473-1484. [DOI] [PubMed] [Google Scholar]
- 35.Schwan, T. G. 1996. Ticks and Borrelia: model systems for investigating pathogen-arthropod interactions. Infect. Agents Dis. 5:167-181. [PubMed] [Google Scholar]
- 36.Schwan, T. G., and J. Piesman. 2002. Vector interactions and molecular adaptations of Lyme disease and relapsing fever spirochetes associated with transmission by ticks. Emerg. Infect. Dis. 8:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skare, J. T., C. I. Champion, T. A. Mirzabekov, E. S. Shang, D. R. Blanco, H. Erdjument-Bromage, P. Tempst, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1996. Porin activity of the native and recombinant outer membrane protein Oms28 of Borrelia burgdorferi. J. Bacteriol. 178:4909-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skare, J. T., T. A. Mirzabekov, E. S. Shang, D. R. Blanco, H. Erdjument-Bromage, J. Bunikis, S. Bergstrom, P. Tempst, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1997. The Oms66 (p66) protein is a Borrelia burgdorferi porin. Infect. Immun. 65:3654-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skare, J. T., E. S. Shang, D. M. Foley, D. R. Blanco, C. I. Champion, T. Mirzabekov, Y. Sokolov, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1995. Virulent strain associated outer membrane proteins of Borrelia burgdorferi. J. Clin. Investig. 96:2380-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steere, A. C., J. Coburn, and L. Glickstein. 2004. The emergence of Lyme disease. J. Clin. Investig. 113:1093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steere, A. C., R. L. Grodzicki, A. N. Kornblatt, J. E. Craft, A. G. Barbour, W. Burgdorfer, G. P. Schmid, E. Johnson, and S. E. Malawista. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733-740. [DOI] [PubMed] [Google Scholar]
- 42.Tokarz, R., J. M. Anderton, L. I. Katona, and J. L. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 72:5419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallace, B. A., J. G. Lees, A. J. Orry, A. Lobley, and R. W. Janes. 2003. Analyses of circular dichroism spectra of membrane proteins. Protein Sci. 12:875-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallich, R., S. E. Moter, M. M. Simon, K. Ebnet, A. Heiberger, and M. D. Kramer. 1990. The Borrelia burgdorferi flagellum-associated 41-kilodalton antigen (flagellin): molecular cloning, expression, and amplification of the gene. Infect. Immun. 58:1711-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, G., R. Iyer, S. Bittker, D. Cooper, J. Small, G. P. Wormser, and I. Schwartz. 2004. Variations in Barbour-Stoenner-Kelly culture medium modulate infectivity and pathogenicity of Borrelia burgdorferi clinical isolates. Infect. Immun. 72:6702-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, G., C. Ojaimi, R. Iyer, V. Saksenberg, S. A. McClain, G. P. Wormser, and I. Schwartz. 2001. Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect. Immun. 69:4303-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141-143. [DOI] [PubMed] [Google Scholar]
- 48.Wimley, W. C. 2003. The versatile beta-barrel membrane protein. Curr. Opin. Struct. Biol. 13:404-411. [DOI] [PubMed] [Google Scholar]