Abstract
Salmonella enterica degrades 1,2-propanediol (1,2-PD) in a coenzyme B12-dependent manner. Previous enzymatic assays of crude cell extracts indicated that a phosphotransacylase (PTAC) was needed for this process, but the enzyme involved was not identified. Here, we show that the pduL gene encodes an evolutionarily distinct PTAC used for 1,2-PD degradation. Growth tests showed that pduL mutants were unable to ferment 1,2-PD and were also impaired for aerobic growth on this compound. Enzyme assays showed that cell extracts from a pduL mutant lacked measurable PTAC activity in a background that also carried a pta mutation (the pta gene was previously shown to encode a PTAC enzyme). Ectopic expression of pduL corrected the growth defects of a pta mutant. PduL fused to eight C-terminal histidine residues (PduL-His8) was purified, and its kinetic constants were determined: the Vmax was 51.7 ± 7.6 μmol min−1 mg−1, and the Km values for propionyl-PO42− and acetyl-PO42− were 0.61 and 0.97 mM, respectively. Sequence analyses showed that PduL is unrelated in amino acid sequence to known PTAC enzymes and that PduL homologues are distributed among at least 49 bacterial species but are absent from the Archaea and Eukarya.
A number of bacterial genera including Salmonella, Klebsiella, Shigella, Yersinia, Listeria, Lactobacillus, and Lactococcus include members that grow on 1,2-propanediol (1,2-PD) in a coenzyme B12-dependent fashion. 1,2-PD is a major product of the fermentation of rhamnose and fucose, which are common sugars in plant cell walls, bacterial exopolysaccharides, and the glycoconjugates of intestinal epithelial cells. Accordingly, the ability to degrade 1,2-PD is thought to provide a selective advantage in anaerobic environments such as the large intestines of higher animals, sediments, and the depths of soils. Recent studies with Salmonella enterica have shown that 1,2-PD degradation is one of the most complex metabolic processes known (6, 14, 15, 18, 19, 31). The degradation of this small molecule requires catabolic enzymes, a system for recycling inactive cobalamins to coenzyme B12, and an unusual polyhedral body-microcompartment that is composed of metabolic enzymes encased within a multiprotein shell.
A pathway for 1,2-PD degradation by S. enterica has been proposed based on enzymatic studies of crude cell extracts and genetic analyses (25, 37). In the first step of the pathway, 1,2-PD is converted to propionaldehyde via coenzyme B12-dependent diol dehydratase (1). Propionaldehyde is then converted to 1-propanol and propionic acid by a reaction series thought to involve 1-propanol dehydrogenase, coenzyme A (CoA)-dependent propionaldehyde dehydrogenase, phosphotransacylase (PTAC), and propionate kinase (25, 37). This pathway generates one ATP, an electron sink, and a three-carbon intermediate (propionyl-CoA), which feeds into central metabolism via the methylcitrate pathway (16).
The genes specifically required for 1,2-PD utilization (pdu) by S. enterica form a single contiguous cluster, the pdu locus. DNA sequence analyses indicate that this locus includes 23 genes (6, 7, 10). Based on experimental and/or bioinformatic analyses, six pdu genes are thought to encode enzymes needed for the 1,2-PD degradative pathway (6), two are involved in transport and regulation (5, 10), two are probably involved in diol dehydratase reactivation (6), two are used for the conversion of vitamin B12 to coenzyme B12 (18, 31), four are of unknown function, and seven share similarity to genes involved in the formation of carboxysomes, a polyhedral body found in certain cyanobacteria and chemoautotrophs (6, 10). Although the methylcitrate pathway is required for the growth of S. enterica on 1,2-PD as a sole carbon source, the genes for this pathway map outside the pdu locus (16).
It was recently shown that S. enterica forms polyhedral bodies (PhBs) during growth on 1,2-PD (6, 15, 36). These bodies are extremely large macromolecular complexes that are 100 to 150 nm in cross section and consist of metabolic enzymes encased within a protein shell. Purification of these structures showed that they are composed of at least 14 different polypeptides (PduABB′CDEGHJKOPTU) (14). This includes four enzymes and as many as seven shell proteins, all of which are encoded by the pdu locus. The first two steps of 1,2-PD degradation are thought to occur in the lumen of the PhBs, and the remaining steps are thought to occur in the cytoplasm of the cell (Fig. 1) (14). The function of PhBs is uncertain. Previous studies suggested that they act as microcompartments that sequester propionaldehyde (an intermediate of 1,2-PD degradation) in order to minimize toxicity and/or prevent carbon loss (9, 14, 15, 28). Interestingly, recent genomic analyses tentatively indicated that there are seven functionally distinct polyhedral bodies distributed among over 40 genera of bacteria (4). Thus, polyhedral bodies may be a general mechanism of metabolic organization in the Bacteria.
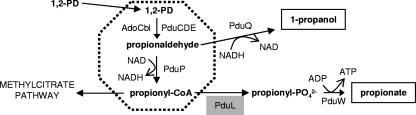
FIG. 1.
Model for 1,2-propanediol degradation by S. enterica. Coenzyme B12 (AdoCbl), coenzyme B12-dependent diol dehydratase (PduCDE), propionaldehyde dehydrogenase (PduP), phosphotransacylase (PduL), propionate kinase (PduW), and 1-propanol dehydrogenase (PduQ) are depicted. The first two steps of 1,2-PD degradation (conversion of 1,2-PD to propionyl-CoA) are proposed to occur within a polyhedral body-microcompartment. The dashed line indicates the shell of the polyhedral body, which is composed of up to seven different polypeptides. ATP is generated during the conversion of propionyl-PO42− to propionate. Given a suitable terminal electron acceptor such as O2, propionyl-CoA is degraded via the methylcitrate pathway, where biosynthetic precursors and additional energy are produced. Two additional enzymes associated with the polyhedral bodies (but not shown in the figure) are a putative diol dehydratase reactivase (PduGH) and an ATP:cob(I)alamin adenosyltransferase (PduO) involved in B12 recycling.
As described above, previous enzymatic analysis of crude cell extracts indicated that a PTAC is used for 1,2-PD degradation (25, 37). However, the enzyme involved was not identified. Here, we show that the pduL gene encodes a conserved, evolutionarily distinct PTAC that functions in 1,2-PD degradation by S. enterica.
MATERIALS AND METHODS
Chemicals and reagents.
