Abstract
The expression of the inner membrane protein NlpA is repressed by the enterotoxigenic Escherichia coli (ETEC) virulence regulator Rns, a member of the AraC/XylS family. The Rns homologs CfaD from ETEC and AggR from enteroaggregative E. coli also repress expression of nlpA. In vitro DNase I and potassium permanganate footprinting revealed that Rns binds to a site overlapping the start codon of nlpA, preventing RNA polymerase from forming an open complex at nlpAp. A second Rns binding site between positions −152 and −195 relative to the nlpA transcription start site is not required for repression. NlpA is not essential for growth of E. coli under laboratory conditions, but it does contribute to the biogenesis of outer membrane vesicles. As outer membrane vesicles have been shown to contain ETEC heat-labile toxin, the repression of nlpA may be an indirect mechanism through which the virulence regulators Rns and CfaD limit the release of toxin.
Enterotoxigenic E. coli (ETEC) is a major cause of profuse watery diarrhea and in underdeveloped nations is a significant cause of infant and childhood mortality (23). There are two types of ETEC exotoxins that produce diarrheal disease, a heat-stable toxin (ST) and a heat-labile toxin (LT). Whereas some ETEC strains express both toxins, other strains produce only one toxin, as either toxin is sufficient to cause diarrhea. The STs are small polypeptides consisting of 18 amino acids (STIa), 19 amino acids (STIb), or 48 amino acids (STII). In the host cell, STI activates guanylate cyclase, which leads to a dramatic increase in cyclic GMP levels in the cytoplasm. The mechanism of action of STII has not been determined yet. Internalization of LT ultimately leads to activation of adenylate cyclase and overproduction of cyclic AMP. LT is a large (>84-kDa) multimeric complex consisting of five B subunits and one A subunit. Although ST and LT have different structures and cellular targets, their effects are similar: the transport of ions across the intestinal epithelium is altered, and water is lost to the intestinal lumen.
The ST and LT preproteins are transported across the bacterial inner membrane by the signal peptide-dependent general export pathway. In the periplasm, the preproteins are processed and the heteromeric LT is assembled. Both STI and STII cross the outer membrane through a tunnel created by a trimer of TolC (5, 33). Despite TolC's ability to export a variety of molecules across the outer membrane, it does not transport LT (10).
Early studies of LT secretion seemed to produce contradictory findings. Some studies showed that as much as 55% of a cell's total LT was bound to the outer membrane, presumably after transport (29). Other studies showed that nearly all of the LT produced by a bacterium was contained in its periplasm and that the LT was not membrane bound (10). It has since been shown that a type II secretion system is required for the transport of LT across the outer membrane (28). The lack of this system in E. coli K-12 explains why studies using K-12 strains showed that LT accumulates in the periplasm (10), while studies using ETEC strains showed that LT is transported and bound to the outer membrane (29). An interaction between the B subunit of LT and lipopolysaccharide anchors the toxin to the outer membrane (11).
Eventually, membrane-bound LT becomes a surface component of outer membrane vesicles (OMVs) as they are shed from the bacterium (12). The biogenesis of OMVs is not fully understood, but a recent study has shown that disruption of nlpA decreases the production of OMVs (16). NlpA is a periplasmic protein that is attached to the inner membrane (34). In this study we found that transcription of nlpA is repressed by the ETEC virulence regulators Rns and CfaD.
MATERIALS AND METHODS
Strains and plasmids.
Plasmid pHKLac1 is a promoterless lacZ reporter and integration plasmid with a pir-dependent origin of replication. It was constructed by cloning a 5.5-kb BamHI-MfeI fragment from pRS550 carrying lacZYA (27) into pAH144 (accession no. AY048731) (8). Transcriptional terminators flank lacZYA in the resulting plasmid. It also carries attPHK022 for IntHK022-mediated integration at attBHK022 in Δpir hosts and aadA for selection with spectinomycin and streptomycin.
