Abstract
The MarR family of transcriptional regulators of bacteria are involved in the regulation of many cellular processes, including pathogenesis. In this work, we have demonstrated genetically that hpaR (hpa, hrp associated), which encodes a putative MarR family regulator, is involved in the hypersensitive response (HR), pathogenicity, and extracellular protease production of the phytopathogenic bacterium Xanthomonas campestris pathovar campestris. A mutation in hpaR resulted in complete loss of virulence in the host plant cabbage, a delayed and weakened HR in the nonhost plant pepper ECW-10R, and an increase in extracellular protease production. Detection of the β-glucuronidase activity of a plasmid-driven hpaR promoter-gusA reporter revealed that the expression of hpaR is positively controlled by HrpG and HrpX and is suppressed in rich medium while being strongly induced in minimal and hrp-inducing media and inside the host. These findings indicate that hpaR belongs to the hrpG and hrpX regulon and that HrpX regulates the extracellular protease production via hpaR in X. campestris pv. campestris.
The MarR (multiple antibiotic resistance regulator) protein, which was originally characterized as the repressor of the multiple antibiotic resistance operon marRAB in Escherichia coli, is a prototypical member of the MarR family of transcriptional regulators that are widely found in bacteria and archaea (2, 13, 47). The MarR members regulate a variety of biological functions, including resistance to multiple antibiotics and other toxic chemicals such as organic solvents, household disinfectants, and oxidative stress agents (1, 2). They also regulate adaptation to different environments and the expression of virulence factors of both plant and animal pathogens (20, 63).
To date, six MarR members have been demonstrated to play an important role in microbial pathogenesis. They are AphA in Vibrio cholerae (28), Hor in Erwinia carotovora (56), MgrA in Staphylococcus aureus (24), PecS in Erwinia chrysanthemi (42), RovA in Yersinia enterocolitica and Yersinia pseudotuberculosis (34, 41), and SlyA in Salmonella enterica serovar Typhimurium as well as Salmonella enterica (14, 31, 37). It has been demonstrated that the role of these MarR members in pathogenesis is to control the expression of virulence-related genes or virulence-associated traits, and their regulatory targets vary in different pathogens (10, 24, 28, 32, 34, 41, 42, 43). Although certain regulatory targets of all these MarR members have been identified, the regulation of the expression of these marR genes has been studied only in Yersinia and Salmonella. In Y. pseudotuberculosis, the expression of rovA requires its own product, RovA, and environmental signals such as temperature and pH are also involved in the regulation of rovA expression (19, 34). In S. enterica serovar Typhimurium, the expression of slyA is also regulated by its own product, SlyA (48), and is induced during the stationary phase as well as during the infection of macrophages (10). The slyA expression of S. enterica is positively regulated by the two-component regulatory system PhoP/PhoQ (37).
Xanthomonas campestris pathovar campestris is the causal agent of cruciferous plant black rot disease (22) and has been used as a model bacterium to study microbe-plant interactions for over two decades. Recently, the entire genomes of X. campestris pv. campestris strains ATCC 33913 and 8004 have been sequenced (17, 40). It has been reported that a transposon insertion mutant of strain 8004, A240D02, in which a transposon is inserted into the open reading frame (ORF) XC_2827, encoding a putative MarR family transcriptional regulator, loses the ability to induce disease on the host plant cabbage (Brassaca oleracae) (40). In this paper, we present genetic evidence to demonstrate that XC_2827 is essential for the pathogenicity on the host plant cabbage and is required for the hypersensitive response (HR) on the nonhost plant pepper ECW-10R. XC_2827 is under the positive control of the two key hrp gene regulators HrpG and HrpX, and therefore we have designated XC_2827 hpaR (for hrp [hypersensitive response and pathogenicity]-associated regulator). These results also demonstrate that hpaR has a negative effect on the extracellular protease production by X. campestris pv. campestris and that HrpX represses extracellular protease production through activating the expression of hpaR.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacteria and plasmids used in this work are listed in Table 1. Escherichia coli strains were grown in LB medium (33) at 37°C. X. campestris pv. campestris strains were grown in NYG medium (15), MMX medium (16), or XVM2 medium (60) at 28°C. Antibiotics were used at the following final concentrations as required: rifampin, 50 μg/ml; kanamycin, 25 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 100 μg/ml; and tetracycline, 15 μg/ml for E. coli and 5 μg/ml for X. campestris pv. campestris.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli JM109 | recA1 endA1 gyrA96 thi supE44 relA1 Δ(lac-proAB)/F′ [traD36 lacIqlacZ ΔM15] | 67 |
| X. campestris pv. campestris strains | ||
| 8004 | Wild type; Rifr | |
| 8004* | 8004 derivative containing an E45K conversion in HrpG; Rifr | 15 |
| 8004* ΔhrpX | As 8004* but hrpX deleted; Rifr Kanr | 27 |
| NK2827 | As 8004 but hpaR::pK18mob; Rifr Kanr | 27 |
| NK2827* | As 8004* but hpaR::pK18mob; Rifr Kanr | This work |
| CNK2827 | NK2827 harboring pXC2827; Rifr Kanr Tcr | This work |
| 8004G | 8004 ΔhrpG; Rifr Kanr | This work |
| 8004X | 8004 ΔhrpX; Rifr Kanr | Laboratory collection |
| 8004pG28 | 8004 harboring pGUS2827; Rifr Kanr Tcr | Laboratory collection |
| 8004GpG28 | 8004G harboring pGUS2827; Rifr Kanr Tcr | This work |
| 8004XpG28 | 8004X harboring pGUS2827; Rifr Kanr Tcr | This work |
| 8004RpG28 | NK2827 harboring pGUS2827; Rifr Kanr Tcr | This work |
| 161E12 | As 8004 but XC_2826::Tn5gusA5; Rifr Kanr Spcr Gmr | This work |
| NK2828 | As 8004 but XC_2828::pK18mob; Rifr Kanr | Laboratory collection |
| Plasmids | ||
| pLAFR6 | Broad-host-range cloning vector; Tcr | 23 |
| pRK2073 | Helper plasmid; Tra+ Mob+ ColE1 Spcr | 30 |
| pK18mob | Suicide plasmid in X. campestris pv. campestris; Kanr | 45 |
| pT18mob | Tetracycline-resistant derivative of pK18mob; Tcr | Laboratory collection |
| pK2827 | pK18mob containing a 267-bp internal fragment of the hpaR gene; Kanr | This work |
| pT2827 | pT18mob containing a 267-bp internal fragment of the hpaR gene; Kanr | This work |
| pXC2827 | pLAFR6 containing a 758-bp fragment including the hpaR gene; Tcr | This work |
| pGUS2827 | pLAFR6 containing a hpaR promoter-gusA fusion fragment; Tcr | This work |
| pGUShrpA | pLAFR6 containing a hrpA operon promoter-gusA fusion fragment; Tcr | This work |
| pGUShrpB | pLAFR6 containing a hrpB operon promoter-gusA fusion fragment; Tcr | Laboratory collection |
| pGUShrpC | pLAFR6 containing a hrpC operon promoter-gusA fusion fragment; Tcr | Laboratory collection |
| pGUShrpD | pLAFR6 containing a hrpD operon promoter-gusA fusion fragment; Tcr | Laboratory collection |
| pGUShrpE | pLAFR6 containing a hrpE operon promoter-gusA fusion fragment; Tcr | Laboratory collection |
| pGUShrpF | pLAFR6 containing a hrpF operon promoter-gusA fusion fragment; Tcr | Laboratory collection |
| pGUShrpG | pLAFR6 containing a hrpG promoter-gusA fusion fragment; Tcr | Laboratory collection |
| pGUShrpX | pLAFR6 containing a hrpX promoter-gusA fusion fragment; Tcr | Laboratory collection |
Rifr, Kanr, Spcr, Gmr, and Tcr, rifampin, kanamycin, spectinomycin, gentamicin, and tetracycline resistant, respectively.