Antibiotics, vitamin B12, and acetyl-PO42− were obtained from Sigma Chemical Company (St. Louis, MO). Propionyl-PO42− was synthesized as described previously (27). Isopropyl-β-d-thiogalactopyranoside (IPTG) was obtained from Diagnostic Chemicals Limited (Charlotteville, PEI, Canada). Restriction enzymes and T4 DNA ligase were obtained from New England Biolabs (Beverly, MA). Coomassie brilliant blue R-250, EDTA, ethidium bromide, 2-mercaptoethanol, and sodium dodecyl sulfate (SDS) were obtained from Bio-Rad (Hercules, CA). Other chemicals were obtained from Fisher Scientific (Pittsburgh, PA).
Bacterial strains, media, and growth conditions.
The bacterial strains used in this study are listed in Table 1. The rich medium used was Luria-Bertani/Lennox (LB) medium (Difco, Detroit, MI) (23). The minimal medium used was no-carbon-E (NCE) medium containing supplements indicated in the text and figure legends (3, 38). MacConkey-1,2-PD indicator medium contained MacConkey agar base (Difco), 1% 1,2-PD, and 200 ng ml−1 of vitamin B12.
TABLE 1.
Bacterial strains used in this study
| Species | Strain | Genotype |
|---|---|---|
| S. enterica serovar Typhimurium LT2 | BE47 | thr-480::Tn10dCam |
| BE188 | ΔpduL670 | |
| BE281 | pta-406::Tn10 | |
| BE284 | ΔpduL670/pLAC22-pduL (Ampr) | |
| BE285 | ΔpduL670/pLAC22-no insert (Ampr) | |
| BE286 | pLAC22-pduL (Ampr) | |
| BE287 | pLAC22-no insert (Ampr) | |
| BE291 | ΔpduL670 pta-209::Tn10 | |
| BE527 | pta-209::Tn10 | |
| BE529 | pta-209::Tn10/pLAC22-no insert (Ampr) | |
| BE530 | pta-209::Tn10/pLAC22-pduL (Ampr) | |
| BE548 | pta-209::Tn10/pBE522-no insert (Kanr) | |
| BE549 | pta-209::Tn10/pBE522-pta (Kanr) | |
| E. coli | BL21(DE3)RIL | (E. coli B) F−ompT hsdS (rB− mB−) dcm+ Tetrgal l (DE3) endA Hte (argU ileY leuW Camr) |
| BE554 | BL21(DE3)RIL/pTA925-pduL-His8 (T7 expression vector containing a minimal clone for production of PduL-his8, Kanr) | |
| BE119 | BL21(DE3)RIL/pTA925-no insert (Kanr) | |
| S17.1λpir | recA(RP4-2-Tc::Mu) λpir |
General molecular methods.
Agarose gel electrophoresis was performed as described previously (30). Plasmid DNA was purified by the alkaline lysis procedure (30) or by using QIAGEN (Chatsworth, CA) products according to the manufacturer's instructions. Following restriction digestion or PCR amplification, DNA was purified using Promega Wizard PCR Preps (Madison, WI) or QIAGEN gel extraction kits. Restriction digests were carried out using standard protocols (30). For ligation of DNA fragments, T4 DNA ligase was used according to the manufacturer's directions. Electroporation was carried out as previously described (6).
Protein methods.
Polyacrylamide gel electrophoresis (PAGE) was performed using Bio-Rad Redigels and Bio-Rad Mini-Protean II electrophoresis cells according to the manufacturer's instructions. Following gel electrophoresis, Coomassie brilliant blue R-250 was used to stain proteins. The protein concentration of solutions was determined using Bio-Rad protein assay reagent (Bio-Rad).
P22 transduction.
Transductional crosses were performed using P22 HT105/1 int-210 (12), a mutant phage that has high transducing ability (32), as described previously. Transductants were tested for phage contamination and sensitivity by streaking on green plates against P22 H5.
Construction of plasmids for production of PduL and PduL-His8.
PCR was used to amplify the pduL coding sequence from template pMGS2 (15). The primers used for amplification were 5′-GCCGCCAGATCTATGGATAAAGAGCTTCTGCAATCA-3′ and 5′-GCCGCCAAGCTTATTATCGCGGGCCTACCAGCCG-3′. These PCR primers introduced BglII and HindIII restriction sites that were used for cloning into vector pLAC22 (39). Following ligation, clones were introduced into Escherichia coli DH5α by electroporation, and transformants were selected by plating onto LB agar supplemented with 100 μg ml−1 ampicillin (Amp) (18). Pure cultures were prepared from selected transformants. The presence of insert DNA was verified by restriction analysis or PCR, and the DNA sequence of selected pduL clones was determined. Clones with the expected DNA sequence were used for further study.
A similar procedure was used to clone PduL fused to eight C-terminal histidine residues (PduL-His8) with the following differences. The primers used for PCR amplification were 5′-GCCGCCAGATCTATGGATAAAGAGCTTCTGCAATCA-3′ and 5′-GCCGCCAAGCTTATTAATGATGATGATGATGATGATGATGTCGCGGGCCTACCAGCCG-3′. The PCR product was ligated into T7 expression vector pTA925 (18). Transformation was done by electroporation, and LB plates supplemented with 25 μg ml−1 kanamycin (Kan) were used to select for transformants.
Growth of the PduL-His8 production strain.
A T7 expression plasmid (pTA925) was used for the production of PduL-His8. The host used for protein expression was E. coli BL21(DE3)RIL (Stratagene). This strain expresses T7 RNA polymerase as well as rare tRNAs for arginine, isoleucine, and leucine. Expression and control strains (BE554 and BE119) were grown in 400 ml LB broth containing 25 μg/ml Kan and 10 μg/ml chloramphenicol. Cultures were incubated at 15°C with shaking at 275 rpm in a 1-liter baffled Erlenmeyer flask. Cells were grown to an optical density at 600 nm of 0.6 to 0.8, and protein expression was induced by the addition of 1 mM IPTG. Cells were incubated for an additional 16 h and harvested by centrifugation at 8,000 × g for 10 min and at 4°C using a Beckman JA-10 rotor and an Avanti J-25 centrifuge.
Purification of PduL-His8.