The nlpA-yicS intergenic region was amplified from ETEC strain H10407 (CfaD+ CFA/I+) (4) with primers nlpA-F1 (GCAGGATCCCACGCTTCATCTGATAATGACG) and nlpA-r1 (GCAGAATTCCGGCCAGCAATAATG) or with primers nlpA-f3 (GCGGGATCCATAAGTGTAGTTTGCTTT) and nlpA-R1 (GCAGAATTCCGGCCAGCAATAATG) (underlining in primer sequences indicates primer-template mismatches that add sites for restriction endonucleases). The nlpA-yicS intergenic region was amplified from enteroaggregative E. coli (EAEC) strain 17-2ΔaggR (22) with primers nlpA-F2 (GCGGGATCCTGATAATGACGCCTGTGGCGTG) and nlpA-r2 (GTCGCAACCTGCCAGCAGAATTC). The PCR products were digested with BamHI and EcoRI and then ligated into the same sites of pHKLac1 to construct pNlpALac1 [nlpApETEC(−391 to +58)::lacZ], pNlpALac2 [nlpApEAEC(−366 to +58)::lacZ], and pNlpALac3 [nlpApETEC(−186 to +58)::lacZ] (numbering is relative to the start codon of nlpA). Each reporter plasmid was integrated into the chromosome of MC4100 [F− araD139 Δ(argF-lac)U169 rpsL150 relA1 flhD5301 deoC1 ptsF25 rbsR] (3) as previously described (8), resulting in strains GPM1080 (attBHK022::pNlpALac1), GPM1092 (attBHK022::pNlpALac2), and GPM1095 (attBHK022::pNlpALac3). Colony PCR was used to verify that each strain contained only a single plasmid integrant, as previously described (8).
Plasmid pGPMRns expresses rns from lacp. It was constructed by ligating a 0.9-kb RsaI fragment from pEU2030 (6) into the SmaI site of pNEB193 (New England Biolabs). To construct pMBPRns1, which expresses an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible maltose binding protein (MBP)-Rns fusion protein from tacp, rns was amplified from pEU2005 (1) with primers RnsNcoI (AGGTATACCATGGACTTTAAATACACTGA) and rnsBam-rv2 (GCGGATCCTGTTTTTATCCACCTTTAAA). The 0.8-kb PCR product was then digested with BamHI and ligated into pMalC2 (New England Biolabs) previously digested with BamHI and XmnI. Other plasmids used in this study include pNTP503, which expresses CfaD (26, 31), and pJPN52, which expresses AggR (22).
Purification of RNA.
Strain GPM1080 was cultured aerobically in 10 ml of Luria-Bertani (LB) medium at 37°C. After the absorbance at 550 nm reached 1.0, 2 ml of PE buffer (5% [vol/vol] phenol, 95% [vol/vol] ethanol) was added to the culture, and the cells were pelleted. The cell pellet was suspended in 10 ml of RNA wash buffer (0.75% [vol/vol] NaCl, 0.8% [vol/vol] phenol, 15.8% [vol/vol] ethanol) and then centrifuged. The resulting pellet was resuspended in 500 μl of 0.9% (vol/vol) NaCl and shaken at room temperature for 30 min after addition of 500 μl water-saturated, nonbuffered phenol. Subsequently, 50 μl of a chloroform-isoamyl alcohol (24:1) solution was added, and the preparation was incubated for an additional 15 min. The solution was then chilled on ice for 5 min and centrifuged in a microcentrifuge at the maximum setting for 5 min. The supernatant was recovered, and the RNA was ethanol precipitated and then resuspended in 22 μl of RNase-free water.
Primer extension.
Two picomoles of 32P-end-labeled oligonucleotide nlpA-r1 (GCAGAATTCCGGCCAGCAATAATG) was combined with 82 μg of total RNA and deoxynucleoside triphosphates at a concentration of 0.8 mM. The solution was heated to 65°C for 5 min and then chilled on ice for 2 min. The annealed primer was then extended with SuperScript III reverse transcriptase used according to the supplier's protocol (Invitrogen). Heat-denatured aliquots were separated on DNA sequencing gels alongside dideoxy chain-terminated sequencing ladders (24).
Purification of MBP-Rns.