DNA manipulation.
DNA manipulation was performed following the procedures described by Sambrook et al. (44). Conjugation between the X. campestris pv. campestris and E. coli strains was performed as described by Turner et al. (59). Restriction enzymes and DNA ligase were used in accordance with the manufacturer's instructions (Promega, Shanghai).
Construction of a nonpolar mutant of hpaR in the X. campestris pv. campestris strains 8004 and 8004*.
The nonpolar mutant of hpaR in the X. campestris pv. campestris strains 8004 (15) and 8004* (27) was constructed by using homologous suicide plasmid integration as described by Windgassen et al. (65), using pK18mob as the vector (45). A 267-bp internal fragment of hpaR was amplified using the total DNA of strain 8004 as the template and the primer pair 2827MF/22827MR (Table 2), which was designed according to the XC_2827 (hpaR) sequence (40). After confirmation by sequencing, the amplified DNA fragment was cloned into the suicide plasmid pK18mob to create the recombinant plasmid pK2827 (Table 1). To ensure the creation of a nonpolar mutant, the 267-bp fragment was inserted such that the transcription orientation of the fragment was the same as that of the lac promoter in the vector. The plasmid pK2827 was transformed into E. coli strain JM109 (67) and then introduced into strains 8004 and 8004* by triparental conjugation using pRK2073 (30) as the helper plasmid. Transconjugants were screened on NYG agar plates supplemented with rifampin and kanamycin, and the obtained transconjugants with a mutation in the hpaR gene were confirmed by PCR. Confirmation PCR was performed using the total DNA of the transconjugants as the template and the primer pair P18conF/2827conR (Table 2) (P18conF is located in pK18mob, and 2827conR is located downstream of the 267-bp internal fragment of hpaR). The expected PCR products were further confirmed by sequencing. One of the confirmed mutants for each parent strain, NK2827 (from 8004) or NK2827* (from 8004*) (Table 1), was used for further study.
TABLE 2.
Primer pairs used in this work
| Purpose | Primer pair | Sequence (5′ to 3′)a | Product length (bp) |
|---|---|---|---|
| Mutagenesis | 2827MF/2827MR | CCCGGATCCGATCCAACCGCTAAACGCGTCG/CCCAAGCTTTCGTCCGGGCTGCTGGCGCGCGC | 267 |
| Mutant confirmation | P18conF/2827conR | GCCGATTCATTAATGCAGCTGGCAC/CACCTGACGGCTGCTCGCTACG | 752 |
| Complementation | 2827CF/2827CR | CCCGAATTCGGACGGCACGCAAACGGCCGAG/CCCAAGCTTCACCTGACGGCTGCTCGCTACG | 758 |
| Reporter construction | 2827PF/2827PR | CCCGAATTCCCGGCCGGCTGGGAGCAGATCGGG/TACTTGTGTATAAGAGTCAGGGCACCGTTGATTACATTACTGAATGG | 420 |
| GusAF/GusAR | CTGACTCTTATACACAAGTAGCG/CCCGGATCCGGCTTTCCCCCCCCCCCC | 1,800 |
Added restriction sites are underlined, and the 20 nucleotides that complemented the first 20 nucleotides of the gusA fragment are in boldface.
Complementation of NK2827.
For complementation of the hpaR mutant NK2827, a 758-bp DNA fragment containing the entire hpaR gene (from 150 bp upstream of the start codon to 100 bp downstream of the stop codon) was amplified by PCR using the total DNA of the wild-type strain 8004 as the template and the primer pair 2827CF/2827CR (Table 2). After being confirmed by sequencing, the amplified DNA fragment was cloned into pLAFR6 (Table 1) to generate the recombinant plasmid pXC2827. The plasmid pXC2827 was transferred into the mutant NK2827 by triparental conjugation. The transconjugants carrying pXC2827 were screened on NYG agar plates containing rifampin, kanamycin, and tetracycline. A confirmed representative transconjugant was named CNK2827 (Table 1) and chosen for further study.
Test of extracellular enzyme activity and extracellular polysaccharide (EPS) production.
A radial diffusion assay (52) was used to test the activity of the extracellular enzymes protease, endoglucanase, and amylase. Two microliters of overnight culture (optical density at 600 nm [OD600] ≈ 1.0) of each X. campestris pv. campestris strain was spotted onto NYG agar plates containing 0.5% (wt/vol) skim milk (for protease) (Sangon, Shangshai, China), 0.25% (wt/vol) carboxymethylcellulose (for endoglucanase) (Sangon, Shangshai, China), or 0.1% (wt/vol) starch (for amylase) (Sangon, Shangshai, China) and incubated at 28°C for 24 h (endoglucanase and amylase) or 48 h (protease). Plates were stained where necessary as described by Tang et al. (53). Zones of clearance around the spot due to the degradation of the substrate were photographed. Three plates were inoculated in each experiment, and each experiment was repeated three times. The relative activity of the enzyme was indicated by the diameter of the clear zone. To quantitatively estimate the extracellular protease activity, the method described by Swift et al. (50) was used.
To estimate EPS production, strains were cultured in 100 ml NYG liquid medium containing 2% (wt/vol) glucose at 28°C with shaking at 200 rpm for 3 days. EPS was precipitated from the culture supernatant with ethanol, dried, and weighed as described by Tang et al. (53).
Virulence assay and determination of bacterial load in planta.
Virulence was tested on potted cabbage Jingfeng no. 1 (Brassaca oleracae cultivar Jingfeng no. 1) grown in a greenhouse with 12-h day-night cycle illuminations with a fluorescent lamp at temperatures of 25 to 28°C. Seedlings with four fully expanded leaves were used for inoculation. Bacterial cells were grown in NYG liquid medium at 28°C with shaking at 200 rpm for 15 h (at the exponential phase of growth). The concentrations of bacterial inocula were adjusted to an optical density of 600 nm of 0.1. Two leaves per plant were inoculated by the leaf-clipping method (18). Sixty leaves were inoculated for each independent experiment. Each treatment was repeated three times. After maintenance in 100% humidity for 24 h, the inoculated plants were maintained in the growth conditions described above. Lesion length was measured at 10 days postinoculation.
