Abstract
When envelope biogenesis is compromised or damage to envelope components occurs, bacteria trigger signaling cascades, which lead to the production of proteins that combat such extracytoplasmic stresses. In Escherichia coli, there are three pathways known to deal with envelope stresses: the Bae, Cpx, and σE responses. Although the effectors of the Bae and Cpx responses are not essential in E. coli, the effector of the σE response, the sigma factor RpoE (σE), is essential for viability. However, mutations that suppress the lethality of an rpoE-null allele can be easily obtained, and here we describe how we have isolated at least four classes of these suppressors. We present the first description of one such suppressor class, loss-of-function mutations in ydcQ, a gene encoding a putative DNA-binding protein. In wild-type rpoE+ strains, ydcQ mutants have two distinct phenotypes: extracytoplasmic stress responses are significantly downregulated, and the production of outer membrane vesicles is severely reduced. We present a model in which σE is not essential per se but, rather, we propose that rpoE mutant cells die, possibly because they overreact to the absence of this σ factor by triggering a cell death signal.
The cell envelope of gram-negative bacteria surrounds the cytoplasm and is composed of the inner and outer membranes, an aqueous intermembrane space known as the periplasm, and the peptidoglycan layer, which resides in the periplasm. In addition to performing essential physiological functions such as energy generation, the cell envelope protects the cell from the environment. It is therefore not surprising that gram-negative bacteria have developed signaling pathways that sense and respond to perturbations of the cell envelope (i.e., envelope stress). Collectively, these pathways are known as extracytoplasmic or envelope stress responses.
In Escherichia coli, three signaling systems are known to monitor envelope stress: the Bae, Cpx, and σE pathways (reviewed in references 1, 4, 35, and 42). All three systems possess components in the inner membrane that are thought to sense and transduce specific signals to their respective effectors in the cytoplasm; these effectors, when activated, direct the transcription of genes encoding envelope biogenesis factors such as chaperones, proteases, and enzymes involved in envelope biosynthesis. Each pathway is activated by a variety of signals, such as misfolded envelope proteins, ethanol, high temperatures, high pH, and toxic compounds.
Both the Bae and the Cpx pathways are two-component systems in which their respective histidine kinases, BaeS and CpxA, are inner membrane proteins with periplasmic domains that sense envelope perturbations (33, 34, 39). When activated by envelope stress, these kinases are thought to autophosphorylate and then transfer their phosphate to their cognate response regulators, BaeR and CpxR, respectively. Once phosphorylated, these DNA-binding response regulators activate transcription of their regulon members (14, 31).
The effector of the σE pathway is the alternative sigma factor RpoE, or σE, which is encoded by the rpoE gene (41). In the absence of activating signals, σE is sequestered in an inactive state at the inner membrane by the anti-sigma factor RseA (2, 3, 8, 13, 30). When the appropriate signals are present, a proteolytic cascade that involves the DegS and RseP proteases is activated, which leads to the degradation of RseA (5, 22, 23). As a result, σE is released into the cytoplasm, where it acts as a sigma factor to activate transcription of more than 80 genes (40).
The σE pathway differs from the Bae and Cpx systems not only because of differences in the mechanism of signal transduction but also because it is the only known envelope stress response that is essential for viability in E. coli (12). However, strains lacking σE can be obtained because one or more suppressors of the lethality conferred by an rpoE-null allele frequently arise (12). Still, such rpoE mutants exhibit phenotypes that reflect envelope defects, such as sensitivity to temperatures above 30°C, hydrophobic antibiotics, and toxic small molecules that disrupt the outer membrane (41).
DegS and RseP are also essential proteins, since strains lacking these proteases are depleted of σE activity (6, 21, 22). A recent study has revealed that overexpression of the small RNA RseX, which negatively regulates the expression of the outer membrane proteins OmpA and OmpC, suppresses degS- and rseP-null mutations (15). Furthermore, RseP is not essential in mutants lacking both OmpC and OmpA. However, neither overexpression of RseX nor the loss of OmpA and OmpC suppress the lethality conferred by rpoE-null mutations (15).
To understand why σE is essential, we decided to identify any suppressor(s) of the lethality conferred by an rpoE-null allele. Phenotypic characterization of 20 transductants that acquired an rpoE::cam allele by P1 transduction revealed that there are at least four different types of suppressors of σE essentiality. We describe here the identification of one such type of suppressor, null alleles of ydcQ, a gene encoding a putative DNA-binding protein. Furthermore, we found that in an rpoE+ background the lack of YdcQ downregulates both outer membrane vesicle production and the basal levels of envelope stress responses. Remarkably, downregulation of the Cpx response by the loss of YdcQ is CpxR independent.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Strains used in the present study are listed in Table 1. Standard microbial techniques were used for strain construction (43). Luria-Bertani (LB), M63 minimal, and lactose MacConkey agar media were prepared as described previously (43). Experiments with strains harboring the rpoE::cam allele were performed at 30°C unless indicated otherwise. All other experiments were performed under aeration at 37°C. Growth was monitored by measuring the optical density at 600 nm (OD600). Unless indicated, all experiments were done by growing cells in LB broth. Tetracycline and kanamycin were used at a concentration of 25 μg/ml. Chloramphenicol and ampicillin were used at concentrations of 20 and 125 μg/ml, respectively.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 9 |
| CAG22216 | MC1061 λ(rpoHP3′-lacZ+) rpoE::cam supCAG | 41 |
| NR670 | MC4100 λ(degP′-lacZ+) | This study |
| JEB9 | NR670 rpoE::cam sup9 | This study |
| JEB10 | NR670 rpoE::cam sup10 | This study |
| JEB75 | NR670 rpoE+nadB::Tn10 sup10 | This study |
| JEB75.1 | NR670 rpoE+nadB+sup10 | This study |
| JEB93 | NR670 rpoE+nadB::Tn10 sup9 | This study |
| JEB93.1 | NR670 rpoE+nadB+sup9 | This study |
| NR668 | MC4100 λ(cpxP′-lacZ+) | This study |
| NR669 | MC4100 λ(rpoHP3′-lacZ+) | This study |
| NR677 | MC4100 λ(spy′-lacZ+) | This study |
| NR754 | MC4100 ara+ | This study |
| NR905 | MC4100 ydcQ::kan | This study |
| NR906 | MC4100 ydcR::kan | This study |
| NR915 | NR677 ydcQ::kan | This study |
| NR916 | NR677 ydcR::kan | This study |
| NR918 | NR668 ydcQ::kan | This study |
| NR919 | NR668 ydcR::kan | This study |
| NR921 | NR669 ydcQ::kan | This study |
| NR922 | NR669 ydcR::kan | This study |
| NR924 | NR670 ydcQ::kan | This study |
| NR925 | NR670 ydcR::kan | This study |
| NR927 | NR754 ydcQ::kan | This study |
| NR986 | MC4100 λ(cpxR′-lacZ+) | This study |
| NR987 | NR986 ydcQ::kan | This study |
| NR989 | NR986 cpxR::spec | This study |
| NR990 | NR989 ydcQ::kan | This study |
Construction of pBADydcQ.