Strain KS1000/pRare2/pMBPRns1 was used for expression of MBP-Rns. The gene encoding Prc (Tsp) protease is disrupted in KS1000 [F′ lacIq lac+ pro+/araΔ(lac-pro) Δprc::kan eda51::Tn10 gyrA rpoB thi-1 argI(Am)] (New England Biolabs). Plasmid pRARE2 (Novagen) provides seven rare tRNAs to supplement the rare codon usage of rns. We found that this strain produced higher yields of MBP-Rns fusion proteins than the previously described strain JM83/pEU750 produced (19). KS1000/pRare2/pMBPRns1 was grown aerobically at 37°C in LB broth containing 0.2% (wt/vol) glucose, 30 μg/ml chloramphenicol, and 100 μg/ml ampicillin. After cells reached the mid-log phase, the culture was transferred to a 30°C shaking water bath, and expression of MBP-Rns was induced by addition of IPTG to a final concentration of 300 μM. After several hours of induction, cells were harvested at 4°C and concentrated >100-fold in ice-cold lysis buffer (10 mM Tris-Cl [pH 7.6 at room temperature], 200 mM NaCl, 1 mM EDTA, 0.5 mM CaCl2, 10 mM β-mercaptoethanol, 100 μg/ml DNase I). Cells were lysed by two passages through a French press. Insoluble material was removed by centrifugation of the lysate at 17,000 × g for 20 min at 4°C. The supernatant was then loaded onto an amylose column and extensively washed with buffer A (10 mM Tris-Cl [pH 7.6 at room temperature], 200 mM NaCl, 1 mM EDTA, 15% [vol/vol] glycerol, 10 mM β-mercaptoethanol). The MBP-Rns fusion protein was eluted from the amylose column with buffer B (buffer A with 10 mM maltose). If necessary, the fusion protein was purified further on a heparin column as previously described (19).
DNase I footprinting.
MBP-Rns was equilibrated with 32P-end-labeled nlpA promoter DNA for 30 min at 37°C in footprinting buffer [10 mM Tris-Cl [pH 7.6 at room temperature], 50 mM KCl, 1 mM dithiothreitol, 0.4 mM MgCl2, 0.2 mM CaCl2, 2 ng/μl poly(dI-dC), 10 μg/ml bovine serum albumin]. After equilibration, DNase I was added to a final concentration of 100 ng/μl for 1 min at 37°C. The cleavage reaction was terminated by addition of 10 volumes of DNase I stop buffer (570 mM ammonium acetate, 50 μg/ml tRNA, 80% [vol/vol] ethanol), briefly vortexed, and then precipitated on dry ice for at least 10 min. The DNA in each sample was pelleted, washed with 70% (vol/vol) ethanol, dried, and then resuspended in 4 μl of loading buffer (80% [vol/vol] formamide, 50 mM Tris-borate [pH 8.3], 1 mM EDTA, 0.1% [wt/vol] xylene cyanol, bromophenol blue). The samples were then heat denatured and separated on sequencing gels. GA and TC sequence ladders were generated by the Maxam-Gilbert method (15).
Potassium permanganate footprinting.
MBP-Rns and 32P-end-labeled nlpA promoter DNA were equilibrated for 20 min at 37°C in permanganate buffer [10 mM Tris-Cl (pH 7.6 at room temperature), 50 mM KCl, 2 ng/μl poly(dI-dC), 0.2 mM MgCl2, 10 μg/ml bovine serum albumin]. RNA polymerase (RNAP), preequilibrated at 37°C, was then added, and the solutions were incubated for an additional 20 min. KMnO4 was then added to a final concentration of 2 mM for 2 min at 37°C. The reaction was quenched by addition of 0.5 volume of permanganate stop buffer (750 mM sodium acetate, 500 mM β-mercaptoethanol, 50 μg/ml tRNA). The DNA was precipitated by addition of 5 volumes of 95% (vol/vol) ethanol, pelleted, washed with 70% (vol/vol) ethanol, and dried. Piperidine cleavage and subsequent steps were performed as previously described (20).
Enzymatic assays.
Reporter strains GPM1080, GPM1092, and GPM1095 transformed with pGPMRns (Rns+ bla), pNTP503 (CfaD+ bla), pJPN52 (AggR+ tet), and vector control plasmids were grown aerobically at 37°C to the stationary phase in LB medium with 100 μg/ml ampicillin or 30 μg/ml tetracycline as appropriate. Cells were lysed, and the β-galactosidase activity was assayed as previously described (17).