The growth of bacteria in cabbage leaf tissue was measured by homogenizing a group of leaves (five leaves for each sampling) in 9 ml sterile water. Diluted homogenates were plated on NYG agar plates supplemented with rifampin (for the wild type) or rifampin plus kanamycin (for mutants). Bacterial CFU were counted after incubation at 28°C for 3 days.
HR test.
The HR was tested on the pepper plant ECW-10R (Capsicum annuum cv. ECW-10R), which is one of the nonhosts commonly used to test the HR of X. campestris pv. campestris (11, 35). The pepper leaves were inoculated by infiltrating an approximately 5-μl bacterial suspension (1 × 107 CFU/ml or 1 × 109 CFU/ml) in 10 mM sodium phosphate buffer (5.8 mM Na2HPO4 and 4.2 mM NaH2PO4, pH 7.0) into the abaxial leaf surface by using a blunt-end plastic syringe. The inoculated plants were maintained in a greenhouse with 12-h day-night cycle illuminations with a fluorescent lamp and a constant temperature of 28°C, and the HR symptoms were observed and photographed at 8, 16, and 24 h after inoculation. At least three plants were inoculated in each experiment, and each experiment was repeated at least two times.
Construction of pGUS2827.
The hpaR reporter plasmid pGUS2827 was constructed by fusing the promoter region of the hpaR gene to the promoterless β-glucuronidase (gusA) gene with its ribosome binding site. The 0.42-kb region upstream of the hpaR ATG (excluding ATG) start codon was amplified by PCR using the total DNA of the wild-type strain 8004 as the template and the primer pair 2827PF/2827PR (Table 2). 2827PR differs from the hpaR sequence by the addition of a 20-nucleotide tag, which is complementary to the first 20 nucleotide of the promoterless gusA fragment, to its 5′ end (Table 2). The 1.8-kb DNA fragment containing the promoterless gusA gene with its ribosome binding site was amplified by PCR using pLAFR1::Tn5gusA5 as the template and the primer pair GusAF/GusAR (Table 2). The 0.42-kb promoter fragment and the 1.8-kb gusA fragment were ligated by fusion PCR (29) to generate the hpaR promoter and promoterless gusA reporter construct. This reporter construct was cloned into the vector pLAFR6 to create the reporter plasmid pGUS2827 (Table 1).
GUS activity assay.
X. campestris pv. campestris strains were cultured in NYG, MMX, and XVM2 for 24 and 48 h. β-Glucuronidase (GUS) activities were determined by measurement of the OD415, using ρ-nitrophenyl β-d-glucuronide as the substrate, as described by Jefferson et al. (25). Histochemical GUS staining was performed using 5-bromo-4-chloro-3-indolylglucuronide (X-Gluc) (Promega, Madison, WI) as a substrate, essentially as described by Jefferson et al. (26).
RESULTS
hpaR is required for the hypersensitive response and pathogenicity of X. campestris pv. campestris.
The transposon EZ::TN〈KAN-2〉 Tnp insertion mutant A240D02 has a single transposon inserted in the ORF XC_2827 (named hpaR [see the introduction) (GenBank accession number YP_243896) (40). The ORF is annotated as a “marR family transcriptional regulator” in the genome of X. campestris pv. campestris strain 8004, and the mutant is unable to cause disease on host plant cabbage (40). The genomic data for strain 8004 reveals that the nearest ORF upstream of hpaR is XC_2828, which is annotated as a “Xanthomonas conserved hypothetical protein” (40). The reverse transcription-PCR result revealed that hpaR and XC_2828 are separately transcribed (data not shown), although they are in the same transcription direction and separated by only 65 bp. The nearest ORF downstream of hpaR is XC_2826, which is annotated as a “peptidyl-dipeptidase.” The transcription direction of XC_2826 is opposite that of hpaR. The result of plant tests by the leaf-clipping method (18) showed that the XC_2826 mutant 161E12 and the XC_2828 mutant NK2828 (Table 1) displayed a wild-type virulence phenotype on the cabbage Jingfeng no. 1 (data not shown), indicating that XC_2826 and XC_2828 might not be involved in the virulence of the pathogen. There is a 228-bp spacer between hpaR and XC_2826 (40). ORF prediction (using Vector NTI software [Invitrogen]) revealed that a small ORF encoding a putative protein of 32 amino acid residues sharing 45% identity and 65% similarity to a region of a hypothetical protein (NCBI accession number XP_758456) in Ustilago maydis exists in the spacer region in the same direction as hpaR. The expression and the biological function of this newly found ORF are not clear.
It has been demonstrated that insertion of the transposon EZ::TN〈KAN-2〉 Tnp can cause a polar effect on the resulting mutant (57). To exclude the possibility that the phenotypes of the EZ::TN〈KAN-2〉 Tnp insertion mutant A240D02 resulted from such a polar effect, we constructed the nonpolar mutant NK2827 and the complemented strain CNK2827 by homologous suicide plasmid integration and introduction of the recombinant plasmid pXC2827, which contains an entire wild-type hpaR gene, into the mutant NK2827 (Table 1) (see Materials and Methods for details). The pathogenicities of the hpaR nonpolar mutant NK2827 and the complemented strain CNK2827 were determined on the host plant cabbage cultivar Jingfeng no. 1 by the leaf-clipping method (18). At 10 days postinoculation, no black-rot disease symptoms were observed on the cabbage leaves inoculated with the mutant strain NK2827, while a typical V-shaped black-rot symptom was observed on the leaves inoculated with the wild-type strain 8004 and the complemented strain CNK2827 (Fig. 1A). The mean lesion lengths caused by the complemented strain CNK2827 and the wild-type strain 8004 were not significantly different (P = 0.01 by t test) (Fig. 1B). To investigate the role of hpaR in the growth of X. campestris pv. campestris in the host, the populations of the hpaR mutant NK2827, the complemented strain CNK2827, and the wild-type strain 8004 in cabbage leaves were determined. The bacterial number of the hpaR mutant recovered from the infected leaves was approximately 10-fold lower than that of the wild-type strain at 1 day postinoculation and thereafter (Fig. 1C), although the hpaR mutant NK2827 and the wild-type strain 8004 grew identically in rich and minimal media (data not shown). The bacterial number of the mutant recovered from the infected leaves was significantly lower than that of the wild type at each of the test points (P = 0.01 by t test). The growth capacity of the mutant strain in planta could be completely restored by hpaR in trans (Fig. 1C). These results demonstrate that hpaR is required for the virulence and in planta growth of X. campestris pv. campestris.
FIG. 1.
