Abstract
The D1:1 protein and its potentially occurring degradation products were overexpressed in Escherichia coli. Protein-DNA interaction is shown for the promoter region of psbAI. The D1:1 degradation products may be involved in transcription regulation of psbAI by binding in the promoter region. Additionally, C-terminal fragments of the D1 protein bind to a sequence with similarity to isiB, a gene which encodes a flavodoxin-like protein.
Oxygenic photosynthesis is the energy conversion mechanism in cyanobacteria, algae, and plants. The light-driven processes in the thylakoid membrane require two photosystems, which are linked in series. Photosystem II (PS II) is the place of water splitting and, thus, oxygen evolution. The integral components of PS II reaction centers are the D1 and D2 proteins, which carry the cofactors of charge separation. In the unicellular cyanobacterium Synechococcus sp. strain PCC 7942, the D1 protein is encoded by a gene family of three psbA genes (8). The transcription of the psbA genes is light dependent. Under low-light conditions, the main transcript is psbAI, encoding the D1:1 form of the protein. psbAII and psbAIII are transcribed under high-light conditions and encode the D1:2 form (7, 15). The two D1 forms differ at 25 amino acid residues. Intact D1 and D2 are required for charge separation and electron transport in PS II. Because of oxygen evolution at PS II, the D1 and D2 proteins are targets of reactive oxygen species (ROS). ROS are responsible for carbonylation of protein side chains (21, 22, 25). This was also shown for the D1 protein (26). ROS-induced modification of the proteins may lead to a high turnover rate of the reaction center proteins. D1 breakdown products were previously shown for cyanobacterial thylakoid membranes (13). In the case of cyanobacterial D1, according to the model of Nixon and coworkers (19), FtsH is responsible for the initial degradation steps (19, 24). Protein cleavage by FtsH can occur from both the N and the C termini (4, 30). After proteolysis of D1, the breakdown products are translocated through the central pore of the hexameric FtsH holoenzyme in an ATP-driven process from PS II into the cytoplasm (19). Since the catalytic Zn2+ binding site of FtsH is exposed on the stromal side of the thylakoid membrane, proteolysis at the N terminus of the D1 protein is probable (19). Kihara and coworkers have shown the dislocation of membrane proteins in FtsH-mediated proteolysis (14).
Additionally, there is a proteolytic event in the QB-binding pocket between helices D and E (13). These two possible endoproteolytic steps result in five potential degradation products, which are released from the membrane, probably by FtsH, into the cyanobacterial cytoplasm. As mentioned above, transcription of the three psbA genes in Synechococcus strain PCC 7942 is regulated by light. Promoter analysis showed that the psbAI promoter is one of the strongest in Synechococcus strain PCC 7942 and contains an unusual −10 element (18). The smallest fragment, promoting the transcription of a psbAI::luxAB fusion protein, starts at −54 and ends at +1 (start codon in reference 18 at +53). In addition, at least one still unidentified protein from Synechococcus strain PCC 7942 binds at the +1-to-+43 upstream region of psbAI (18). Light-regulated psbA transcription, which is mediated by the 5′ upstream region, was shown previously in other cyanobacteria (6, 7). In the present investigation, recombinant D1:1 and its potential breakdown products from Synechococcus strain PCC 7942 were expressed individually in Escherichia coli and subjected to an electrophoretic mobility shift assay. The DNA probes were derived from the psbAI upstream region.
MATERIALS AND METHODS
D1 immunodetection.
Cells were broken by French press treatment (76 MPa). The homogenate was centrifuged for 2 h at 39,000 × g at 4°C. The resulting supernatant was concentrated by precipitation with ammonium sulfate (0 to 35% saturation), and after dialysis, the precipitate was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Electroblotting of the polypeptides was performed with a Protran BA 85 nitrocellulose membrane (Schleicher & Schuell). Immunodecoration was done with αD1N, kindly provided by R. Barbato, which was raised against amino acids 2 to 238 of the D1 protein from wheat (3). Development was done with an Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore) followed by detection with a Diana II bioimager (Raytest).