RESULTS
Characterization of nlpAp.
The virulence of some ETEC strains is dependent upon Rns, which positively regulates its own expression and the expression of CS1, CS2, and CS3 pili. Rns recognizes the same binding sites as CfaD (18), a regulator typically found in strains of ETEC that express CFA/I pili (2). We tried to identify additional genes in the Rns/CfaD regulon by searching the genomes of E24377A (Rns+ CS1+ CS3+ ST+ LT+; accession no. AAJZ00000000), H10407 (CfaD+ CFA/I+ ST+ LT+), and MG1655 (K-12; accession no. U00096) for sites similar to known Rns binding sites (19, 20). In silico sequence analysis predicted a Rns binding site (GATAAAAAAAT) partially in the 221-bp intergenic region between the divergently encoded yicS and nlpA genes. Potentially, yicS encodes an 11-kDa secreted protein of unknown function. A 28-kDa inner membrane lipoprotein is encoded by nlpA (32). The last two nucleotides of the putative Rns binding site are the first two nucleotides of nlpA's ATG start codon. The nucleotide sequence of the nlpA-yicS locus is completely conserved in strains H10407, E24377A, and MG1655.
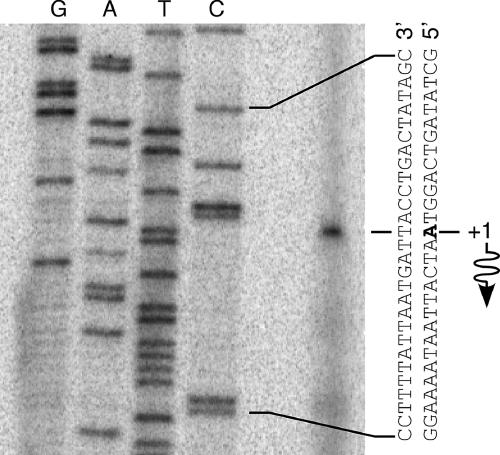
In order to evaluate the significance of the putative Rns binding site overlapping the start codon of nlpA, it was necessary to determine the location of nlpAp. We determined the transcription start site (TSS) of nlpA in the absence of Rns by primer extension and found that the nlpA message initiates 24 bp upstream of the nlpA start codon (Fig. 1). We did not attempt to map the TSS of nlpA in the presence of Rns because, as shown below, Rns represses nlpAp at the transcription initiation step.
FIG. 1.
Identification of the nlpA transcription start site. Primer extension was used to map the Rns-independent transcription start site of nlpA with reverse transcriptase and RNA harvested from strain GPM1080 growing logarithmically in LB medium. A dideoxy chain-terminated sequencing ladder was generated with the same primer. The first nucleotide of the nlpA mRNA is indicated by bold type, and the wavy arrow indicates the direction of transcription.
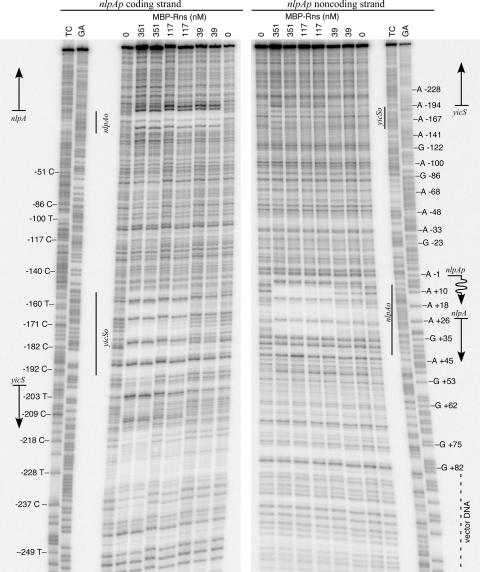
We next confirmed that the predicted Rns binding site overlapping the start codon of nlpA is an actual binding site by in vitro DNase I footprinting with an MBP-Rns fusion protein (Fig. 2). For in vitro work it is necessary to use MBP-Rns instead of Rns because it is severalfold more soluble than Rns. Fortunately, MBP does not interfere with the activity of Rns in vivo (19). We found that MBP-Rns protects a region from +5 to +40 (numbering relative to the TSS of nlpAp), thus encompassing the predicted Rns binding site from +16 to +26. The DNase I footprint of MBP-Rns is larger than the core sequence of highly conserved nucleotides comprising the predicted binding site because steric occlusion prevents DNase I cleavage within several nucleotides of bound MBP-Rns.