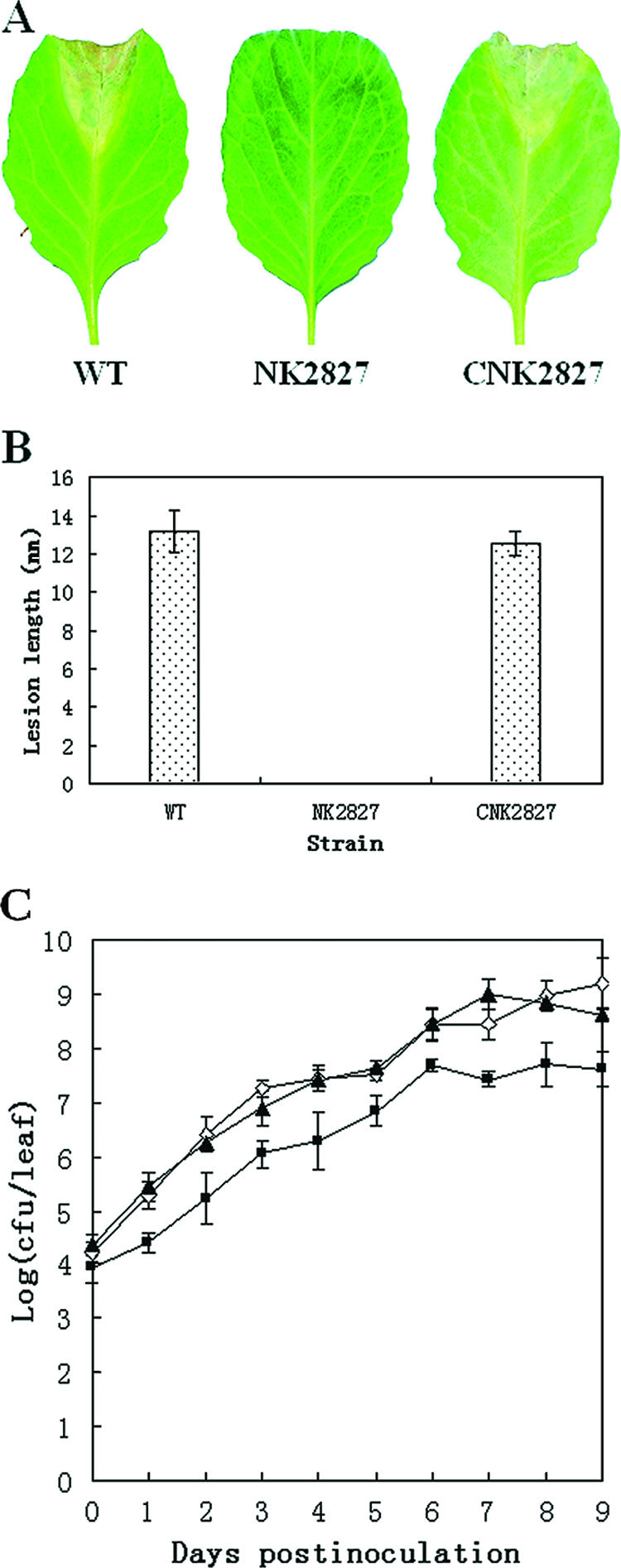
hpaR is essential for the virulence and in planta growth of X. campestris pv. campestris. (A) Black rot symptoms caused by X. campestris pv. campestris strains on inoculated leaves of cabbage (Brassaca oleracae cultivar Jingfeng no. 1). Photographs were taken on day 10 postinoculation. (B) Average lesion lengths caused by the X. campestris pv. campestris strains. Values are the means ± standard deviations from three repeats, each with 60 leaves. (C) In planta growth of X. campestris pv. campestris. Bacteria were recovered from the inoculated leaves every day for a period of 10 days postinoculation. Data are the means ± standard deviations from three repeats. ▴, wild-type (WT) strain 8004; ▪, hpaR mutant NK2827; ⋄, complemented strain CNK2827.
To determine whether hpaR is required for the HR of the pathogen, the hpaR mutant NK2827 was inoculated on the pepper cultivar ECW-10R, a nonhost plant typically used to test the HR of X. campestris pv. campestris (11, 35). The result shows that NK2827 elicited a delayed and weakened HR compared to the wild type, and this reduced HR could be restored to the wild-type level by hpaR in trans (Fig. 2). These findings reveal that hpaR is required for X. campestris pv. campestris to cause a full HR in the nonhost plant pepper ECW-10R.
FIG. 2.
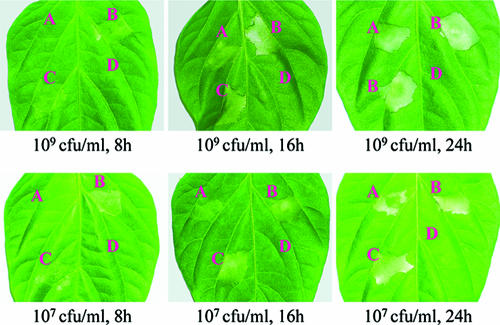
Symptoms induced in pepper leaves (Capsicum annuum cv. ECW-10R) by the X. campestris pv. campestris strains. Approximately 5 μl bacterial culture suspended in 10 mM sodium phosphate buffer was infiltrated into the leaf mesophyll tissue with a blunt-end plastic syringe. Pictures of the pepper leaf were taken at 8, 16, and 24 h after infiltration. Three replications were done in each experiment, and each experiment was repeated three times. The results presented are from a representative experiment, and similar results were obtained in all other independent experiments. The cell concentrations used for inoculation and the observation time (postinoculation) are indicated. (A) hpaR mutant NK2827; (B) complemented strain CNK22827; (C), wild-type strain 8004; (D) type III-deficient strain (hrcV mutant; negative control).
The expression of hpaR is positively regulated by HrpG and HrpX.
In many gram-negative phytopathogenic bacteria, hrp (hypersensitive response and pathogenicity) genes are required for pathogenicity on host plants and for the HR on nonhost plants (3, 4, 5, 8, 64) and are repressed in nutrient-rich media but induced in nutrient-deficient media and inside the host (9, 46, 54, 58, 60, 66). In xanthomonads, hrp genes are regulated by HrpG and HrpX (39, 54, 61, 62). To study the regulatory relationship between hpaR and hrp as well as the expression of hpaR under different growth conditions, a plasmid-driven hpaR promoter-gusA transcriptional fusion reporter, pGUS2827, was constructed and introduced into the hrpX mutant 8004X, the hrpG mutant 8004G, and the wild-type strain 8004 to create the reporter strains 8004XpG28, 8004GpG28, and 8004pG28, respectively (Table 1) (see Materials and Methods). The GUS activities of these strains were measured in the rich medium NYG, the minimal medium MMX, and the hrp-inducing medium XVM2. As shown in Table 3, the GUS activity of the hpaR-gusA reporter pGUS2827 in the hrpX or hrpG mutation background (strains 8004XpG28 and 8004GpG28) is significantly lower than that in the wild-type background (strain 8004pG28) in both XVM2 and MMX media, indicating that hrpG and hrpX are required for the expression of hpaR in these conditions. To determine if hrpG and hrpX are also required for the expression of hpaR inside the host plant, the reporter strains were introduced into the cabbage leaves by leaf clipping, and the GUS activities in the infected tissues were detected at 2 days postinoculation. As shown in Fig. 3, the infected tissue of the leaves inoculated with 8004pG28 became dark blue, while no blue color was observed in the leaves inoculated with 8004GpG28 or 8004XpG28, although the numbers of living cells of the three strains inside the infected leaves were similar. The plasmid stability test revealed the reporter plasmid pGUS2827 to be stable inside the host whether in the wild-type, hrpG mutation, or hrpX mutation background (data not shown). These results demonstrate that the expression of hpaR also depends on hrpG and hrpX when the pathogen is inside its host.