Subcloning and overexpression of the pD1 protein and its “degradation” products, B1 to B5.
Synechococcus strain PCC 7942 genomic DNA was used as a template for PCR amplification. The oligonucleotides used are shown in Table 1. Sequences overlapped by three nucleotides.
TABLE 1.
PCR primers used for this study
| Primer name | Restriction enzyme | Primer sequencea |
|---|---|---|
| p97fwd | XbaI | TTAATCTAGAAGCTAAAAATTAAGGGTTTT |
| p518fwd | BamHI | ATATTGGATCCAGGGCGTAAGCGGAGA |
| (D1, B1, B4)fwd | EcoRI | ATTAGAATTCATGACCAGCATTCTTCGC |
| (B2, B5)fwd | NcoI | AATTCCATGGCTTATCCGATTTGGGAAGCC |
| B3fwd | EcoRI | ATTAGAATTCTACAAATTTGGTCAAGAG |
| p97rev | XbaI | TTAATCTAGAGAGGTTGTAAAGGGCAAG |
| p518rev | BamHI | TATCAGGATCCTGTCAATCAGTCCACC |
| B1rev | XhoI | AATTCTCGAGATAGAAATGCAGGCCGATG |
| (B2, B4)rev | XhoI | AATTCTCGAGGTAGCCGTAGTTTTGGCTCT |
| (D1, B3, B5)rev | XhoI | AATTCTCGAGACCGTGAATTGAAGGCGCAG |
Underlining indicates the restriction site.
The fragments were cloned into the EcoRI-XhoI or NcoI-XhoI sites of the expression vector pET32a (Novagen) to generate Trx-His-S fusion proteins and transformed into Escherichia coli ADA494(DE3)/pLysS (Novagen). The resulting clones were named pETD1 and pETB1 to pETB5 (Fig. 1). In order to show that binding activity was not caused by any E. coli proteins, the expression vector pET32a was transformed into the above-mentioned strain as a negative control. The Trx-His-S tag was linked to the N terminus of the D1 protein. The Trx tag was shown to enhance the solubility of hydrophobic polypeptides (27). Recombinant proteins were expressed in E. coli ADA494(DE3)/pLysS (Novagen). Liquid cultures (500 ml) were grown in Luria-Bertani broth in the presence of 50 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol at 37°C. Expression was induced at an optical density at 600 nm of 0.6 by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. The cells were shaken overnight at 21°C.
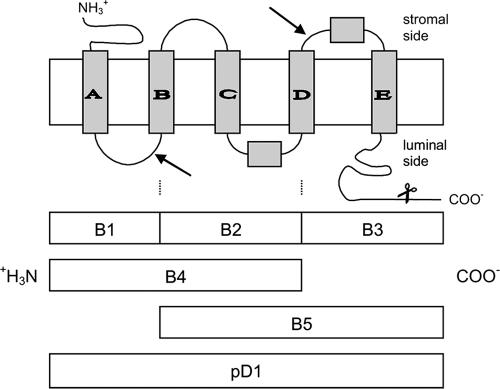
FIG. 1.
D1 protein structure in the thylakoid membrane. Gray boxes A to E represent the five transmembrane helices. The arrows indicate the protease cleavage sites. The recombinant proteins, representing the naturally occurring degradation products, are displayed below. The scissors show the point of C-terminal processing during maturation.
Protein purification.
E. coli ADA494(DE3)/pLysS cells were harvested by centrifugation at 5,000 × g for 5 min (20°C) and resuspended in 5 ml His-binding buffer (including 6 M urea for the insoluble fraction). Cells were disrupted by sonication (10 times for 10 s each [30 W]) on ice. From the soluble as well as the insoluble protein fractions, His-tagged proteins were purified according to the manufacturer's instructions (Novagen). The majority of recombinant protein was found in the soluble fraction in the case of pD1, B2, B4, and B5 and in inclusion bodies in the case of B1 and B3 (Fig. 1).