The PduL-His8 production strain (BE554) was grown as described above. One gram of cells (wet weight) was suspended in 3 ml of buffer containing 50 mM Tris-HCl (pH 7.2), 200 mM (NH4)2SO4, and 0.4 mM AEBSF [4-(2-aminoethyl)benzenesulfonylfluoride·HCl]. Cells were broken using a French pressure cell at 20,000 lb/in2, and the resulting cell extract was centrifuged at 20,000 × g using a Beckman JA-17 rotor. The supernatant fraction was filtered through a 0.45-μm syringe filter. A chromatography column containing 1 ml of Ni-nitrilotriacetic acid resin (QIAGEN) was equilibrated with 5 ml of buffer A [50 mM Tris-HCl (pH 7.7), 25 mM KCl, 300 mM NaCl, 200 mM (NH4)2SO4, 10 mM imidazole, and 5 mM 2-mercaptoethanol]. Filtered cell extract (2.5 ml, about 50 mg protein) was applied to the column. The column was washed with 15 ml of buffer A supplemented with 10% glycerol and 80 mM imidazole. The column was eluted with 10 ml of buffer A (in two of 5-ml steps) supplemented with 10% glycerol and 400 mM imidazole.
Preparation of cell extracts of S. enterica.
Cells were grown under conditions that induce the pdu operon (5). Cell paste was suspended in 25 mM Tris-HCl (pH 7.2) containing 0.4 mM protease inhibitor AEBSF (3 ml of buffer per gram cells [wet weight]). Cells were broken using a French pressure cell (Thermo Electron Corp., Waltham, MA) at 20,000 lb/in2.
PTAC assays.
PTAC assays measured the conversion of acyl-PO42− plus coenzyme A (HS-CoA) to acyl-CoA and were performed as described previously (21). Standard assay mixtures contained 50 mM Tris-HCl (pH 7.2), 20 mM KCl, 0.2 mM HS-CoA, and 1 mM acyl-PO42− in a total volume of 1 ml. For kinetic studies, the concentrations of certain assay components varied as indicated in the text. Activity was determined by monitoring the absorbance at 232 nm over time and by using an E232 of 5.5 mM−1 cm−1 for calculations.
Construction of a nonpolar PduL deletion.
Bases 22 to 612 of the pduL coding sequence were deleted via a PCR-based method (24). The deletion was designed to leave all predicted translational start and stop signals of pdu genes intact. The following primers were used for PCR amplification of the flanking regions of the pduL gene: 5′-GCTCTAGAGCCGAAATCAGCCTAATCGATGGCG-3′ (primer 1), 5′-CGTTCATCGCGGGCCTACCAGCCGATCCATTACGCTTCACCTCGC-3′ (primer 2), 5′-CGGCTGGTAGGCCCGCGATGAACG-3′ (primer 3), and 5′-CGAGCTCGCCAGATGCATGATTTACTC-3′ (primer 4). Primers 1 and 2 were used to amplify a 498-bp region upstream of the pduL gene, and primers 3 and 4 were used to amplify 527 bases downstream of the pduL gene. The upstream and downstream amplification products were purified and then fused by a PCR that included 1 ng/μl of each product and primers 1 and 4. The fused product was digested with XbaI and SacI (these sites were designed into primers 1 and 4, respectively) and ligated into suicide vector pCVD442 that had been similarly digested. The ligation mixture was used to transform E. coli S17.1 by electroporation, and transformants were selected on LB medium supplemented with Amp (100 μg/ml). Six transformants were screened by restriction analysis, and all transformants released an insert of the expected size (1,058 bp). One of these transformants was used to introduce the pduL deletion into the S. enterica chromosome using the procedure described previously by Miller and Mekalanos (24), with the following modifications. For the conjugation step, strain BE47 was used as the recipient, and exconjugants were selected by plating onto LB agar supplemented with Amp (100 μg/ml) and chloramphenicol (20 μg/ml). Deletion of the pduL coding sequence was verified by PCR using chromosomal DNA as a template. Finally, the thr-480 dCAM insertion used for selection of exconjugants was “crossed off” by P22 transduction using a phage lysate prepared with the wild-type strain and by selecting for prototrophy on NCE glucose minimal medium.
Aerobic growth curves.
Growth media are described in the figure legends. For strains carrying pLAC22, media were also supplemented with 100 μg/ml Amp and 0.2 mM IPTG. To prepare the inoculum, LB cultures (2 ml) were incubated overnight at 37°C, and cells were then collected by centrifugation and resuspended in growth curve medium. Media were inoculated to a density of 0.15 absorbance units, and growth was monitored by measuring the optical density at 600 nm using a BioTek Synergy microplate reader with 48-well flat-bottom plates (Falcon), 0.5 ml of growth medium per well at 37°C, and shaking set at level 4. A relatively low volume of growth medium (0.5 ml) in a 48-well microplate was necessary for adequate aeration, and controls showed that growth under these conditions was similar to growth in shake flasks (data not shown).
Anaerobic growth curves.
A procedure similar to that for aerobic growth curves was used to measure the fermentation of 1,2-PD but with the following differences. Cultures were inoculated to an initial density of 0.1 absorbance units. Microplates were sealed with polyolefin membranes (Fisher Scientific) inside an anaerobic growth chamber (Coy Laboratory Products, Grass Lake, MI). To monitor whether anaerobic conditions were maintained within the microplate, NCE glycerol minimal medium was added to several wells and inoculated with wild-type S. enterica. S. enterica cannot ferment glycerol; therefore, growth on glycerol would indicate the presence of oxygen. For the experiments described in this study, no growth in wells containing glycerol minimal medium was observed, indicating that the headspace contained minimal amounts of oxygen (data not shown).
DNA sequencing and analysis.
DNA sequencing was carried out at the Iowa State University (ISU) DNA Facility using automated sequencing equipment from Applied Biosystems Inc. The template for DNA sequencing was plasmid DNA purified using QIAGEN 100 tips or QIAGEN Mini-Prep kits. Blast software was used for sequence similarity searching (2).
High-pressure liquid chromatography (HPLC).
A Microsorb C18 column (150 by 4.6 mm) was used with a Varian ProStar system that included a model 230 solvent delivery module, a model 430 autosampler, and a model 325 UV-Vis detector (Varian, Palo Alto, CA). Buffers A and B contained 10 mM NH4 formate (pH 4.6) and 10% or 90% methanol, respectively. The flow rate was 1 ml min−1, and the buffer composition varied as follows (minute:percent buffer): 0:0, 5:0, 17:100, 22:100, 23:0, and 28:0.
HPLC electrospray ionization mass spectrometry (HPLC-ESI-MS).