FIG. 2.
Identification of two Rns binding sites in the nlpA-yicS intergenic region. DNase I footprinting of MBP-Rns bound to the coding and noncoding strands of nlpA revealed two Rns binding sites in the nlpA-yicS intergenic region. The Rns binding sites are designated nlpAo and yicSo based on their proximity to nlpA and yicS. Straight arrows indicate the relative positions of nlpA and yicS. The numbering is relative to the transcription start site of nlpA, which is indicated by a wavy arrow. Lanes GA and TC contained Maxam-Gilbert sequence ladders.
Although sequence analysis predicted only one Rns binding site in the nlpA-yicS intergenic region, an additional MBP-Rns binding site was observed from −152 to −195 relative to the TSS of nlpAp (Fig. 2). This second site was not identified by our in silico analysis because it is the most divergent of all known Rns binding sites. For simplicity, the two Rns binding sites are referred to below as nlpAo and yicSo based on their proximity to the open reading frames. Qualitatively, it appears that the affinity of Rns for nlpAo is higher than the affinity of Rns for yicSo because nlpAo is saturated by lower concentrations of MBP-Rns than yicSo. As shown below, only nlpAo is required for the repression of nlpAp.
Repression of nlpA by Rns, CfaD, and AggR.
To assess whether Rns regulates the expression of nlpA, we constructed nlpAp-lacZ fusions that were integrated into the chromosome of K-12 strain MC4100. In the absence of Rns, CfaD, and AggR, β-galactosidase was expressed at moderate levels from nlpApETEC(−367 to +82)::lacZ (numbering relative to the TSS) (Table 1). In contrast, we observed nearly complete repression of nlpAp in the presence of Rns (Table 1). Although a slight decrease in Rns-independent expression from nlpApETEC(−162 to +82)::lacZ was observed compared to the expression from nlpApETEC(−367 to +82)::lacZ, repression of nlpAp was not dependent upon yicSo since yicSo was deleted from nlpApETEC(−162 to +82)::lacZ (Table 1). These results demonstrate that Rns functions as a repressor of nlpA transcription and that the Rns binding site nlpAo is sufficient for repression.
TABLE 1.
Repression of nlpAp by Rns, CfaD, and AggR
| nlpAp | Position relative to:
|
ß-Galactosidase activity (Miller units) in the presence ofa:
|
||||
|---|---|---|---|---|---|---|
| TSS | Open reading frame | No regulator | Rns+ | CfaD+ | AggR+ | |
| nlpApETEC::lacZ | −367 to +82 | −391 to +58 | 479 ± 10 | 9 ± 6 | 20 ± 10 | 15 ± 2 |
| nlpApETEC::lacZ | −162 to +82 | −186 to +58 | 348 ± 15 | 17 ± 4 | NDb | 25 ± 7 |
| nlpApEAEC::lacZ | ND | −366 to +58 | 223 ± 6 | 18 ± 10 | ND | 18 ± 1 |
The values are means and standard deviations (n = 3).
ND, not determined.
In transcriptional activation assays, Rns was shown to be functionally interchangeable with CfaD, which activates the expression of CFA/I pili in some strains of ETEC, and the enteroaggregative virulence regulator AggR (18). As shown in Table 1, both CfaD and AggR also repressed transcription from nlpApETEC. Although the nlpA-yicS loci are identical in K-12 and ETEC strains, this locus is not as well conserved in the prototypical EAEC strain 042. To determine if nlpA is regulated in EAEC, an additional reporter strain was constructed using the nlpA promoter cloned from EAEC strain 17-2ΔaggR (22). Strain 17-2ΔaggR is a derivative of EAEC strain 17-2 from which AggR was originally cloned (21, 22). As shown in Table 1, nlpApEAEC was repressed by AggR, Rns, and CfaD as tightly as nlpApETEC was repressed. Thus, nlpA is repressed by Rns or CfaD in ETEC and by AggR in EAEC.
Mechanism of repression.