TABLE 3.
GUS activities of the hpaR promoter-gusA reporter in different genetic backgrounds under different growth conditions
| Strain | GUS activity (U)a in:
|
||
|---|---|---|---|
| NYG | MMX | XVM2 | |
| 8004pG28 | 0.043 ± 0.01a | 2.74 ± 0.445a | 1.19 ± 0.06a |
| 8004XpG28 | 0.025 ± 0.007a | 0.25 ± 0.041c | 0.055 ± 0.003b |
| 8004GpG28 | 0.040 ± 0.003a | 0.44 ± 0.106b | 0.09 ± 0.046b |
| 8004R pG28 | 0.051 ± 0.008a | 2.53 ± 0.314a | 1.23 ± 0.12a |
GUS activities were determined after growth of X. campestris pv. campestris in NYG for 24 h and in MMX and XVM2 for 48 h. Data are the means ± standard deviations from triplicate measurements. Different letters indicate significant differences (P = 0.05). The experiment was repeated twice, and similar results were obtained.
FIG. 3.
hrpG and hrpX are essential for the expression of hpaR in the infected tissue. 8004pG28, 8004GpG28, and 8004XpG28 were inoculated into the cabbage (Brassaca oleracae cultivar Jingfeng no. 1) leaves by leaf clipping. Because the cells of the hrpG and hrpX mutants grow very poorly but the wild-type cells grow quite well inside the host plant (61, 62), to ensure that the cell numbers of 8004pG28, 8004GpG28, and 8004XpG28 were similar inside the inoculated leaves at the time of GUS staining, the cell concentrations of 8004pG28, 8004GpG28, and 8004XpG28 were adjusted to OD600s of 0.1, 1.3, and 1.3, respectively. At 2 days postinoculation, the infected leaves were stained using an in situ GUS staining method to measure the β-glucuronidase activity, and the cell numbers inside the infected leaves were measured in a parallel experiment. Twenty leaves were inoculated (three leaves were chosen for GUS staining and five leaves for measurement of the cell number) in each experiment, and each experiment was repeated twice. The average bacterial numbers inside the tested leaves are indicated.
The GUS activities of strain 8004pG28 in the rich medium NYG were 63-fold and 27-fold lower than those in the minimal medium MMX and the hrp-inducing medium XVM2, respectively (Table 3), implying that the expression of hpaR is suppressed in rich medium and strongly induced in minimal and hrp-inducing media. The histochemical GUS staining result reveals that hpaR is also strongly expressed inside the host (Fig. 3). These results are consistent with the general conclusion that the expression of hrp and hrp-associated genes is highly regulated and generally suppressed in complex media but is induced in planta and in certain nutrient-poor synthetic media (9, 46, 54, 58, 60, 66).
To investigate whether hpaR regulates the expression of hrp genes, promoter-gusA transcriptional fusion reporters of the six hrp units (hrpA to hrpF) (8, 61, 62), as well as the reporters of hrpG and hrpX genes of X. campestris pv. campestris (Table 1), were introduced into the hpaR mutant NK2827 and the wild-type strain 8004. The GUS activities of the resulting reporter strains were detected in the minimal medium MMX. The results showed that there is no significant difference among these strains (data not shown), suggesting that none of these hrp genes is regulated by hpaR under the tested conditions.
To determine whether the expression of hpaR is self-regulated, the hpaR-gusA reporter plasmid pGUS2827 was introduced into the hpaR mutant NK2827 to generate the strain 8004RpG28, and the GUS activities of the strain were measured in the rich medium NYG, the minimal medium MMX, and the hrp-inducing medium XVM2. The results showed that the GUS activities of 8004RpG28 and 8004pG28 are identical in each of the media (Table 3), indicating that the X. campestris pv. campestris hpaR is not self-regulated.
Inactivation of hpaR results in overproduction of extracellular protease.
To gain insights into the nature of the role of hpaR in the virulence of X. campestris pv. campestris, the effect of hpaR mutation on the known virulence-associated traits were examined. It has been demonstrated that EPS is an important virulence factor and that extracellular enzymes, including protease, endoglucanase, and amylase, collectively contribute to the virulence of X. campestris pv. campestris (12). To determine whether a mutation in hpaR has any effect on these factors, the production of EPS and the three extracellular enzymes of the hpaR mutant was measured (see details in Materials and Methods). The results showed that the production of EPS as well as extracellular endoglucanase and amylase by the hpaR mutant was similar to that of the wild type in both rich and minimal media (data not shown). However, the hpaR mutant produced significantly higher extracellular protease activity than the wild type in the minimal medium MMX (Table 4), although the two strains produced similar levels of extracellular protease activity in the rich medium NYG (Table 4). The increased protease activity of the hpaR mutant in MMX could be lowered to the wild-type level by hpaR in trans (Table 4). These results suggest that in X. campestris pv. campestris, HpaR negatively regulates, directly or indirectly, the production of extracellular protease under hrp-inducing conditions.
TABLE 4.
Extracellular protease activities of X. campestris pv. campestris strains in rich medium and minimal medium
| Strain | Extracellular protease activity (A366)a in:
|
|
|---|---|---|
| NYG | MMX | |
| 8004 (wild type) | 0.286 ± 0.039a | 0.144 ± 0.029a |
| NK2827 | 0.264 ± 0.020a | 0.253 ± 0.010b |
| CNK2827 | 0.270 ± 0.031a | 0.165 ± 0.011a |
| 8004* | 0.108 ± 0.035b | NTb |
| 8004* ΔhrpX | 0.276 ± 0.010a | NT |
| NK2827* | 0.276 ± 0.010a | NT |
Data are the means ± standard deviations of triplicate measurements; different letters indicate significant differences (P = 0.05). The experiment was repeated twice, and similar results were obtained.
NT, not tested.