A plasmid expressing ydcQ under the control of the arabinose promoter was constructed by inserting the ydcQ open reading frame into the EcoRI and XbaI sites of pBAD18 (17). The ydcQ open reading frame was amplified by PCR using MC4100 genomic DNA as a template and the primers 5ydcQEcoRI (5′-AAAGAATTCGAGTTAATCGCCAATTAA; the EcoRI site is underlined) and 3ydcQXbaI (5′-AAATCTAGATATCAGTTGTTAAAAATG; the XbaI site is underlined).
Genetic mapping.
To map sup9 and sup10, we used bacteriophage P1 mapping as follows. We generated a P1 lysate from a pool of mutants carrying randomly inserted mini-Tntet cassettes in the chromosome of our wild-type strain MC4100 (24). This pool of mutants was used as a donor of wild-type alleles in P1 transductions where the recipients were rpoE+ strains carrying the degP′-lacZ+ fusion and either sup9 or sup10. We then screened for transductants that showed increased LacZ activity on lactose MacConkey agar. We found that a mini-Tntet insertion in ydcS was >95% linked to sup9 and sup10. PCR amplification and DNA sequencing of the region upstream of ydcS revealed that sup9 and sup10 are IS1E insertions in ydcQ.
Outer membrane vesicle preparations.
Cultures were grown overnight in LB broth at 37°C to an OD600 of ∼4. Samples from different cultures were standardized by OD600, and they were pelleted at 10,000 × g for 10 min at 4°C. The culture supernatant was filtered through a 0.45-μm-pore-size low protein-binding cellulose acetate membrane filter (Corning) and then centrifuged for 1 h at 40,000 × g at 4°C. The pellet, which contained outer membrane vesicles, was resuspended in 50 mM HEPES (pH 6.8). Before being loaded onto a 15% polyacrylamide gel containing sodium dodecyl sulfate (SDS), the samples were mixed with SDS sample buffer (26) and boiled for 10 min.
Western blot analysis.
Cultures were grown overnight in LB broth at 37°C, and 1-ml samples were pelleted. To standardize the samples, the pellets were resuspended in a volume (in milliliters) of SDS sample buffer equal to an OD600/10. The samples were boiled for 10 min, and equal volumes were subjected to electrophoresis in 15% polyacrylamide gels containing SDS as described by Laemmli (26). Proteins were transferred to nitrocellulose membranes (Schleicher & Schuell), and Western blot analysis was performed with rabbit polyclonal sera against DegP, Spy, LamB (which also recognizes OmpA), MBP, and RpoS (from our laboratory collection) as primary antibodies at dilutions of 1:30,000, 1:2,500, 1:30,000, 1:30,000, and 1:6,000, respectively. Donkey anti-rabbit immunoglobulin G (IgG) horseradish peroxidase conjugate (Amersham Pharmacia Biotech) was used as secondary antibody at a 1:8,000 dilution. For lipopolysaccharide (LPS), a monoclonal antibody to LPS core (HyCult Biotechnology, The Netherlands) and goat anti-mouse IgG-horseradish peroxidase conjugate (Bio-Rad) were used at a 1:5,000 dilution. For visualization of bands, the ECL antibody detection kit (Amersham Pharmacia Biotech) and X-Omat film (Kodak) were used.
β-Galactosidase assays.
Cultures were grown overnight in LB broth at 37°C. β-Galactosidase assays were performed by using a microtiter plate assay as described previously (44). The β-galactosidase activity of each strain is presented relative to the wild-type strain, which was assigned 100% activity. For each experiment, every sample was assayed three times, and the average activity and standard deviation are shown. The data presented were derived from a single experiment representative of at least three independent experiments.
RESULTS
Isolation of mutations that suppress rpoE essentiality.
To understand why σE is essential (12), we identified and characterized mutations that suppress the lethality conferred by the rpoE::cam null allele (41). The strategy that we used is described in Fig. 1. We introduced the rpoE::cam allele into our wild-type strain MC4100 carrying the degP'-lacZ+ reporter fusion, which is under the control of both σE and Cpx. Typically, after 48 h at 30°C, we obtained 0 to 22 transductants in each cross. As expected, these transductants exhibited very low levels of LacZ activity on lactose MacConkey indicator agar, which reflects the fact that expression of degP′-lacZ+ is partially dependent on σE.