Protein analysis.
In order to exclude protein degradation by E. coli, sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed according to the method of Laemmli (16), using 10% to 15% acrylamide (acrylamide:bis-acrylamide, 30:0.8) in the separating gel. A semidry Western blot analysis on a Protran BA 85 nitrocellulose membrane (Schleicher & Schuell) was used to determine the correct sizes of the overexpressed proteins. An S protein-horseradish peroxidase conjugate (Novagen) raised against the S tag was used at a dilution of 1:5,000, followed by enhanced chemiluminescence detection (data not shown).
Construction of DNA probes.
Probes p97 and p518 were amplified by PCR. The primer sequences are shown in Table 1. Synechococcus strain PCC 7942 genomic DNA was used as a template for amplification. Restriction with XbaI (p97) and BamHI (p518) was performed in order to generate 5′ overhanging ends. The DNA probes were labeled with [α-32P]dCTP, using Klenow polymerase, according to the manufacturer's instructions.
Electrophoretic mobility shift assays.
Purified protein (168 ng to 2.535 μg) was incubated with 0.5 μg poly(dI-dC) · poly(dI-dC) and an unlabeled DNA probe (see figure legends for further information) for 20 min at room temperature in binding buffer (4 mM Tris-HCl, pH 8.0, 12 mM HEPES-OH, pH 7.9, 12% glycerol, 0.5 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol) in a final volume of 23 μl. After this prebinding step, 2 μl (2 ng) of radioactive DNA probe (3.7 kBq) was added. Binding was finished after 1 h of incubation at 4°C. The samples were loaded on a 3.5% polyacrylamide (acrylamide:bis-acrylamide, 30:0.8) gel and subjected to electrophoresis at a constant voltage of 125 V at 4°C for 2 h. Gels were dried and subsequently analyzed by autoradiography (Hyperfilm MP; Amersham).
RESULTS
Detection of a soluble D1 protein degradation product by Western blot analysis.
The occurrence of one D1 breakdown product in the cytoplasm was shown by immunodecoration (Fig. 2). The sizes of the polypeptides detected with an antibody raised against the N terminus (amino acids 2 to 238) of the D1 protein from wheat (3) were 25 kDa and 83 kDa.
FIG. 2.
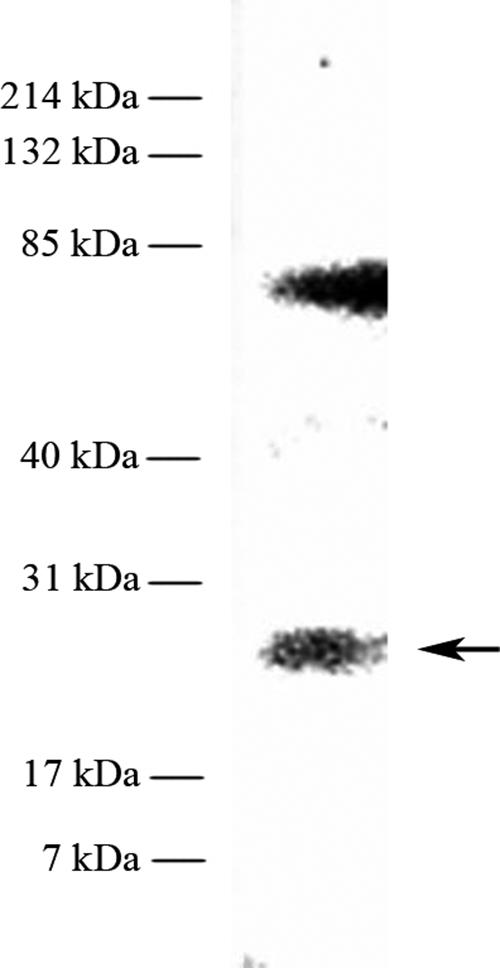
Immunological detection of a soluble D1 degradation product with αD1N. The lane contains 294 μg cytoplasmic protein extract from Synechococcus (ammonium sulfate saturation from 0 to 35%).