During plant-pathogen interactions, Xanthomonas is exposed to plant-generated H2O2, a growth inhibition and killing factor for bacteria (49). It has been reported that the marR family regulator slyA of S. enterica serovar Typhimurium is required for the resistance of the pathogen to oxidative stress (14, 31). To investigate whether a mutation in hpaR of X. campestris pv. campestris has any effects on H2O2 sensitivity, we determined the growth rate of the hpaR mutant in the rich medium NYG and the minimal medium MMX supplemented with different levels of H2O2. The results showed that the hpaR mutant and the wild-type strain displayed similar H2O2 sensitivity levels (data not shown), indicating that hpaR is not required for the resistance of X. campestris pv. campestris to oxidative stress. Parallel experiments also revealed that the mutation in hpaR did not affect the normal function of the gluconeogenic pathway (data not shown), which has been demonstrated to be required for the full virulence of X. campestris pv. campestris (51).
HrpX represses extracellular protease production via hpaR.
It has been reported that 8004*, a hrp gene constitutive expression mutant of X. campestris pv. campestris strain 8004, displays reduced extracellular protease production in the NYG medium and that the deletion of hrpX in the strain 8004* restores the extracellular protease production (27). According to this evidence and the observation that in the MMX medium hpaR has a negative effect on extracellular protease production and the expression of hpaR is activated by HrpX (Tables 3 and 4), we hypothesized that HrpX might repress extracellular protease production via hpaR. To test this hypothesis, we inactivated hpaR of strain 8004* and examined the extracellular protease activity of the resulting mutant (designated NK2827*) (see Materials and Methods) in the hrp-repressing medium NYG. As expected, in the NYG medium, the hrp constitutive expression mutant 8004* exhibited a significant reduction in extracellular protease activity compared to the wild-type strain 8004, and deletion of hrpX (8004* ΔhrpX) or disruption of hpaR (NK2827*) in 8004* restored the extracellular protease activity to the wild-type level (Table 4) (27). These results indicate that X. campestris pv. campestris HrpX negatively regulates the extracellular protease production through controlling the expression of the MarR family transcriptional regulator hpaR.
DISCUSSION
The MarR transcriptional regulators are members of a large regulatory protein family that are widely distributed in bacteria and involved in the regulation of varied cellular processes, including the pathogenesis of both plant and human pathogens (1, 2). In this work, we have genetically demonstrated that hpaR, a gene which encodes a putative MarR family transcriptional regulator, is required for the hypersensitive response and pathogenicity of the plant pathogen X. campestris pv. campestris. The data also reveal that the expression of hpaR is under the positive control of the two key hrp regulatory genes hrpG and hrpX and is involved in the negative regulation of the extracellular protease production by X. campestris pv. campestris.
The hrp genes of phytopathogenic bacteria encode a type III secretion system (TTSS) which is essential in inducing pathogenicity in their host plants and in triggering a hypersensitive response in resistant or nonhost plants (3, 5, 64). In xanthomonads, the hrp gene cluster comprises six operons (hrpA to hrpF) and is under the positive control of HrpG and HrpX (8, 61, 62). It has long been considered that HrpX is the regulator of genes with a TTSS-associated function, such as other hrp genes and the TTSS effector genes (6, 7, 39, 61). However, in this study, we have demonstrated that X. campestris pv. campestris hpaR, a gene which regulates the production of extracellular protease secreted by the type II secretion system, is also under the control of HrpG and HrpX. These results led to the discovery of a novel regulatory cascade in which HrpX positively regulates the expression of hpaR and HpaR negatively controls the extracellular protease production by X. campestris pv. campestris. The hpaR mutant of X. campestris pv. campestris produced a wild-type level of extracellular endoglucanase and amylase activities, indicating that hpaR is not involved in the type II secretion system. Determination of how HpaR regulates the extracellular protease production shall require further study.
It has been reported that, based on a cDNA AFLP analysis, HrpX of the Xanthomonas campestris pv. vesicatoria strain 85-10 positively regulates a gene encoding a MarR family transcriptional regulator and negatively regulates a gene encoding an extracellular protease (36). Recently, the whole genome of Xanthomonas campestris pv. vesicatoria 85-10 has been sequenced (55), and a genome BLAST search showed that the gene corresponding to the gene encoding a MarR family regulator is ORF XCV1512 (GenBank accession number CAJ23144). Sequence comparison revealed that hpaR and XCV1512 share 84% identity at the amino acid level. Interestingly, like the situation in X. campestris pv. campestris, a hrp gene constitutive expression mutant (85*) of X. campestris pv. vesicatoria strain 85-10 also displays reduced extracellular protease production, and deletion of hrpX in 85* (85* ΔhrpX) restores the production of extracellular protease to the wild-type level (36). These findings suggest that an hpaR homologue and a similar regulatory cascade, in which HrpX represses extracellular protease production through activation of hpaR, might also exist in X. campestris pv. vesicatoria.
This work suggests that HrpX of X. campestris may be a global regulator that regulates not only the type III secretion system but also extracellular protease production associated with the type II secretion system. Recently, Furutani et al. (21) demonstrated that HrpXo regulates the expression of cysP2, which encodes a type II secretion protein in Xanthomonas oryzae pv. oryzae, and they thus proposed that HrpXo may act as a global regulator. Based on microarray results, Occhialini et al. (38) found that the HrpB (the homologue of HrpX) of Ralstonia solanacearum regulates genes governing chemotaxis, biosynthesis, or catabolism of various low-molecular-weight chemical compounds and siderophore production and uptake. They concluded that HrpB is a master regulator and a regulatory switch controlling multiple virulence pathways (38).
Inactivation of hpaR resulted in a complete loss of the virulence of X. campestris pv. campestris in the host cabbage Jingfeng no. 1, indicating that HpaR plays a very important role in normal pathogenesis. However, besides the observed overproduction of extracellular protease, a mutation in hpaR did not affect the expression of any hrp genes at the transcription level or known virulence-associated traits such as the production of EPS and extracellular endoglucanase and amylase. The mutation also did not affect sensitivity to H2O2 or the gluconeogenic pathway. Because extracellular protease has a positive effect on the virulence of X. campestris pv. campestris (12), it is unlikely that the overproduction of extracellular protease would lead to a complete loss of pathogenic virulence, although it is possible that extracellular protease overproduction may have harmful effects on the pathogen. Based on the facts that hpaR is under the positive control of HrpX and encodes a putative regulatory protein and that a mutation in hpaR affects the HR and virulence of the pathogen, we suspect that an important subset of TTSS effectors and/or genes involved in an unknown process essential for the pathogenesis of X. campestris pv. campestris is under the positive control of hpaR. Studies to validate this hypothesis are under way in our laboratory.
Acknowledgments
We are grateful to J. Maxwell Dow for critical suggestions and helpful discussions on the manuscript.
This work was supported by the 973 Program of the Ministry of Science and Technology of China (2006CB101902) and the 863 Program of the Ministry of Science and Technology of China.