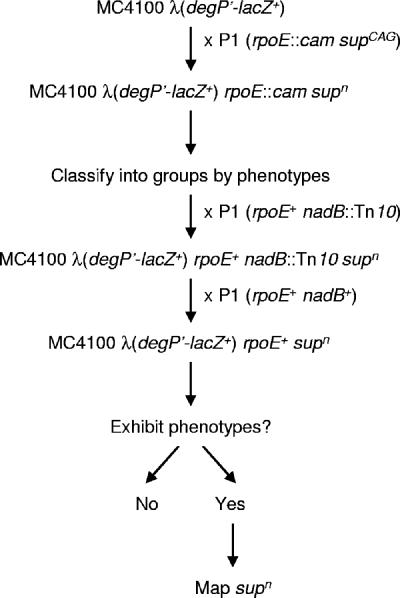
FIG. 1.
Strategy used to map suppressors of rpoE::cam lethality. A detailed description of this strategy is given in Results and Materials and Methods. The rpoE::cam P1 donor strain used was CAG22216, which carries the unidentified suppressor supCAG.
To determine whether all of the isolated rpoE mutants carried the same suppressor (supn), we monitored several phenotypes (degP′-lacZ+ activity, growth at various temperatures and on different media, sensitivity to antibiotics and detergents; Fig. 1 and representative results shown in Table 2). We concluded that there are at least four different phenotypic classes among the rpoE::cam supn transductants that we analyzed.
TABLE 2.
Representative phenotypic classes of rpoE suppressors
| Strain genotypea | Growthb |
|||||
|---|---|---|---|---|---|---|
| SDS/EDTA | 22°C | 37 oC | 42 oC | Glucose | Glycerol | |
| rpoE+ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| rpoE::cam sup9 | + | + | +++ | + | ++ | ++ |
| rpoE::cam sup1 | + | +++ | ++++ | +++ | − | − |
| rpoE::cam sup5 | − | + | +++ | ++ | ++ | ++ |
| rpoE::cam sup21 | ++ (M) | + (M) | ++++ (M) | + | ++ (M) | + |
rpoE+ refers to NR670. Shown are data derived from a representative suppressor supn for each of the four suppressor classes.
Growth of suppressor strains is reported relative to that of an rpoE+ strain, which was arbitrarily given a “++++” value. No growth is reported as “−”. “(M)” refers to a mucoid growth phenotype. SDS/EDTA = LB agar + 0.5% SDS 0.5 mM EDTA; 22°C, 37°C, and 42°C refer to LB agar incubated at the indicated temperatures; glucose refers to glucose M63 minimal agar; glycerol refers to glycerol M63 minimal agar.
To facilitate genetic mapping, we sought suppressors that conferred a tractable phenotype in the presence of a wild-type rpoE allele. Therefore, we replaced the rpoE::cam allele with a wild-type rpoE allele by cotransduction with the nadB::Tn10 allele, which is located immediately downstream of rpoE (Fig. 1). To eliminate any possible effects caused by the nadB::Tn10 allele in the resulting rpoE+ supn strains, we introduced a nadB+ allele by crossing them with an rpoE+ nadB+ donor (Fig. 1). We then searched for any phenotypes that would allow us to map the suppressor mutation (degP′-lacZ+ activity, growth at various temperatures and on different media, and sensitivity to antibiotics and detergents).
The only tractable phenotypic difference that we could detect was that two of the strains still exhibited reduced expression of degP′-lacZ+ in the presence of the rpoE+ allele. We confirmed that these two strains carried suppressors of rpoE essentiality by crossing them with an rpoE::cam donor. Unlike the wild-type, these strains are efficient recipients for rpoE::cam in a transduction cross (>200 rpoE::cam transductants). This manuscript deals with the characterization of these two suppressor alleles, which we will refer to as sup9 and sup10.
sup9 and sup10 are insertion mutations in ydcQ.
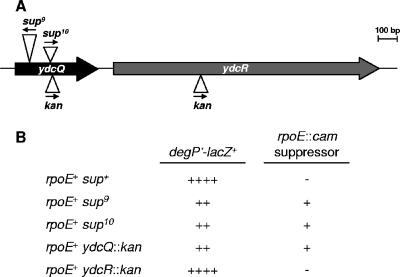
As described in Materials and Methods, standard genetic and molecular methods showed that sup9 and sup10 are two independent IS1E element insertions oriented in opposite directions in ydcQ (Fig. 2A), a gene specifying a protein of unknown function that is composed of 145 amino acids and includes a predicted helix-turn-helix DNA-binding domain in its C terminus, as well as an uncharacterized domain found in small proteins (UPF0150 domain, found at http://www.sanger.ac.uk/Software/Pfam/). We introduced either sup9 or sup10 into a wild-type strain carrying the degP′-lacZ+ fusion by cotransduction with a closely linked marker (ydcS::miniTn10) which, on its own, does not affect either LacZ fusion activity or suppression (data not shown). Both resulting strains had decreased LacZ activity, and genetic tests demonstrated that they carried suppressors of the lethality conferred by rpoE::cam (Fig. 2B). These results confirm that both the lowered expression of degP′-lacZ+ and the suppression of rpoE essentiality are caused by the disruption of ydcQ.
FIG. 2.
Disruption of ydcQ suppresses lethality of rpoE::cam and downregulates degP transcription. (A) Genetic organization of the ydcQ locus and localization of various insertion elements and antibiotic resistance markers. Arrows indicate the direction of transcription. (B) Disruption of ydcQ but not of ydcR reduces expression of degP′-lacZ+ and suppresses lethality of rpoE::cam. Relative levels of LacZ expression from the degP′-lacZ+ fusion on lactose MacConkey agar are indicated with “+” signs. If a mutation is able to suppress the lethality of rpoE::cam, a “+” is shown on the right column, but if a mutation is not able to suppress lethality, a “−” is shown.