An Agilent (Palo Alto, CA) ion trap model 1100 mass spectrometer was operated in the positive mode. The ion source parameters were optimized for the formation of [M + H]+ ions with a source temperature of 310°C, a capillary voltage of 3.2 kV, and a cone voltage of 25 V. Nitrogen was used as the nebulizing gas and as the drying gas at flow rates of 15 and 400 liters h−1, respectively.
RESULTS
The pta gene is nonessential for 1,2-PD degradation.
Prior biochemical studies indicated that PTAC was needed for 1,2-PD degradation by S. enterica (25, 37). In the proposed biochemical pathway, this enzyme catalyzed the conversion of propionyl-CoA to propionyl-phosphate (transfer of a three-carbon acyl group) (Fig. 1). An enzyme with homology to known PTAC enzymes is not encoded by the pdu locus (6). However, S. enterica produces a phosphotransacetylase (Pta) encoded by the pta gene, which maps outside the pdu operon at centisome 50 (20). Pta is used for growth on acetate and inositol and plays a key role in the interconversion of acetyl-CoA and acetyl-PO42− (transfer of a two-carbon acyl group) (20). In vitro, Pta not only mediates the interconversion of acetyl-CoA and acetyl phosphate, it also catalyzes the interconversion of propionyl-CoA and propionyl-PO42− (20, 35). This raised the question of whether pta plays a role in 1,2-PD degradation. Two independent pta mutants (BE281 and BE527) were examined for growth on 1,2-PD. Aerobically, both grew slightly faster than wild-type S. enterica. The doubling times for the wild type were typically 7.5 to 8.5 h, and those for the pta mutants were 6 to 7 h. Both pta mutants were reconstructed via P22 transduction to ensure that they were isogenic with the wild type and that both still exhibited a small increase in the growth rate on 1,2-PD (not shown). These results confirmed previous studies that showed that pta is not essential for aerobic growth of S. enterica on 1,2-PD minimal medium (26).
Similarly, pta was unnecessary for the fermentation of 1,2-PD. Strains with pta mutations and wild-type S. enterica both had doubling times of 5 to 6 h during 1,2-PD fermentation. Phenotypic test showed that the pta mutants used in the above-mentioned studies were impaired for growth on acetate or inositol minimal medium and lacked detectable pta activity in cell extracts, as is expected for pta mutants (20). In addition, the growth defects on acetate and inositol were corrected by a pta minimal clone, and the chromosomal location of both pta insertions was verified by PCR (data not shown). Hence, we conclude that the pta gene is not essential for B12-dependent 1,2-PD degradation under either aerobic or anaerobic growth conditions.
Strains with pduL mutations produce less propionic acid on MacConkey-1,2-propanediol indicator medium.
The finding that the pta gene was unnecessary for 1,2-PD degradation suggested that S. enterica expresses an additional PTAC that is sufficient to mediate this process. As part of ongoing studies of 1,2-PD degradation by S. enterica, we constructed a series of precise pdu deletion mutations using PCR-based methods (11, 24). Tests with MacConkey-1,2-PD medium (17) indicated that three independent pduL deletion mutants each produced less propionic acid from 1,2-PD than did wild-type S. enterica. The enzymes that convert 1,2-PD to propionic acid were proposed to include coenzyme B12-dependent diol dehydratase, propionaldehyde dehydrogenase, propionate kinase, and PTAC (Fig. 1) (25, 37). Of these, only the gene for PTAC was not identified (7, 19, 26). Hence, these results tentatively suggested that the pduL gene encodes a PTAC enzyme.
Enzyme assays indicate that pduL encodes a PTAC enzyme.
Wild-type S. enterica as well as pduL and pta null mutants were grown under conditions that induce the pdu operon (5). Cell extracts were prepared and PTAC activity was measured using an assay that monitors the conversion of acyl-phosphate plus HS-CoA to acyl-CoA and inorganic phosphate (the reverse reaction with respect to 1,2-PD degradation). Both propionyl-PO42− and acetyl-PO42− were used as substrates. Cell extract from the wild-type strain had 8.9 μmol min−1 mg−1 PTAC activity with propionyl-PO42− (Table 2). Extracts from pta or pduL mutants had partial activity (6.0 and 2.6 μmol min−1 mg−1, respectively) (Table 2). PTAC activity was undetectable in cell extracts from the double mutant (pta pduL). The simplest interpretation of these results is that pduL and pta each encode PTAC enzymes, which was previously shown for pta (20). Furthermore, the pta pduL double mutant lacked detectable PTAC activity, demonstrating that the activity in Pta− PduL+ cell extracts required PduL.
TABLE 2.
Phosphotransacylase activity in cell extracts from selected strains of S. entericaa
| Strain (relevant genotype) | PTAC activity (μmol min−1 mg−1)b
|
||
|---|---|---|---|
| Propionyl-PO42− | Acetyl-PO42− | Propionyl-PO42− preferencec | |
| Wild type (PduL+ Pta+) | 8.9 | 3.5 | 2.5 |
| BE188 (PduL− Pta+) | 2.6 | 3.2 | 0.8 |
| BE527 (PduL+ Pta−) | 6.0 | 0.4 | 15.0 |
| BE291 (PduL− Pta−) | ND | ND | |
Cells were grown on NCE succinate minimal medium supplemented with 1,2-PD to ensure induction of the pdu operon.
PTAC assay mixtures contained 0.2 mM HS-CoA and 1 mM propionyl-PO42− or acetyl-PO42− and assay buffer. Activity was determined by monitoring the absorbance of reaction mixtures at 232 nm. ND, not detected (the lower detection limit of the assay is estimated to be 0.03 μmol min−1 mg−1).
Propionyl-PO42− preference = (activity with propionyl-PO42−)/(activity with acetyl-PO42−).
Enzyme assays also showed that the PTAC in Pta− PduL+ cell extracts was 15-fold more active with propionyl-PO42− than with acetyl-PO42− (6 μmol min−1 mg−1 compared to 0.4 μmol min−1 mg−1) (Table 2). This is consistent with a role for PduL in 1,2-PD degradation. On the other hand, the PTAC activity in Pta+ PduL− cell extracts was 1.6-fold more active with acetyl-PO42−, consistent with the role of Pta in acetyl group metabolism (20).
For the above-described enzyme assays, controls showed that detectable PTAC activity required acyl-phosphate and HS-CoA, and in all cases, PTAC activity was linear with the enzyme concentration (data not shown).
PduL mutants are impaired for aerobic growth on 1,2-PD.