Specific protein-DNA binding activities of the D1:1 protein and its cleavage products at the psbAI promoter region (p97).
To demonstrate the interaction of the D1:1 protein with its own promoter region, the recombinant protein and the labeled probe p97 were subjected to electrophoretic mobility shift assays. To determine whether specific binding occurs, both nonspecific [poly(dI-dC) · poly(dI-dC)] and specific (unlabeled DNA probe in an excess of up to 170×) competitors were used in the assays. The overexpressed tags, which were subjected to the same purification procedure as the tagged proteins, were used as a negative control (Fig. 3). In the case of the pD1:1 protein, strong protein-DNA binding could be shown as a retardation shift after electrophoresis (Fig. 4). The signal strength decreased with increasing concentrations of the unlabeled DNA probe p97. The DNA binding of the different D1:1 breakdown products varied. As concluded from the signal intensity, the highest binding affinity was shown by the B2 protein, followed by B3 (Fig. 5 and 6). The C-terminal pD1:1 protein fragment B5 did not show any binding affinity for probe p97 (Fig. 6). In all cases of retardation, specific binding was once more shown by decreasing shift signals upon the addition of unlabeled DNA probe.
FIG. 3.
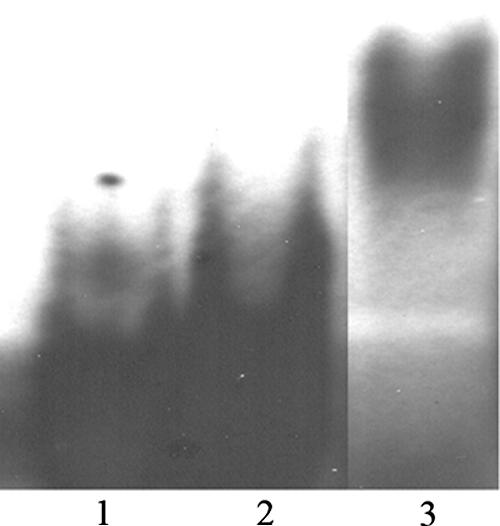
Electrophoretic mobility shift assay as a negative control for the probe p97. Lane 1 contains the assay mix without protein. Lane 2 contains 1.615 μg of overexpressed tags. Lane 3 contains 840 ng of pD1 protein as a positive control.
FIG. 4.
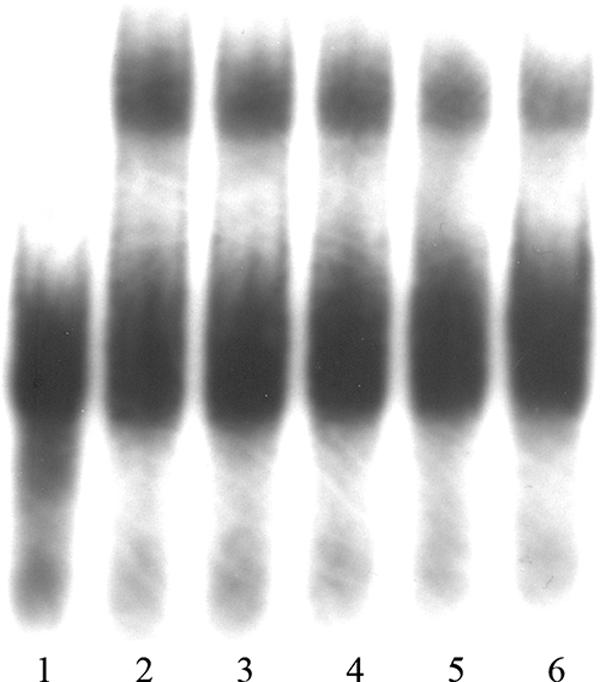
Electrophoretic mobility shift assay of probe p97 with recombinant pD1:1 protein. The sample in lane 1 contained the assay mix without protein. The sample in lane 2 contained no competitor DNA. In lanes 2 to 6, 168 ng of recombinant pD1 protein was added. The excess of unlabeled probe p97 for specific competition in lanes 3 to 6 was 20× (lane 3), 50× (lane 4), 100× (lane 5), and 170× (lane 6).