Footnotes
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410-413. [DOI] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 3.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arlat, M., C. L. Gough, C. E. Barber, C. Boucher, and M. J. Daniels. 1991. Xanthomonas campestris contains a cluster of hrp genes related to the larger hrp cluster of Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 4:593-601. [DOI] [PubMed] [Google Scholar]
- 5.Arnold, D. L., A. Pitman, and R. W. Jackson. 2003. Pathogenicity and other genomic islands in plant pathogenic bacteria. Mol. Plant Pathol. 4:407-420. [DOI] [PubMed] [Google Scholar]
- 6.Astua-Monge, G., J. Freitas-Astua, G. Bacocina, J. Roncoletta, S. A. Carvalho, and M. A. Machado. 2005. Expression profiling of virulence and pathogenicity genes of Xanthomonas axonopodis pv. citri. J. Bacteriol. 187:1201-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Astua-Monge, G., G. V. Minsavage, R. E. Stall, M. J. Davis, U. Bonas, and J. B. Jones. 2000. Resistance of tomato and pepper to T3 strains of Xanthomonas campestris pv. vesicatoria is specified by a plant-inducible avirulence gene. Mol. Plant-Microbe Interact. 13:911-921. [DOI] [PubMed] [Google Scholar]
- 8.Bonas, U., R. Schulete, S. Fenselau, G. V. Minsavage, B. J. Staskawicz, and R. E. Stall. 1991. Isolation of a gene cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol. Plant-Microbe Interact. 4:81-88. [Google Scholar]
- 9.Brito, B., M. Marenda, P. Baberis, C. Boucher, and S. Genin. 1999. prhJ and hrpG: two new components of the plant signal-dependent regulatory cascade controlled by PrhA in Ralstonia solanacearum. Mol. Microbiol. 31:237-251. [DOI] [PubMed] [Google Scholar]
- 10.Buchmeier, N., S. Bossie, C. Y. Chen, F. C. Fang, D. G. Guiney, and S. J. Libby. 1997. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65:3725-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castaneda, A., J. D. Reddy, B. El-Yacoubi, and D. W. Gabriel. 2005. Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity, but one avr gene affected race specificity. Mol. Plant-Microbe Interact. 18:1306-1317. [DOI] [PubMed] [Google Scholar]
- 12.Chan, J. W., and P. H. Goodwin. 1999. The molecular genetics of virulence of Xanthomonas campestris. Biotechnol. Adv. 17:489-508. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, S. P., S. B. Levy, J. Foulds, and J. L. Rosner. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a Mar-independent pathway. J. Bacteriol. 175:7856-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels, J. J. D., I. B. Autenrieth, A. Ludwig, and W. Goebel. 1996. The gene slyA of Salmonella typhimurium is required for destruction of M cells and intracellular survival but not for invasion or colonization of the murine small intestine. Infect. Immun. 64:5075-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniels, M. J., C. E. Barber, P. C. Turner, M. K. Sawczyc, R. J. W. Byrde, and A. H. Fielding. 1984. Clonging of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad-host-range cosmid pLAFR1. EMBO J. 3:3323-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniels, M. J., C. E. Barber, P. C. Turner, W. G. Cleary, and M. K. Sawczyc. 1984. Isolation of mutants of Xanthomonas campestris pathovar campestris showing altered pathogenicity. J. Gen. Microbiol. 130:2447-2455. [Google Scholar]
- 17.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 18.Dow, J. M., L. Crossman, K. Findlay, Y.-Q. He, J.-X. Feng, and J.-L. Tang. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 100:10995-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellison, D. W., M. B. Lawrenz, and V. L. Miller. 2004. Invasin and beyond: regulation of Yersinia virulence by RovA. Trends Microbiol. 12:296-300. [DOI] [PubMed] [Google Scholar]
- 20.Ellison, D. W., and V. L. Miller. 2006. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9:1-7. [DOI] [PubMed] [Google Scholar]
- 21.Furutani, A., S. Tsuge, K. Ohnishi, Y. Hikichi, T. Oku, K. Tsuno, Y. Inoue, H. Ochiai, H. Kaku, and Y. Kubo. 2004. Evidence for HrpXo-dependent expression of type II secretory proteins in Xanthomonas oryzae pv. oryzae. J. Bacteriol. 186:1374-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayward, A. C. 1993. The host of Xanthomonas, p. 51-54. In J. G. Swings and E. L. Civerolo (ed.), Xanthomonas. Chapman & Hall, London, United Kingdom.
- 23.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374-1377. [DOI] [PubMed] [Google Scholar]
- 24.Ingavale, S., W. van Wamel, T. T. Luong, C. Y. Lee, and A. L. Cheung. 2005. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jefferson, R. A., S. M. Burges, and D. Hirsh. 1986. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83:8447-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jefferson, R. A., T. A. Kavanagh, and M. W. Bevan. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, B., R.-Q. Xu, X.-Z. Li, H.-Y. Wei, F.-A. Bai, X. Hu, Y.-Q. He, and J.-L. Tang. 2006. Construction and characterization of a hrpG mutant rendering constitutive expression of hrp genes in Xanthomonas campestris pv. campestri. Prog. Nat. Sci. 16:480-485. [Google Scholar]
- 28.Kovacikova, G., W. Lin, and K. Skorupski. 2004. Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol. Microbiol. 53:129-142. [DOI] [PubMed] [Google Scholar]
- 29.Kuwayama, H., S. Obara, T. Morio, M. Katoh, H. Urushihara, and Y. Tanaka. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leong, S. A., G. S. Ditta, and D. R. Helinski. 1982. Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta-aminolevulinic acid synthetase from Rhizobium meliloti. J. Biol. Chem. 257:8724-8730. [PubMed] [Google Scholar]
- 31.Libby, S. J., W. Goebel, A. Ludwig, N. Buchmeier, F. Bowe, F. C. Fang, D. G. Guiney, J. G. Songer, and F. Heffron. 1994. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc. Natl. Acad. Sci. USA 91:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linehan, S. A., A. Rytkonen, X. J. Yu, M. Liu, and D. W. Holden. 2005. SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infect. Immun. 73:4354-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Nagel, G., A. Lahrz, and P. Dersch. 2001. Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol. Microbiol. 41:1249-1269. [DOI] [PubMed] [Google Scholar]
- 35.Newman, M. A., E. von Roepenack-Lahaye, A. Parr, M. J. Daniels, and J. M. Dow. 2001. Induction of hydroxycinnamoyl-tyramine conjugates in pepper by Xanthomonas campestris, a plant defense response activated by hrp gene-dependent and hrp gene-independent mechanisms. Mol. Plant-Microbe Interact. 14:785-792. [DOI] [PubMed] [Google Scholar]