Compared to wild-type S. enterica, a pduL mutant (BE188) was impaired for aerobic growth on 1,2-PD (Fig. 2A). The doubling time for S. enterica was 7.9 ± 0.5 h, and that of a pduL mutant was 12.6 ± 0.6 h. Further impairment of growth was not seen in the pduL pta double mutant even though this mutant lacked measurable PTAC activity (data not shown). These findings are consistent with the idea that PduL is a PTAC involved in 1,2-PD degradation. Aerobically, growth on 1,2-PD is expected to proceed in the absence of PTAC, since propionyl-CoA (which is formed prior to the PTAC reaction) can be metabolized via the methylcitrate pathway (Fig. 1) (16). The observed growth impairment of the pduL mutant was likely due to reduced ATP synthesis resulting from a block in the oxidative branch of the 1,2-PD degradative pathway, although other explanations cannot be ruled out.
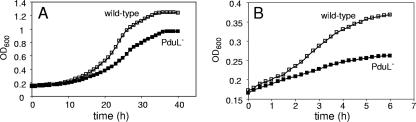
FIG. 2.
Growth of a pduL null mutant on 1,2-PD. (A) Aerobic growth of wild-type S. enterica and a pduL mutant (BE188) on 1,2-PD minimal medium. (B) Fermentation of 1,2-PD. BE188 carries a precise deletion of the pduL gene made by a PCR-based method. The observed growth defects of BE188 were corrected by a pduL minimal clone. Each growth curve was performed three times in quadruplicate. Representative curves are shown. Log-phase doubling times with standard deviations are given in the text. The aerobic growth medium was NCE supplemented with 52 mM 1,2-PD, 200 ng ml−1 vitamin B12, and 0.3 mM valine, isoleucine, leucine, and threonine. The fermentation medium was NCE supplemented with 52 mM 1,2-PD, 200 ng ml−1 vitamin B12, and 0.2% yeast extract. The inoculation and incubation procedures are described in Materials and Methods. OD600, optical density at 600 nm.
pduL mutants are unable to obtain energy from 1,2-PD under fermentative conditions.
In the absence of an exogenous electron acceptor, the methylcitrate pathway is inoperative, and S. enterica is unable to grow on 1,2-PD as a sole carbon source (29). Under these conditions, S. enterica converts 1,2-PD to 1-propanol and propionate (Fig. 1). This stimulates growth on minimal medium supplemented with a small amount of yeast extract by providing a source of ATP (29). The production of ATP via the conversion of 1,2-PD to propionate is expected to require PTAC (Fig. 1). Therefore, we tested the effect of a pduL mutation on this process. The doubling times of wild-type S. enterica in the presence and absence of 1,2-PD under fermentative conditions were 5.5 ± 0.35 h and 10.7 ± 0.29 h, respectively (Fig. 2B); the fermentation of 1,2-PD stimulated the growth rate about twofold, which is consistent with previous studies (29). The doubling times of the pduL mutant (BE188) under fermentative conditions in the presence and absence of 1,2-PD were 13.1 ± 1.0 and 13.3 ± 0.95 h, respectively. This indicated that energy production via 1,2-PD fermentation was completely eliminated by the pduL mutation. This is consistent with a role for PduL in 1,2-PD degradation.
The pduL mutant used in the above-described studies contained a wild-type pta gene, indicating that Pta did not substitute for PduL under the conditions used to measure 1,2-PD fermentation. Furthermore, a pta null mutation (alone or in combination with a pduL null mutation) did not affect the fermentation of 1,2-PD (data not shown). These results indicate that the Pta enzyme is insufficient to support the fermentation of 1,2-PD and that PduL is required.
The observed phenotypes of a pduL mutant are complemented by a pduL minimal clone.
Growth tests showed that the aerobic and anaerobic growth phenotypes described above for a pduL mutant (BE188) were fully corrected by the expression of a pduL minimal clone using a tightly regulated expression vector, pLAC22. Under aerobic conditions, the doubling times for BE287 (wild-type S. enterica/pLAC22-no insert) and BE286 (wild-type S. enterica/pLAC22-pduL) were 5.4 ± 0.12 h and 6.0 ± 0.32 h, respectively. The doubling times for strains for BE285 (pduL/pLAC22-no insert) and BE284 (pduL/pLAC22-pduL) were 8.2 ± 0.71 and 5.5 ± 0.25 h, respectively. Under anaerobic conditions, the doubling times for S. enterica/pLAC22-no insert, S. enterica/pLAC22-pduL, pduL/pLAC22-no insert, and pduL/pLAC22-pduL were 5.0 ± 0.22, 5.2 ± 0.37, 11.1 ± 0.19, and 5.3 ± 0.40 h, respectively. Thus, under aerobic and anaerobic conditions, a pduL minimal clone complemented the growth defect of the pduL mutation. On the other hand, vectors without an insert did not correct the observed growth defects of the pduL mutant. These results show that the observed phenotypes of the pduL mutant (BE188) resulted from the deletion of the pduL gene but not from polarity or a mutation acquired during strain construction.
PduL substitutes for Pta in vivo during acetate utilization.
Growth tests were performed to determine whether PduL could substitute for Pta in vivo. Previous studies showed that pta mutants grow very slowly on minimal medium with >30 mM acetate (20). Here, we show that ectopic expression of PduL corrects this defect (Fig. 3). The doubling times of the Pta+ strain and the pta mutant on acetate minimal medium were 4.4 ± 0.41 and 28.6 ± 1.4 h, respectively. The doubling time of a pta mutant producing PduL from expression vector pLAC22 was 3.7 ± 0.1 h, which was slightly faster than the control (4.4 ± 0.41 h). Hence, ectopic expression of pduL fully corrected the growth defect of the pta mutant on acetate minimal medium. Since Pta is a well-studied PTAC enzyme, this finding indicates that PduL also has PTAC activity.
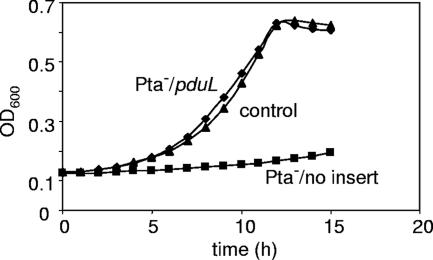
FIG. 3.