Because ydcQ is located immediately upstream (predicted 79-bp intragenic region) of ydcR, which also specifies a predicted DNA-binding protein, we investigated whether either aforementioned phenotype could be caused by polar effects of the IS1E insertions on ydcR. We obtained kan insertions in both ydcQ and ydcR from the University of Wisconsin E. coli project; in both instances, the kan cassette is oriented in the same direction as ydcQ and ydcR transcription (Fig. 2A). Because ydcR expression is driven by the kan promoter in the ydcQ::kan strain, we believe that this ydcQ mutation is not polar. We found that only the ydcQ::kan allele reduced the expression of degP′-lacZ+ and suppressed rpoE::cam-induced lethality (Fig. 2B), which suggests that YdcR does not contribute to any of the aforementioned phenotypes.
In order to verify that ydcR was not involved, we conducted complementation tests by scoring the ability of the ydcQ::kan allele to suppress the lethality of rpoE::cam in diploids overexpressing ydcQ in trans from the pBAD18-based vector, pBADycdQ. In a wild-type strain in the presence or absence of the inducer arabinose neither pBAD18 nor pBADydcQ had an effect on the yield of rpoE::cam transductants (Table 3). However, induction of expression of the plasmid-borne ydcQ in a ydcQ::kan strain dramatically reduced the numbers of rpoE::cam transductants compared to the vector and uninduced pBADydcQ controls (Table 3). Thus, it is the loss of YdcQ function that suppresses the lethality conferred by the rpoE::cam allele. From hereafter we will characterize ydcQ mutants in a wild-type rpoE background.
TABLE 3.
Complementation of suppression by ydcQ
| Straina | No. of rpoE::cam transductantsb |
|
|---|---|---|
| Without arabinose | With arabinose | |
| NR754(pBAD18) | 0 | 0 |
| NR754(pBADydcQ) | 1 | 4 |
| NR927(pBAD18) | 705 | >1,000 |
| NR927(pBADydcQ) | 490 | 29 |
NR754 = ydcQ+ ara+; NR927 = ydcQ::kan ara+.
After 48 h at 30° C.
Loss of YdcQ downregulates extracytoplasmic stress responses.
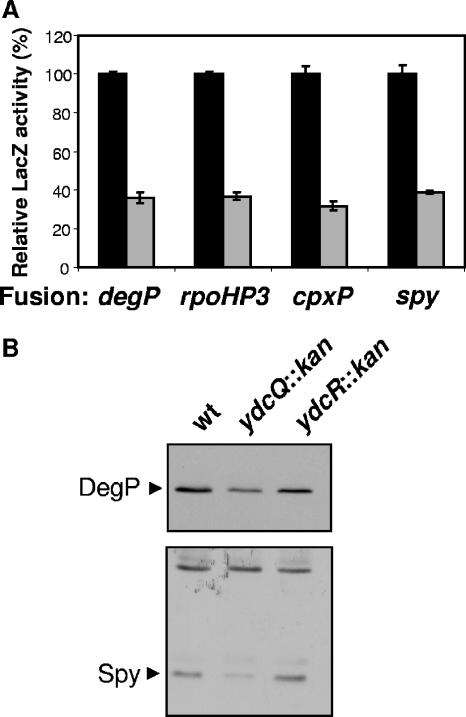
Expression of degP′-lacZ+ is reduced to ca. 35% in a ydcQ::kan rpoE+ background compared to the wild-type strain (Fig. 3A). Since degP is under the control of both σE and Cpx, we explored which of these two extracytoplasmic stress responses was downregulated by the loss of YdcQ. To do so, we monitored the effects of the ydcQ::kan allele on the expression reporter fusions that are solely regulated by either σE or Cpx using the rpoHP3′-lacZ+ (29) and cpxP′-lacZ+ (10) fusions, respectively.
FIG. 3.
Loss of YdcQ downregulates envelope stress responses. (A) LacZ activity of various reporter fusions (shown in the x axis) controlled by envelope stress responses in a wild-type (▪) and a ydcQ::kan (░⃞) strain. The degP′-lacZ+ fusion is controlled by both σE and Cpx, rpoHP3′-lacZ+ by σE, cpxP′-lacZ+ by Cpx, and spy′-lacZ+ by Cpx and Bae. Relative LacZ activities are shown where 100% corresponds to the LacZ activity of the wild-type strain carrying each fusion. Shown is an experiment representative of at least three independent experiments. (B) Western blotting of whole-cell samples showing reduced levels of DegP and Spy in a ydcQ::kan mutant (NR905) with respect to the wild-type (wt) MC4100 and ydcR::kan mutant (NR906) strains.
Similar to the effects observed with the degP′-lacZ+ fusion, the ydcQ::kan allele reduced expression of the rpoHP3′-lacZ+ fusion by ca. 65% (Fig. 3A), which indicates that the ydcQ::kan allele downregulates the σE response. Surprisingly, we also detected a comparable reduction in the expression of the Cpx-regulated fusion cpxP′-lacZ+ (Fig. 3A). Furthermore, we found that the ydcQ::kan allele reduces LacZ activity in strains carrying the spy′-lacZ+ fusion (36), which is controlled by the Bae stress pathway (34). However, because spy is also regulated by the Cpx system, we cannot distinguish whether this downregulation of spy′-lacZ+ is dependent on Bae, Cpx, both, or neither.
We also confirmed that ydcQ::kan reduces the cellular levels of DegP and Spy by comparing their steady-state levels in wild-type and ydcQ mutant strains by Western blot analysis (Fig. 3B). In addition, as expected from the results presented above (Fig. 2B), the presence of the ydcR::kan allele did not alter the levels of either DegP or Spy (Fig. 3B), which thereby confirms that the phenotypes reported here are indeed caused solely by the loss of YdcQ.