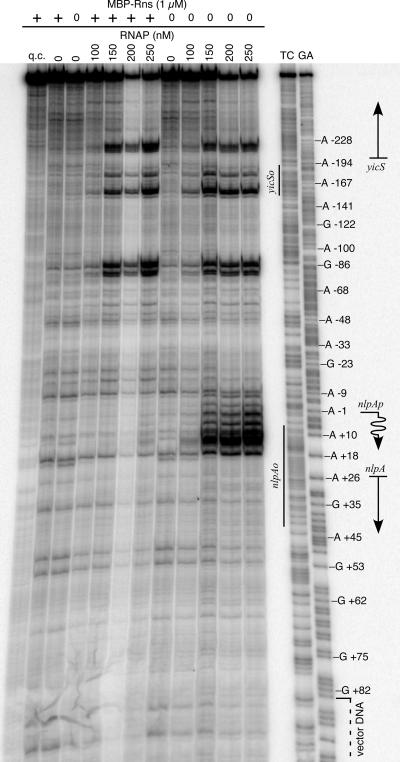
Potassium permanganate, a reagent that is reactive with unpaired thymine nucleotides in transcription bubbles (9, 25), was used to determine if Rns interferes with the formation of RNAP-open (RPo) complexes or a later step of transcription, such as transcript elongation. In the absence of Rns, potassium permanganate reactivity was detected from −8 to +17 (numbering relative to the TSS of nlpA) (Fig. 3). This region of reactivity corresponds to single-stranded DNA within an RPo complex at nlpAp as it is dependent upon RNAP. Although the region of reactivity is consistent with the transcription start site of nlpA (Fig. 1), we noted that it extends further downstream than the usual 4 nucleotides reported for other promoters (25). One possible explanation for the extended permanganate footprint is the AT richness (85%) from position 5 to position 17, which might contribute to DNA melting beyond the expected region. Regardless, potassium permanganate reactivity at nlpAp was not observed in the presence of MBP-Rns (Fig. 3). These results demonstrate that Rns represses nlpAp at the initiation step of transcription because it prevents the formation of an RPo complex. Given the proximity of nlpAo to nlpAp, it seems likely that Rns bound at nlpAo sterically occludes RNAP from nlpAp. These results also demonstrate that Rns is sufficient for repression of nlpAp and that other factors are not required.
FIG. 3.
Rns represses nlpAp by preventing the formation of an open complex at nlpAp. Potassium permanganate footprinting of RNAP bound to the noncoding strand of nlpA in the presence and absence of MBP-Rns is shown. A straight arrow indicates the position of nlpA. The numbering is relative to the transcription start site of nlpA, which is indicated by a wavy arrow. Lanes GA and TC contained Maxam-Gilbert sequence ladders. Lane q.c. was not treated with potassium permanganate and was a control for the quality of the DNA template.
In addition to the RPo complex observed at nlpAp, other RNAP-dependent regions of potassium permanganate reactivity were observed in the nlpA-yicS intergenic region (Fig. 3). One or more of the RPo complexes may be at the promoter or promoters for yicS. Alternatively, they may be the result of RNAP binding to pseudopromoter sequences. In either case, the additional RPo complexes were not investigated further since they were not affected by MBP-Rns and do not correspond to the known TSS of nlpA (Fig. 1).
DISCUSSION
Our results demonstrate that the virulence regulators Rns and CfaD of ETEC and AggR of EAEC repress the transcription of nlpA. Two Rns binding sites were identified by DNase I footprinting; these sites were nlpAo from +5 to +40 and yicSo from −152 to −195 (numbering relative to the TSS of nlpA). The nlpAo binding site was sufficient for repression of nlpA in vivo, and Rns binding to this site prevented RNAP from forming an open complex at nlpAp. We have not yet determined the function, if any, of yicSo, but additional studies are planned. It is possible that Rns binding to yicSo affects the expression of yicS, a gene upstream of and divergently encoded from nlpA. NlpA is undoubtedly repressed in all strains of ETEC that carry Rns or CfaD or one of the functional homologs (CsvR, CsfR, or CswR) with which they are interchangeable (18, 30). Although Rns is also interchangeable with VirF from Shigella flexneri (18), we did not investigate the regulation of nlpA by VirF because nlpA has been disrupted in each of the three S. flexneri strains for which a genomic sequence is available (strain 2457T [accession no. AE014073], strain 301 [accession no. AE005674], and strain 8401 [accession no. CP000266]).