Ectopic expression of a pduL minimal clone corrects the growth defect of a pta mutant on acetate minimal medium. Data for S. enterica harboring expression vector pLAC22 without an insert (control), a pta mutant carrying pLAC22 without an insert (Pta−/no insert), and a pta mutant harboring a pduL minimal clone in pLAC22 (Pta−/pduL) are shown. The relevant strains are BE287, BE529, and BE530 (Table 1). Controls showed that pLAC22 with or without the pduL insert did not observably affect the growth of the wild-type strain on acetate minimal medium (data not shown). In addition, the growth defect of the pta mutant was fully corrected by a pta minimal clone, showing that the effects of this mutation were not due to polarity (data not shown). The growth medium was NCE supplemented with 0.2% Na acetate and 0.2 mM IPTG. Each growth study was performed twice in quadruplicate. Representative curves are shown. Log-phase doubling times with standard deviations are given in the text. OD600, optical density at 600 nm.
Propionyl-CoA is a product of the PduL reaction.
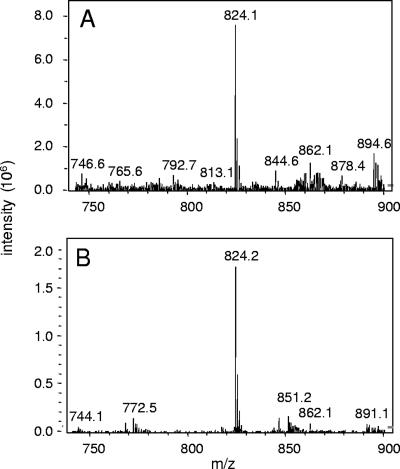
The finding that HS-CoA was required for PduL activity (see above) indicated that an acyl-CoA was formed in assay mixtures. Moreover, the PTAC assay used to measure PduL activity monitored the absorbance at 232 nm, which monitors the formation of thioester bonds (13). To confirm that acyl-CoA was formed in PduL enzyme assays, HPLC and HPLC-mass spectrometry were performed. Assay mixtures contained cell extract from BE527 (Pta− PduL+) or BE291 (Pta− PduL−), HS-CoA, propionyl-PO42−, and standard components; therefore, propionyl-CoA was the expected acyl-CoA product. Reverse-phase HPLC with UV detection at 260 nm was used to identify CoA compounds present in reaction mixtures. A single major compound with a retention time of 10.9 min was detected when Pta− PduL+ cell extracts were used. This compound coeluted with authentic propionyl-CoA following coinjection and produced a mass spectrum characteristic of propionyl-CoA via HPLC-ESI-MS (Fig. 4). The major peaks for authentic propionyl-CoA and the PduL reaction product that eluted at 10.9 min were at m/z 824.1 or 824.2, respectively. These peaks correspond to [M + H]+ for propionyl-CoA. In contrast, propionyl-CoA was undetectable by HPLC in assay mixtures containing cell extract from BE291 (Pta− PduL−), and the major HPLC peak observed corresponded to HS-CoA (retention time, 4.5 min). Our interpretation of the above-described results is that propionyl-CoA is a product of the PduL reaction.
FIG. 4.
Propionyl-CoA is produced by the PduL reaction. (A) HPLC-ESI-MS of propionyl-CoA standard. (B) HPLC-ESI-MS of a PduL assay after 10 min of incubation at 30°C. The assay mixture initially contained 10 μg cell extract from BE527 (Pta− PduL+), 0.4 mM HS-CoA, 1 mM propionyl-PO42−, and buffer.
Production of PduL-His8 protein.
E. coli strain BE554 was constructed to produce high levels of recombinant PduL fused to eight C-terminal histidine residues (PduL-His8). Protein expression by BE554 and a control strain (BE119) was examined by SDS-PAGE (not shown) and enzyme assays. Relatively high amounts of a protein with a molecular mass of nearly 27 kDa were present in the inclusion body fraction from the pduL expression strain, which is near the predicted molecular mass for PduL-His8 (24 kDa). However, a modest amount of soluble protein near this mass was produced.
To better assess the production of PduL-His8, cell extracts were tested for PTAC activity. Soluble extracts from the PduL production strain typically contained about 50 μmol min−1 mg−1 activity, whereas the control strain produced only 2 μmol min−1 mg−1 PTAC activity. Hence, a reasonable amount of PduL-His8 appeared to be soluble and active under the production conditions used. On the other hand, although the inclusion body fraction from the expression strain contained high amounts of PduL-His8 protein, the PTAC activity of this fraction was typically about twofold lower than that of the soluble fraction, indicating that most of the PduL in this fraction was inactive. No PTAC activity was detected in the inclusion body fraction from the control strain. The PTAC assay used measured the conversion of propionyl-PO42− and HS-CoA to propionyl-CoA and HPO42−. No activity could be detected when enzyme, HS-CoA, or propionyl-PO42− was omitted from the assay mixture. In each case, PTAC activity was linear with regard to the enzyme concentration (data not shown).
Purification and kinetic characterization of PduL-His8.
PduL-His8 was purified from cell extracts of strain BE554 by nickel affinity chromatography (Fig. 5). Purified PduL-His8 appeared to be homogenous following SDS-PAGE and Coomassie blue staining (Fig. 5). Kinetic analysis showed that purified PduL-His8 was 3.8-fold more active with propionyl-PO42− than with acetyl-PO42− (Vmax of 51.7 and 13.5 μmol min−1 mg−1, respectively) (Table 3). It also had a lower Km for propionyl-PO42− than for acetyl-PO42− (0.61 ± 0.06 versus 0.97 ± 0.26 mM). No activity was detected when PduL-His8, HS-CoA, or propionyl-PO42− (or acetyl-PO42−) was omitted from the assay mixture. In each case, the PduL-His8 activity was linear with the enzyme concentration (data not shown).
FIG. 5.
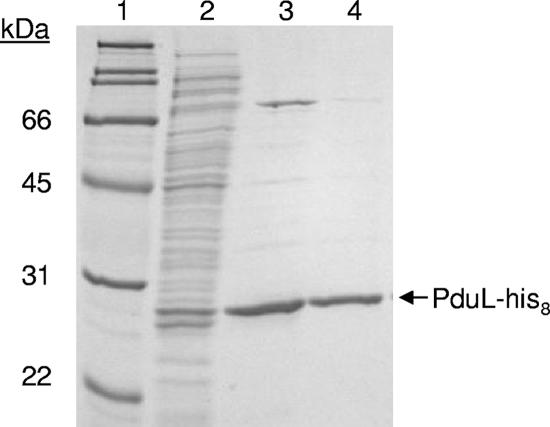
SDS-PAGE analysis of PduL-His8 purification. Lane 1, molecular mass markers; lane 2, soluble crude extracts from the PduL-His8 production strain (BE554) (10 μg protein); lane 3, fraction obtained from the first elution step using buffer with 400 mM imidazole (2 μg protein); lane 4, fraction obtained from the second elution step using buffer with 400 mM imidazole (2 μg protein). A 12% acrylamide gel was stained with Coomassie brilliant blue R-250.