We also determined that the ydcQ-null allele does not have a general negative effect on all LacZ reporter fusion by verifying that the ydcQ::kan allele does not alter the LacZ levels in a strain carrying an rpoS750′-′lacZ fusion (data not shown). Furthermore, since RpoS is the sigma factor controlling the stationary-phase stress response (32), YdcQ does not affect every stress response in E. coli. In addition, we can conclude that the downregulatory effect that the loss of YdcQ has on the expression of genes regulated by envelope stress responses is not the result of a global effect on gene expression. First, ydcQ mutants do not exhibit growth defects as determined by growth curve analysis (data not shown). Moreover, Western blot analyses have revealed that the levels of many proteins that are regulated by different mechanisms are not affected by the loss of YdcQ (data not shown and Western blots shown in Fig. 3 and 4). However, since we do not know how many genes other than the ones reported here are affected by the loss of YdcQ, we cannot conclude at this time whether YdcQ only affects the expression of genes regulated by envelope stress responses.
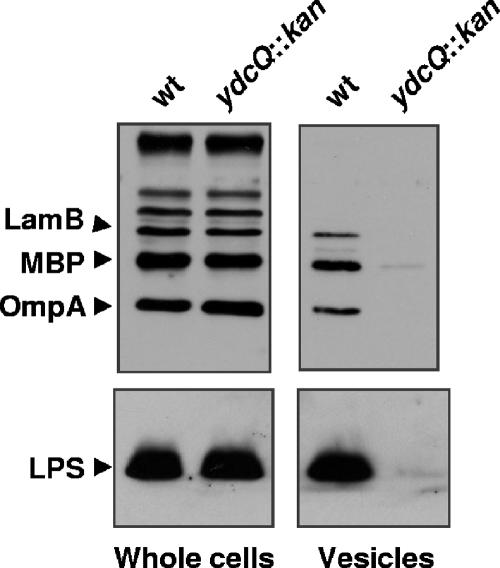
FIG. 4.
Loss of YdcQ downregulates production of outer membrane vesicles. Western blot analysis of the outer membrane proteins OmpA and LamB, periplasmic protein MBP, and outer membrane lipid LPS shows that although the loss of YdcQ function in NR905 does not alter the levels of these envelope components in whole cells, it reduces the amount of outer membrane vesicles present in culture supernatants compared to the wild-type strain MC4100.
Nevertheless, the results presented here show that the loss of YdcQ downregulates extracytoplasmic stress responses. Consistent with the fact that extracytoplasmic stress-combating factors such as DegP are downregulated in cells lacking YdcQ, we have found that ydcQ mutants are more sensitive to envelope stress, as determined by their increased sensitivity to the presence of misfolded PapE or PapG proteins (20) in the periplasm (data not shown).
Loss of YdcQ downregulates the production of outer membrane vesicles.
Recently, the Kuehn laboratory has shown that mutations that alter (upregulate or downregulate) the σE pathway increase the production of outer membrane vesicles and they have suggested that vesiculation is an envelope stress response (28). Accordingly, we hypothesized that increased vesiculation in the ydcQ mutant could be the stress response that compensates for the loss of σE. Therefore, we tested whether the loss of YdcQ increases the release of outer membrane vesicles.
To test the effects of the loss of YdcQ on outer membrane vesicle production, we isolated outer membrane vesicles from supernatants of overnight cultures of both wild-type and ydcQ::kan strains and compared their relative levels. Because outer membrane vesicles are composed of both outer membrane (LPS, phospholipids, and proteins) and periplasm but lack inner membrane and cytoplasmic components, we monitored the quantity of vesicle material released into the growth media by using Western blot analysis to detect levels of LPS, the periplasmic protein MBP, and the outer membrane proteins LamB and OmpA. It is important to note that the levels of these components in whole cells are not affected by the absence of YdcQ (Fig. 4, whole-cell panel). We also confirmed that our vesicle preparations were not contaminated by cell lysis because we could not detect any cytoplasmic RpoS in these samples (data not shown).
Analysis of outer membrane vesicle preparations showed that, in contrast to our prediction, the ydcQ mutant releases significantly lower levels of outer membrane vesicles than the wild-type strain (Fig. 4). The fact that we could detect less protein by Coomassie blue staining after electrophoresis of vesicle preparations further confirmed the reduction of vesiculation (data not shown). Therefore, our data show that the loss of YdcQ reduces the production of outer membrane vesicles, even though the σE response is decreased. In addition, we note that we could not detect any gross differences in envelope composition that could explain this decreased vesiculation phenotype, since the overall protein profiles of cellular envelope fractions of the ydcQ mutant resembled those of wild type (data not shown).
Since it has been recently suggested that vesiculation could be another envelope stress response (28), it is possible that the loss of YdcQ downregulates outer membrane vesicle production in the same way it downregulates the other envelope stress responses. In other words, it is possible that all known extracytoplasmic stress responses, including outer membrane vesicle formation, are downregulated in ydcQ mutants.
Loss of YdcQ downregulates Cpx independently of CpxR.
As described above, extracytoplasmic stress responses are downregulated in a ydcQ-null mutant. A plausible explanation for this global effect of YdcQ on envelope stress responses is that ydcQ mutants have a lower basal level of inducing signals in their extracytoplasmic compartments. Although we do not know of any signal that induces all known envelope stress responses, one could reason, for example, that an unidentified stress response is activated in the absence of YdcQ, which then reduces the level of basal stress (i.e., activating signals) in the cell envelope. As a result, all other extracytoplasmic stress responses would be downregulated. Furthermore, activation of an additional stress response could also explain how the loss of YdcQ suppresses the lethality conferred by rpoE::cam; such a response could compensate for the loss of σE.
If the downregulation of extracytoplasmic stress responses caused by the loss of YdcQ was due to a reduction in the basal level of envelope stress (i.e., reduced inducing signals), this downregulation would be dependent on an intact signaling cascade and the effector of each stress response. That is, using the Cpx pathway as an example, if the absence of YdcQ caused the downregulation of Cpx by lowering envelope stress, this effect would not be detectable in strains that lack the response regulator CpxR. We therefore tested whether the loss of YdcQ reduced the basal level of envelope stress by comparing the effects of the ydcQ::kan null allele in wild-type and cpxR mutant strains that carry the Cpx-regulated cpxR′-lacZ+ reporter fusion (37). Notably, we could not use cpxP′-lacZ+ because, without CpxR, expression from this fusion is undetectable (10).