Until now, Rns, CfaD, and AggR were only known to function as activators, but our results demonstrate that these regulators are bifunctional. However Rns, CfaD, and AggR do not switch functions in response to effector ligands since they repress nlpA under the same conditions that they activate expression of pilin genes. This differentiates them from other AraC/XylS family members that have different activities in response to effector molecules (7). The factor that is most likely to determine the effect of Rns, CfaD, and AggR at a given promoter is the precise position of the regulator's binding site relative to RNAP. Binding sites that occlude RNAP binding to promoter elements function as repressor sites, while other binding sites may have a stimulatory effect. The location of a Rns binding site downstream of the TSS is not sufficient to predict whether it functions as a repressor or activator because Rns has been shown to bind downstream of its own promoter, where it is an activator (20). However, Rns binds further downstream from its own promoter than from nlpAp.
In E. coli NlpA is a periplasmic protein bound to the inner membrane. Original investigations of a nlpA::kan mutant revealed no significant differences in growth, cell morphology, or chemical sensitivity compared to a wild-type strain (32). However, in a recent screen for transposon insertions that affect the production of OMVs, it was observed that a nlpA::Tn5(kan) mutant produces less OMVs than an isogenic wild-type strain of E. coli (16). Although the mechanism by which NlpA contributes to the formation of OMVs is not known, it has been suggested that NlpA may have a direct role in OMV biogenesis (16).
The OMVs produced by ETEC contain nearly all of the pathogen's secreted LT, and OMVs deliver the toxin into eukaryotic cells (11-13). Our studies raise the possibility that the ETEC virulence regulators Rns and CfaD indirectly control the release of LT by repressing the transcription of nlpA, thus limiting the production of OMVs. Since these regulators are not found in all ETEC strains, this may also account, at least in part, for the reported variability of OMV production between strains (14).
Acknowledgments
We thank J. R. Scott, J. P. Nataro, and B. L. Wanner for kindly providing strains and plasmids. Preliminary sequence data for EAEC strain 042 was produced by the Pathogen Sequencing Unit at the Sanger Institute and was obtained from the website http://www.sanger.ac.uk/Projects/Escherichia_Shigella/. Preliminary sequence data for ETEC strain H10407 was produced by the Sanger Institute in collaboration with I. Henderson and M. Pallen and was obtained from http://www.sanger.ac.uk/Projects/E_coli_H10407/.
This work was supported by Public Health Service award AI 057648 from the National Institutes of Health and by the University of Miami Miller School of Medicine.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Caron, J., L. M. Coffield, and J. R. Scott. 1989. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. USA 86:963-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caron, J., and J. R. Scott. 1990. A rns-like regulatory gene for colonization factor antigen I (CFA/I) that controls expression of CFA/I pilin. Infect. Immun. 58:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 4.Evans, D. G., R. P. Silver, D. J. Evans, Jr., D. G. Chase, and S. L. Gorbach. 1975. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect. Immun. 12:656-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foreman, D. T., Y. Martinez, G. Coombs, A. Torres, and Y. M. Kupersztoch. 1995. TolC and DsbA are needed for the secretion of STB, a heat-stable enterotoxin of Escherichia coli. Mol. Microbiol. 18:237-245. [DOI] [PubMed] [Google Scholar]
- 6.Froehlich, B., L. Husmann, J. Caron, and J. R. Scott. 1994. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J. Bacteriol. 176:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayatsu, H., and T. Ukita. 1967. The selective degradation of pyrimidines in nucleic acids by permanganate oxidation. Biochem. Biophys. Res. Commun. 29:556-561. [DOI] [PubMed] [Google Scholar]