TABLE 3.
Kinetic constants for purified recombinant PduL-His8a
| Substrate | Km (mM) | Vmax (μmol min−1 mg−1) | kcat (s−1) | kcat/Km (mM s−1) |
|---|---|---|---|---|
| Propionyl-PO42− | 0.61 ± 0.06 | 51.7 ± 7.6 | 20.7 | 33.9 |
| Acetyl-PO42− | 0.97 ± 0.26 | 13.4 ± 1.8 | 5.4 | 5.6 |
PTAC assays were performed by monitoring the absorbance of reaction mixtures at 232 nm as described in Materials and Methods. The kinetic constants for propionyl-PO42− and acetyl-PO42− were determined using an excess of HS-CoA (0.4 mM).
For PduL-His8, crude cell extracts typically had PTAC activity of about 50 μmol min−1 mg−1. Nickel affinity chromatography yielded purified enzyme with specific activities of about 45 to 55 μmol min−1 mg−1. Visual inspection of SDS-PAGE gels indicated that PduL-His8 comprised about 2 to 5% of the total cell protein (Fig. 5); hence, these results suggest that PduL-His8 lost activity during the course of purification. Indeed, PduL-His8 was unstable in the purified form and lost about 50% of its activity 48 h after purification. The reported kinetic constants are based on assays done as soon as possible after purification. As the activity of PduL-His8 decreased, its Km for HS-CoA and propionyl-PO42− remained unchanged within experimental error. Its activity with propionyl-PO42− and acetyl-PO42− decreased in direct proportion. We infer that the Km values reported here are representative but that the Vmax values are probably underestimated.
DISCUSSION
Here, we presented genetic and biochemical evidence that the pduL gene encodes a PTAC involved in 1,2-PD degradation. Growth tests showed that a pduL mutant was impaired for 1,2-PD degradation under aerobic and anaerobic conditions, indicating a role for pduL in 1,2-PD degradation. Growth studies also showed that ectopic expression of PduL corrected the growth defect of a pta mutant on acetate minimal medium, indicating that PduL has phosphotransacetylase activity in vivo. Enzyme assays of crude cell extracts demonstrated that pduL was necessary for the production of detectable PTAC activity in a genetic background that contained a pta mutation. HPLC-ESI-MS showed that propionyl-CoA was produced from propionyl-PO42− and HS-CoA in PduL enzyme assays. PduL-His8 was purified and found to have a specific activity of 57.4 μmol min−1 mg−1 with propionyl-PO42− as a substrate, indicating the PduL is sufficient for PTAC activity. Thus, genetic and biochemical studies indicate that PduL is a PTAC involved in 1,2-PD degradation.
BLAST and PSI-BLAST analyses (five iterations) showed that PduL lacks significant similarity to known PTAC enzymes. Thus, PduL is evolutionarily distinct. It either evolved independently of known PTAC enzymes or became highly divergent from them over time. Sequence analyses also showed that PduL homologues (expect <7 × 10−30) are found in 49/337 (14.5%) complete microbial genomes present in the GenBank database. No homologues of PduL were found among the Archaea or Eukarya. Hence, PduL is apparently a conserved protein specific to the Bacteria. In addition, BLAST analyses identified four PduL homologues that are fused to sequences with homology to acetate kinases. These may be dual-function enzymes that convert acyl-CoA to its corresponding organic acid with the synthesis of ATP.
Including PduL, there are three known classes of phosphotransacylases. The S. enterica PduL, Pta, and EutD enzymes are representatives of these classes. PduL is comprised of 210 amino acids, while Pta and EutD are 714 and 338 amino acids in length, respectively (8). EutD and the C-terminal region of Pta are homologous. PduL lacks significant similarity to EutD and Pta. The N-terminal region of Pta contains BioD-like and “DRTGG” domains. These two domains are absent from EutD and PduL. The physiological role of these domains is uncertain. In enteric bacteria, acetyl group metabolism is linked to two-component signal transduction systems and DNA damage and repair in a complex manner (22, 34, 40). The N-terminal domains of Pta may be needed to accommodate expanded physiological roles.
Kinetic studies with purified PduL-His8 were performed (Table 3). The Vmax of PduL with propionyl-PO42− as a substrate was 51.7 ± 7.6 μmol min−1 mg−1. Previously purified Pta enzymes were reported to have specific activities between 32 and 9006 μmol min−1 mg−1 when acyl-PO42− was used as a substrate (33). The activity of PduL-His8 is at the low end of this range but may be underestimated due to its instability (see Results).
The Km values of PduL-His8 for HS-CoA and propionyl-PO42− were determined to be 0.61 ± 0.06 and 0.032 ± 0.06 mM, respectively. Reported Km values of Pta homologues for HS-CoA range from 0.03 to 1.7 mM, and those for acetyl-PO42− range from 0.024 to 4.7 mM (33). Thus, Km values of PduL for HS-CoA and propionyl-PO42− appear to be within a physiologically meaningful range. Kinetic studies found that the kcat/Km ratio of Pdu-His8 was about sixfold higher for propionyl-PO42− than for acetyl-PO42−. The selectivity of PduL for propionyl-PO42− is consistent with a role in 1,2-PD degradation. Furthermore, its relatively low activity with acetyl-PO42− may be important to minimize the perturbation of acetyl group metabolism (22, 40).
Two findings in this report were surprising to us: (i) the observation that pduL mutants were significantly impaired for growth on 1,2-PD minimal medium (Fig. 2) and (ii) the finding that a pduL pta double mutant was not further impaired for growth on 1,2-PD. Because Pta is known to be active with propionyl-PO42−, we expected that Pta would partly substitute for PduL during growth of S. enterica on 1,2-PD. However, in the studies reported here, it did not. A simple explanation is that PduL is more efficient at propionyl group metabolism in vivo. Two explanations that we think are more interesting are the differential regulation of pta and pduL as well as the possibility that PduL is specifically adapted to function in concert with the polyhedral body-microcompartment involved in 1,2-PD degradation.