As expected, since the cpxR′-lacZ+ reporter fusion is positively regulated by Cpx (37) and the loss of YdcQ downregulates the Cpx response, the ydcQ::kan allele reduced the expression of this fusion (Fig. 5). Likewise, as previously shown (37), a cpxR::spec null allele (11) reduced the expression of cpxR′-lacZ+ (Fig. 5). We found that the ydcQ cpxR double mutant strain had an even lower level of LacZ activity than either single mutant. In fact, the level of reduction caused by the loss of YdcQ was the same (ca. 30%) whether CpxR was present or not. This additive effect indicates that YdcQ and the effector protein CpxR regulate the Cpx response through independent pathways. We also detected a similar additive effect on DegP and Spy levels upon the loss of both CpxR and YdcQ (data not shown). Together, these results indicate that the ydcQ::kan-dependent downregulation of the Cpx response is not dependent on CpxR, and, therefore, it is not caused by a reduction in the basal level of envelope stress that is monitored by CpxA, which then signals to CpxR. Thus, CpxR and YdcQ regulate the Cpx regulon by two different pathways.
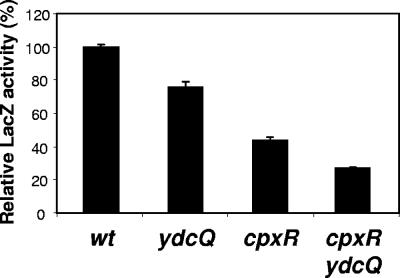
FIG. 5.
YdcQ downregulates Cpx in a CpxR-independent manner. LacZ activity of strains carrying the Cpx-regulated fusion cpxR′-lacZ+. Individually, the ydcQ::kan (ydcQ) and cpxR::spec (cpxR) alleles reduce the expression of cpxR with respect to wild type (wt). Together, both mutations reduce cpxR expression even further, demonstrating that they act in different pathways. Relative LacZ activities are shown where 100% corresponds to the LacZ activity of the wild-type strain carrying each fusion. Shown is an experiment representative of at least three independent experiments. From left to right, strains used were NR986, NR987, NR989, and NR990.
Cpx is not required in the absence of σE and YcdQ.
Although the loss of YdcQ downregulates the Cpx pathway, it is possible that Cpx activity could become essential in the absence of σE, since there is overlap in the genes that both pathways activate (38). However, we were able to easily introduce the rpoE::cam allele into a ydcQ::kan cpxR::spec double mutant strain. Furthermore, we found that constitutive activation of the Cpx pathway by the cpxA* allele cpxA24 (39) does not behave as a suppressor of rpoE::cam-induced lethality. Thus, Cpx is not required in the absence of σE and its activation cannot substitute for the loss of σE.
DISCUSSION
The Bae, Cpx, and σE signaling pathways sense and respond to envelope stress in E. coli. Of these, only the σE pathway is essential in E. coli, although mutations that suppress the lethality of an rpoE-null allele often arise. After isolating and phenotypically characterizing a collection of strains lacking σE, we have found that there are at least four classes of suppressors, which can explain why suppressors are isolated so easily. In the present study, we have determined that one suppressor class consists of null mutations in ydcQ, a gene encoding a putative DNA-binding protein that contains a predicted helix-turn-helix domain. In addition, we have found that, in rpoE+ strains, null mutations in ydcQ downregulate envelope stress responses and outer membrane vesicle production. In the case of Cpx, we have demonstrated that the downregulatory effect of ydcQ mutations occurs through an unknown mechanism independent of CpxR.
The ydcQ mutants provide a useful tool for understanding the essential nature of σE. We have considered two models to explain both the requirement for σE and the suppression of rpoE-null alleles by ydcQ mutations. The first model is based on the idea that in the absence of σE, E. coli cannot produce one or more factors that are essential for viability. Since σE regulates the expression of proteins that are involved in essential functions, such as envelope biogenesis and cell division (40), it is possible that σE is essential in E. coli because in its absence one or more of these factors are underproduced. If so, a suppressor of rpoE essentiality would increase the expression of such factors in the absence of σE. According to this model, YdcQ would directly or indirectly downregulate the expression of those essential factors. Although we cannot rule out this model at the moment, we do not favor it because many of these essential functions and their σE-dependent regulation are conserved in bacteria where σE is not essential (40).
The second model proposes that σE is not essential per se; instead, cell death is caused by the overreaction of E. coli to the lack of σE. In other words, we propose that E. coli reacts to the loss of σE by triggering a lethal response. One such type of suicidal death induced in response to cellular stress has already been described in E. coli. Cell death caused by the activation of the mazEF toxin-antitoxin system can be triggered by different stresses, such as amino acid starvation, DNA damage, and inhibition of transcription and translation (25). Since σE participates in the regulation of many important physiological functions in E. coli, it is possible that the misregulation of these processes, although not fatal on its own, causes the cell to trigger a response that eventually results in death. Disruption of this response would allow cells that lack σE to survive and, in this model, YdcQ would be a positive (direct or indirect) regulator of the lethal response. However, even though YdcQ would be required for this lethal response, it would not be sufficient to cause death since, in rpoE+ cells, overexpression of YdcQ alone does not cause death. Thus, both YdcQ and the loss of σE would be required for cell death.