- 10.Hirst, T. R., L. L. Randall, and S. J. Hardy. 1984. Cellular location of heat-labile enterotoxin in Escherichia coli. J. Bacteriol. 157:637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horstman, A. L., and M. J. Kuehn. 2002. Bacterial surface association of heat-labile enterotoxin through lipopolysaccharide after secretion via the general secretory pathway. J. Biol. Chem. 277:32538-32545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horstman, A. L., and M. J. Kuehn. 2000. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275:12489-12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kesty, N. C., K. M. Mason, M. Reedy, S. E. Miller, and M. J. Kuehn. 2004. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 23:4538-4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasaro, M. A., J. F. Rodrigues, C. Mathias-Santos, B. E. Guth, A. Regua-Mangia, A. J. Piantino Ferreira, M. Takagi, J. Cabrera-Crespo, M. E. Sbrogio-Almeida, and L. C. de Souza Ferreira. 2006. Production and release of heat-labile toxin by wild-type human-derived enterotoxigenic Escherichia coli. FEMS Immunol. Med. Microbiol. 48:123-131. [DOI] [PubMed] [Google Scholar]
- 15.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McBroom, A. J., A. P. Johnson, S. Vemulapalli, and M. J. Kuehn. 2006. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 18.Munson, G. P., L. G. Holcomb, and J. R. Scott. 2001. Novel group of virulence activators within the AraC family that are not restricted to upstream binding sites. Infect. Immun. 69:186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munson, G. P., and J. R. Scott. 1999. Binding site recognition by Rns, a virulence regulator in the AraC family. J. Bacteriol. 181:2110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munson, G. P., and J. R. Scott. 2000. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol. Microbiol. 36:1391-1402. [DOI] [PubMed] [Google Scholar]
- 21.Nataro, J. P., D. Yikang, J. A. Giron, S. J. Savarino, M. H. Kothary, and R. Hall. 1993. Aggregative adherence fimbria I expression in enteroaggregative Escherichia coli requires two unlinked plasmid regions. Infect. Immun. 61:1126-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nataro, J. P., D. Yikang, D. Yingkang, and K. Walker. 1994. AggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli. J. Bacteriol. 176:4691-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qadri, F., A. M. Svennerholm, A. S. Faruque, and R. B. Sack. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18:465-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasse-Dwight, S., and J. D. Gralla. 1989. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J. Biol. Chem. 264:8074-8081. [PubMed] [Google Scholar]
- 26.Savelkoul, P. H., G. A. Willshaw, M. M. McConnell, H. R. Smith, A. M. Hamers, B. A. van der Zeijst, and W. Gaastra. 1990. Expression of CFA/I fimbriae is positively regulated. Microb. Pathog. 8:91-99. [DOI] [PubMed] [Google Scholar]
- 27.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 28.Tauschek, M., R. J. Gorrell, R. A. Strugnell, and R. M. Robins-Browne. 2002. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7066-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wensink, J., H. Gankema, W. H. Jansen, P. A. Guinee, and B. Witholt. 1978. Isolation of the membranes of an enterotoxigenic strain of Escherichia coli and distribution of enterotoxin activity in different subcellular fractions. Biochim. Biophys. Acta 514:128-136. [DOI] [PubMed] [Google Scholar]
- 30.Willshaw, G. A., H. R. Smith, M. M. McConnell, and B. Rowe. 1991. Cloning of regulator genes controlling fimbrial production by enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 66:125-129. [DOI] [PubMed] [Google Scholar]
- 31.Willshaw, G. A., H. R. Smith, and B. Rowe. 1983. Cloning of regions encoding colonisation factor antigen 1 and heat-stable enterotoxin in Escherichia coli. FEMS Microbiol. Lett. 16:101-106. [Google Scholar]
- 32.Yamaguchi, K., and M. Inouye. 1988. Lipoprotein 28, an inner membrane protein of Escherichia coli encoded by nlpA, is not essential for growth. J. Bacteriol. 170:3747-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamanaka, H., T. Nomura, Y. Fujii, and K. Okamoto. 1998. Need for TolC, an Escherichia coli outer membrane protein, in the secretion of heat-stable enterotoxin I across the outer membrane. Microb. Pathog. 25:111-120. [DOI] [PubMed] [Google Scholar]
- 34.Yu, F., S. Inouye, and M. Inouye. 1986. Lipoprotein-28, a cytoplasmic membrane lipoprotein from Escherichia coli. Cloning, DNA sequence, and expression of its gene. J. Biol. Chem. 261:2284-2288. [PubMed] [Google Scholar]