- 36.Noel, L., F. Thieme, D. Nennstiel, and U. Bonas. 2001. cDNA-AFLP analysis unravels a genome-wide hrpG-regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 41:1271-1281. [DOI] [PubMed] [Google Scholar]
- 37.Norte, V. A., M. R. Stapleton, and J. Green. 2003. PhoP-responsive expression of the Salmonella enterica serovar Typhimurium slyA gene. J. Bacteriol. 185:3508-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Occhialini, A., S. Cunnac, N. Reymond, S. Genin, and C. Boucher. 2005. Genome-wide analysis of gene expression in Ralstonia solanacearum reveals that the hrpB gene acts as a regulatory switch controlling multiple virulence pathways. Mol. Plant-Microbe Interact. 18:938-949. [DOI] [PubMed] [Google Scholar]
- 39.Oku, T., A. M. Alvarez, and C. I. Kado. 1995. Conservation of the hypersensitivity-pathogenicity regulatory gene hrpX of Xanthomonas campestris and X. oryzae. DNA Sequence 5:245-249. [DOI] [PubMed] [Google Scholar]
- 40.Qian, W., Y. Jia, S.-X. Ren, Y.-Q. He, J.-X. Feng, L.-F. Lu, Q. Sun, G. Ying, D.-J. Tang, H. Tang, W. Wu, P. Hao, L. Wang, B.-L. Jiang, S. Zeng, W.-Y. Gu, G. Lu, L. Rong, Y. Tian, Z. Yao, G. Fu, B. Chen, R. Fang, B. Qiang, Z. Chen, G.-P. Zhao, J.-L. Tang, and C. He. 2005. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 15:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Revell, P. A., and V. L. Miller. 2000. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol. Microbiol. 35:677-685. [DOI] [PubMed] [Google Scholar]
- 42.Reverchon, S., W. Nasser, and J. Robert-Baudouy. 1994. pecS: a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol. Microbiol. 11:1127-1139. [DOI] [PubMed] [Google Scholar]
- 43.Reverchon, S., C. Rouanet, D. Expert, and W. Nasser. 2002. Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J. Bacteriol. 184:654-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 45.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 46.Schulte, R., and U. Bonas. 1992. A Xanthomonas pathogenicity locus is induced by sucrose and sulfur-containing amino acid. Plant Cell 4:79-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seoane, A. S., and S. B. Levy. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 177:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stapleton, M. R., V. A. Norte, R. C. Read, and J. Green. 2002. Interaction of the Salmonella typhimurium transcription and virulence factor SlyA with target DNA and identification of members of the SlyA regulon. J. Biol. Chem. 277:17630-17637. [DOI] [PubMed] [Google Scholar]
- 49.Sutherland, M. W. 1991. The generation of oxygen radicals during host plant responses to infection. Physiol. Mol. Plant Pathol. 39:79-93. [Google Scholar]
- 50.Swift, S., M. J. Lynch, L. Fish, D. F. Kirke, J. M. Tomas, S. A. B. G. Stewart, and P. Williams. 1999. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect. Immun. 67:5192-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang, D.-J., Y.-Q. He, J.-X. Feng, B.-R. He, B,-L. Jiang, G.-T. Lu, B. Chen, and J.-L. Tang. 2005. Xanthomonas campestris pv. campestris possesses a single gluconeogenic pathway that is required for virulence. J. Bacteriol. 187:6231-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang, J.-L., C. L. Gough, C. E. Barber, J. M. Dow, and M. J. Daniels. 1987. Molecular cloning of protease gene(s) from Xanthomonas campestris pv. campestris: expression in Escherichia coli and role in pathogenicity. Mol. Gen. Genet. 210:443-448. [Google Scholar]
- 53.Tang, J.-L., Y.-N. Liu, C. E. Barber, J. M. Dow, J. C. Wootton, and M. J. Daniels. 1991. Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris. Mol. Gen. Genet. 226:409-417. [DOI] [PubMed] [Google Scholar]
- 54.Tang, X., Y. Xiao, and J. M. Zhou. 2006. Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant-Microbe. Interact. 19:1159-1166. [DOI] [PubMed] [Google Scholar]
- 55.Thieme, F., R. Koebnik, T. Bekel, C. Berger, J. Boch, D. Buttner, C. Caldana, L. Gaigalat, A. Goesmann, S. Kay, O. Kirchner, C. Lanz, B. Linke, A. C. McHardy, F. Meyer, G. Mittenhuber, D. H. Nies, U. Niesbach-Klosgen, T. Patschkowski, C. Ruckert, O. Rupp, S. Schneiker, S. C. Schuster, F. J. Vorholter, E. Weber, A. Puhler, U. Bonas, D. Bartels, and O. Kaiser. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187:7254-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomson, N. R., A. Cox, B. W. Bycroft, G. S. Stewart, P. Williams, and G. P. Salmond. 1997. The rap and hor proteins of Erwinia, Serratia and Yersinia: a novel subgroup in a growing superfamily of proteins regulating diverse physiological processes in bacterial pathogens. Mol. Microbiol. 26:531-544. [DOI] [PubMed] [Google Scholar]
- 57.Torres, A. G., and J. B. Kaper. 2003. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect. Immun. 71:4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsuge, S., A. Furutani, R. Fukunaka, T. Oku, K. Tsuno, H. Ochiai, Y. Inoue, H. Kaku, and Y. Kubo. 2002. Expression of Xanthomonas oryzae pv. oryzae hrp genes in a novel synthetic medium, XOM2. J. Gen. Plant Pathol. 68:363-371. [Google Scholar]
- 59.Turner, P., C. E. Barber, and M. J. Daniels. 1985. Evidence for clustered pathogenicity genes in Xanthomonas campestris pv. campestris. Mol. Gen. Genet. 199:338-343. [Google Scholar]
- 60.Wengelnik, K., C. Marie, M. Russel, and U. Bonas. 1996. Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive reaction. J. Bacteriol. 178:1061-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wengelnik, K., and U. Bonas. 1996. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178:3462-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wengelnik, K., G. Van den Ackerveken, and U. Bonas. 1996. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol. Plant-Microbe Interact. 9:704-712. [DOI] [PubMed] [Google Scholar]
- 63.Wilkinson, S. P., and A. Grove. 2004. HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans. J. Biol. Chem. 279:51442-51450. [DOI] [PubMed] [Google Scholar]
- 64.Willis, D. K., J. J. Rich, and E. M. Hrabak. 1991. hrp genes of phytopathogenic bacteria. Mol. Plant-Microbe. Interact. 4:132-138. [Google Scholar]
- 65.Windgassen, M., A. Urban, and K. E. Jaeger. 2000. Rapid gene inactivation in Pseudomonas aeruginosa. FEMS Microbl. Let. 193:201-205. [DOI] [PubMed] [Google Scholar]
- 66.Xiao, Y., Y. Lu, S. Heu, and S. W. Hutcheson. 1992. Organization and environmental regulation of the Pseudomonas syringae pv. syringae 61 hrp cluster. J. Bacteriol. 174:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]