Since the absence of YdcQ downregulates all known extracytoplasmic stress responses, either these responses are downregulated in the same manner as the cell death pathway or they are themselves involved in triggering death. Thus, it is possible that the remaining envelope stress pathways could be involved in the upregulation of the lethal overreaction response to the absence of σE. Accordingly, in the presence of an rpoE-null allele, envelope stress responses could be positively regulating the system and/or the signal that leads to death, and ydcQ-null alleles would suppress death by downregulating these responses. This model could explain the paradoxical finding that the lethality caused by the loss of an envelope stress response is suppressed by a mutation that downregulates the remaining envelope stress responses. One would have expected that upregulation of these envelope stress responses might compensate for the loss of σE, since there is overlapping regulation of some factors. However, they are clearly not playing a compensatory role in the absence of σE, since Cpx is not required for the survival of an rpoE-null strain. If this overreaction model is correct, we must note that the absence of one of these stress responses is not sufficient to suppress death induced by the loss of σE, since null mutations in cpxR or baeR are not suppressors of rpoE-null alleles (data not shown).
Our studies have revealed that the loss of YdcQ downregulates Cpx in the absence of its effector CpxR. Although we do not yet understand this novel type of regulatory mechanism, we can rule out the possibility that it is caused by a global effect on transcription, since growth rate, the levels of many proteins, and the activity of an rpoS-lacZ fusion are not affected by the loss of YdcQ. Further studies are required, however, to determine whether the loss of YdcQ only affects the regulation of envelope stress responses.
It has been suggested that outer membrane vesicle production is yet another envelope stress response (28). The fact that null mutations in ydcQ affect vesiculation in the same way that they affect the Bae, Cpx, and σE pathways is consistent with this idea. Loss of YdcQ causes a reduction in the production of outer membrane vesicles, just as it downregulates the aforementioned envelope stress responses. However, our finding that ydcQ mutations downregulate both outer membrane vesicle production and the σE pathway at first sight seems to contradict a recent report stating that mutations that either upregulate or downregulate the σE pathway cause an increase in outer membrane vesicle production (28). It is possible that the reported alterations of the σE pathway result in an envelope stress that is detected by an unknown regulatory mechanism that directly controls vesiculation. It is also likely that all of the downregulatory effects of ydcQ mutations on envelope stress pathways, including vesiculation, are independent of their respective effectors and thus the stress signal (i.e., envelope stress). Therefore, outer membrane vesicle production, just like the other envelope stress responses, could be downregulated in a ydcQ mutant independently of the stress status of the envelope and the integrity of the signaling pathway. Alternatively, as previously suggested (28), the reported alterations (upregulation and downregulation) of the σE pathway could increase vesiculation because they both cause accumulation of material in the cell envelope. However, since the loss of YdcQ downregulates multiple envelope stress responses, it is possible that as a result, it decreases the amount of material (i.e., factors positively regulated by the envelope stress responses that are downregulated by the loss of YdcQ) in the cell envelope. This, in turn, could cause a decrease in vesicle formation.
We have found a tantalizing correlation between rpoE essentiality and ydcQ. Unlike σE, YdcQ is not highly conserved among gram-negative bacteria. Using BLAST in the OMNIOME.pep (http://BLAST.wustl.edu), we could only detect YdcQ homologs in a few gram-negative bacteria, among them Shigella, Yersinia, Photorhabdus, Pseudomonas syringae, and Pseudomonas fluorescens. Although we lack a comprehensive study of the essential nature of rpoE among different bacteria, it has been shown that rpoE is essential in E. coli and Yersinia enterocolitica (12, 18), but not in Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, or Neisseria gonorrhoeae (16, 19, 27). Interestingly, we could find YdcQ homologs in both E. coli and several Yersinia species but not in S. enterica serovar Typhimurium, P. aeruginosa, and N. gonorrhoeae. In contrast, we could find homologs of ydcR, the gene downstream of ydcQ, in E. coli, several Yersinia species, serovar Typhimurium, and P. aeruginosa but not in N. gonorrhoeae. Therefore, it is possible that rpoE is not essential in bacteria that naturally lack YdcQ.
We have described YdcQ as a novel player in the regulation of envelope stress responses in E. coli. Furthermore, YdcQ is also involved in the death induced by the loss of σE. Although we do not understand the mechanism by which YdcQ regulates extracytoplasmic stress responses and cell death, we propose that σE is not essential per se. Rather, we suggest that the absence of σE might cause E. coli to trigger a cell death signal that involves YdcQ. The properties of YdcQ are reminiscent of SfiA (SulA), an important cell cycle checkpoint control protein of the SOS response (7). Perhaps there is a cell cycle checkpoint control for envelope stress in gram-negative bacteria.
Acknowledgments
We thank the members of the Silhavy laboratory and Mark Mandel for comments and helpful suggestions.
This study was supported by grant GM34821 (T.J.S.) from the National Institute of General Medical Sciences and by the Blair Senior Thesis Fund (J.E.B.).
Footnotes
Published ahead of print on 15 December 2006.