Acknowledgments
We thank the ISU DNA Sequencing and Synthesis Facility for assistance with DNA analyses. We thank Ann Perera and the ISU Metabolomics Laboratory for help with mass spectrometry.
We thank the ISU Office of Biotechnology for financial support.
Footnotes
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Abeles, R. H., and H. A. Lee. 1961. Intramolecular oxidation-reduction requiring a cobamide coenzyme. J. Biol. Chem. 236:2347-2350. [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz, D., J. M. Hushon, H. J. Whitfield, Jr., J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobik, T. A. 2006. Polyhedral organelles compartmenting bacterial metabolic processes. Appl. Microbiol. Biotechnol. 70:517-525. [DOI] [PubMed] [Google Scholar]
- 5.Bobik, T. A., M. Ailion, and J. R. Roth. 1992. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J. Bacteriol. 174:2253-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobik, T. A., Y. Xu, R. M. Jeter, K. E. Otto, and J. R. Roth. 1997. Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J. Bacteriol. 179:6633-6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinsmade, S. R., and J. C. Escalante-Semerena. 2004. The eutD gene of Salmonella enterica encodes a protein with phosphotransacetylase enzyme activity. J. Bacteriol. 186:1890-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinsmade, S. R., T. Paldon, and J. C. Escalante-Semerena. 2005. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol. 187:8039-8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, P., D. I. Andersson, and J. R. Roth. 1994. The control region of the pdu/cob regulon in Salmonella typhimurium. J. Bacteriol. 176:5474-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics: a manual for genetic engineering. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 13.Dawson, R. M. C., D. C. Elliott, W. H. Elliott, and K. M. Jones (ed.). 1969. Data for biochemical research, 2nd ed. Oxford University Press, Oxford, United Kingdom.
- 14.Havemann, G. D., and T. A. Bobik. 2003. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 185:5086-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Havemann, G. D., E. M. Sampson, and T. A. Bobik. 2002. PduA is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 184:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horswill, A., and J. Escalante-Semerena. 1997. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the σ54 family of activators, and the prpBCDE genes constitute an operon. J. Bacteriol. 179:928-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeter, R. M. 1990. Cobalamin-dependent 1,2-propanediol utilization by Salmonella typhimurium. J. Gen. Microbiol. 136:887-986. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, C. L., E. Pechonick, S. D. Park, G. D. Havemann, N. A. Leal, and T. A. Bobik. 2001. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J. Bacteriol. 183:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leal, N. A., G. D. Havemann, and T. A. Bobik. 2003. PduP is a coenzyme A-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch. Microbiol. 180:353-361. [DOI] [PubMed] [Google Scholar]
- 20.LeVine, S. M., F. Ardeshir, and G. F. Ames. 1980. Isolation and characterization of acetate kinase and phosphotransacetylase mutants of Escherichia coli and Salmonella typhimurium. J. Bacteriol. 143:1081-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundie, L. L., Jr., and J. G. Ferry. 1989. Activation of acetate by Methanosarcina thermophila. J. Biol. Chem. 264:18392-18396. [PubMed] [Google Scholar]
- 22.McCleary, W. R., J. B. Stock, and A. J. Ninfa. 1993. Is acetyl phosphate a global signal in Escherichia coli? J. Bacteriol. 175:2793-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 24.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obradors, N., J. Badia, L. Baldoma, and J. Aguilar. 1988. Anaerobic metabolism of the l-rhamnose fermentation product 1,2-propanediol in Salmonella typhimurium. J. Bacteriol. 170:2159-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palacios, S., V. J. Starai, and J. C. Escalante-Semerena. 2003. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J. Bacteriol. 185:2802-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paoli, P., P. Cirri, L. Camici, G. Manao, G. Cappugi, G. Moneti, G. Pieraccini, G. Camici, and G. Ramponi. 1997. Common-type acylphosphatase: steady-state kinetics and leaving-group dependence. Biochem. J. 327:177-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penrod, J. T., and J. R. Roth. 2006. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J. Bacteriol. 188:2865-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price-Carter, M., J. Tingey, T. A. Bobik, and J. R. Roth. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 31.Sampson, E. M., C. L. Johnson, and T. A. Bobik. 2005. Biochemical evidence that the pduS gene encodes a bifunctional cobalamin reductase. Microbiology 151:1169-1177. [DOI] [PubMed] [Google Scholar]
- 32.Schmieger, H. 1971. A method for detection of phage mutants with altered transducing ability. Mol. Gen. Genet. 110:378-381. [DOI] [PubMed] [Google Scholar]
- 33.Schomburg, I., A. Chang, C. Ebeling, M. Gremse, C. Heldt, G. Huhn, and D. Schomburg. 2004. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 32:D431-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi, I. Y., J. Stansbury, and A. Kuzminov. 2005. A defect in the acetyl coenzyme A↔acetate pathway poisons recombinational repair-deficient mutants of Escherichia coli. J. Bacteriol. 187:1266-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimizu, M., T. Suzuki, K. Y. Kameda, and Y. Abiko. 1969. Phosphotransacetylase of Escherichia coli B, purification and properties. Biochim. Biophys. Acta 191:550-558. [DOI] [PubMed] [Google Scholar]
- 36.Shively, J. M., C. E. Bradburne, H. C. Aldrich, T. A. Bobik, J. L. Mehlman, S. Jin, and S. H. Baker. 1998. Sequence homologs of the carboxysomal polypeptide CsoS1 of the thiobacilli are present in cyanobacteria and enteric bacteria that form carboxysomes-polyhedral bodies. Can. J. Bot. 76:906-916. [Google Scholar]
- 37.Toraya, T., S. Honda, and S. Fukui. 1979. Fermentation of 1,2-propanediol and 1,2-ethanediol by some genera of Enterobacteriaceae, involving coenzyme B12-dependent diol dehydratase. J. Bacteriol. 139:39-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 39.Warren, J. W., J. R. Walker, J. R. Roth, and E. Altman. 2000. Construction and characterization of a highly regulable expression vector, pLAC11, and its multipurpose derivatives, pLAC22 and pLAC33. Plasmid 44:138-151. [DOI] [PubMed] [Google Scholar]
- 40.Wolfe, A. J., D. E. Chang, J. D. Walker, J. E. Seitz-Partridge, M. D. Vidaurri, C. F. Lange, B. M. Pruss, M. C. Henk, J. C. Larkin, and T. Conway. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol. Microbiol. 48:977-988. [DOI] [PubMed] [Google Scholar]