REFERENCES
- 1.Ades, S. E. 2004. Control of the alternative sigma factor σE in Escherichia coli. Curr. Opin. Microbiol. 7:157-162. [DOI] [PubMed] [Google Scholar]
- 2.Ades, S. E., L. E. Connolly, B. M. Alba, and C. A. Gross. 1999. The Escherichia coli σE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 13:2449-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ades, S. E., I. L. Grigorova, and C. A. Gross. 2003. Regulation of the alternative sigma factor σE during initiation, adaptation, and shutoff of the extracytoplasmic heat shock response in Escherichia coli. J. Bacteriol. 185:2512-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 52:613-619. [DOI] [PubMed] [Google Scholar]
- 5.Alba, B. M., J. A. Leeds, C. Onufryk, C. Z. Lu, and C. A. Gross. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the σE-dependent extracytoplasmic stress response. Genes Dev. 16:2156-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alba, B. M., H. J. Zhong, J. C. Pelayo, and C. A. Gross. 2001. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide σE activity. Mol. Microbiol. 40:1323-1333. [DOI] [PubMed] [Google Scholar]
- 7.Autret, S., A. Levine, I. B. Holland, and S. J. Seror. 1997. Cell cycle checkpoints in bacteria. Biochimie 79:549-554. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, E. A., J. L. Tupy, T. M. Gruber, S. Wang, M. M. Sharp, C. A. Gross, and S. A. Darst. 2003. Crystal structure of Escherichia coli σE with the cytoplasmic domain of its anti-sigma RseA. Mol. Cell 11:1067-1078. [DOI] [PubMed] [Google Scholar]
- 9.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 10.Danese, P. N., and T. J. Silhavy. 1998. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danese, P. N., W. B. Snyder, C. L. Cosma, L. J. Davis, and T. J. Silhavy. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9:387-398. [DOI] [PubMed] [Google Scholar]
- 12.De Las Penas, A., L. Connolly, and C. A. Gross. 1997. σE is an essential sigma factor in Escherichia coli. J. Bacteriol. 179:6862-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Las Penas, A., L. Connolly, and C. A. Gross. 1997. The σE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of σE. Mol. Microbiol. 24:373-385. [DOI] [PubMed] [Google Scholar]
- 14.De Wulf, P., A. M. McGuire, X. Liu, and E. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277:26652-26661. [DOI] [PubMed] [Google Scholar]
- 15.Douchin, V., C. Bohn, and P. Bouloc. 2006. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J. Biol. Chem. 281:12253-12259. [DOI] [PubMed] [Google Scholar]
- 16.Du, Y., J. Lenz, and C. G. Arvidson. 2005. Global gene expression and the role of sigma factors in Neisseria gonorrhoeae in interactions with epithelial cells. Infect. Immun. 73:4834-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heusipp, G., M. A. Schmidt, and V. L. Miller. 2003. Identification of rpoE and nadB as host responsive elements of Yersinia enterocolitica. FEMS Microbiol. Lett. 226:291-298. [DOI] [PubMed] [Google Scholar]
- 19.Humphreys, S., A. Stevenson, A. Bacon, A. B. Weinhardt, and M. Roberts. 1999. The alternative sigma factor, sigmaE, is critically important for the virulence of Salmonella typhimurium. Infect. Immun. 67:1560-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isaac, D. D., J. S. Pinkner, S. J. Hultgren, and T. J. Silhavy. 2005. The extracytoplasmic adaptor protein CpxP is degraded with substrate by DegP. Proc. Natl. Acad. Sci. USA 102:17775-17779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehara, K., Y. Akiyama, and K. Ito. 2001. Characterization of the yaeL gene product and its S2P-protease motifs in Escherichia coli. Gene 281:71-79. [DOI] [PubMed] [Google Scholar]
- 22.Kanehara, K., K. Ito, and Y. Akiyama. 2002. YaeL (EcfE) activates the σE pathway of stress response through a site-2 cleavage of anti-σE, RseA. Genes Dev. 16:2147-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehara, K., K. Ito, and Y. Akiyama. 2003. YaeL proteolysis of RseA is controlled by the PDZ domain of YaeL and a Gln-rich region of RseA. EMBO J. 22:6389-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 25.Kolodkin-Gal, I., and H. Engelberg-Kulka. 2006. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J. Bacteriol. 188:3420-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Martin, D. W., B. W. Holloway, and V. Deretic. 1993. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J. Bacteriol. 175:1153-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBroom, A. J., A. P. Johnson, S. Vemulapalli, and M. J. Kuehn. 2006. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mecsas, J., P. E. Rouviere, J. W. Erickson, T. J. Donohue, and C. A. Gross. 1993. The activity of σE, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7:2618-2628. [DOI] [PubMed] [Google Scholar]
- 30.Missiakas, D., M. P. Mayer, M. Lemaire, C. Georgopoulos, and S. Raina. 1997. Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB, and RseC proteins. Mol. Microbiol. 24:355-371. [DOI] [PubMed] [Google Scholar]
- 31.Nishino, K., T. Honda, and A. Yamaguchi. 2005. Genome-wide analyses of Escherichia coli gene expression responsive to the BaeSR two-component regulatory system. J. Bacteriol. 187:1763-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson, C. N., M. J. Mandel, and T. J. Silhavy. 2005. Escherichia coli starvation diets: essential nutrients weigh in distinctly. J. Bacteriol. 187:7549-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pogliano, J., A. S. Lynch, D. Belin, E. C. Lin, and J. Beckwith. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11:1169-1182. [DOI] [PubMed] [Google Scholar]
- 34.Raffa, R. G., and T. L. Raivio. 2002. A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45:1599-1611. [DOI] [PubMed] [Google Scholar]
- 35.Raivio, T. L. 2005. Envelope stress responses and gram-negative bacterial pathogenesis. Mol. Microbiol. 56:1119-1128. [DOI] [PubMed] [Google Scholar]
- 36.Raivio, T. L., M. W. Laird, J. C. Joly, and T. J. Silhavy. 2000. Tethering of CpxP to the inner membrane prevents spheroplast induction of the Cpx envelope stress response. Mol. Microbiol. 37:1186-1197. [DOI] [PubMed] [Google Scholar]
- 37.Raivio, T. L., D. L. Popkin, and T. J. Silhavy. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J. Bacteriol. 181:5263-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raivio, T. L., and T. J. Silhavy. 1999. The σE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2:159-165. [DOI] [PubMed] [Google Scholar]
- 39.Raivio, T. L., and T. J. Silhavy. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 179:7724-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhodius, V. A., W. C. Suh, G. Nonaka, J. West, and C. A. Gross. 2006. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 4:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rouviere, P. E., A. De Las Penas, J. Mecsas, C. Z. Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat-shock sigma factor, σE, in Escherichia coli. EMBO J. 14:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz, N., and T. J. Silhavy. 2005. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr. Opin. Microbiol. 8:122-126. [DOI] [PubMed] [Google Scholar]
- 43.Silhavy, T. J., Berman, M. L., and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 44.Slauch, J. M., and T. J. Silhavy. 1991. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 173:4039-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]