FIG. 5.
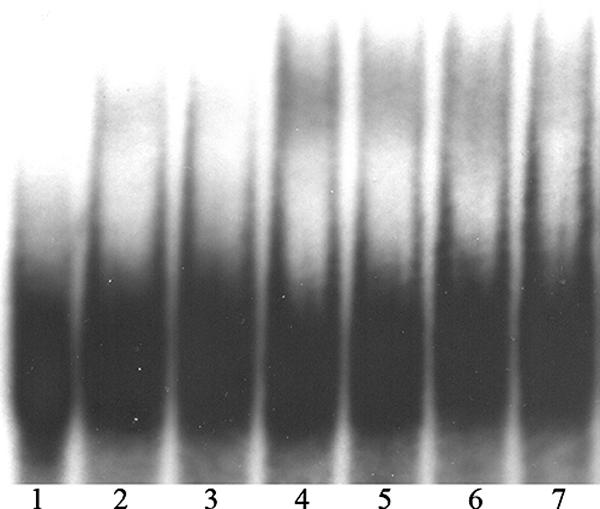
Electrophoretic mobility shift assay of probe p97 with the recombinant D1:1 fragments B1, B2, and B3. The sample in lane 1 contained the assay mix without protein. The samples in lanes 2 and 3 included 450 ng of fragment B1. The protein content in lanes 4 and 5 was 740 ng of fragment B2. In lanes 6 and 7, 580 ng of fragment B3 was applied. In lanes 3, 5, and 7, a 70× excess of unlabeled probe p97 was added.
FIG. 6.
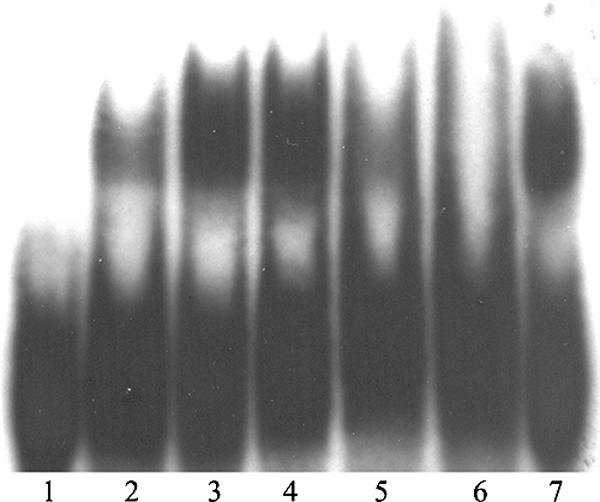
Electrophoretic mobility shift assay of probe p97 with recombinant polypeptides B1 to B5 and the pD1:1 protein. The sample in lane 1 contained the assay mix without protein. The sample in lane 2 contained 450 ng of B1 protein. The sample in lane 3 contained 740 ng of B2 protein. In lane 4, 580 ng of recombinant B3 protein was added. Lane 5 contains 1.155 μg of B4. Lane 6 contains 2.535 μg of fragment B5. In lane 7, 168 ng of pD1:1 was added.
Specific protein-DNA binding activities of the pD1:1 protein and its cleavage products at an isiB-like gene upstream of psbAI (p518).
The determination of specific binding of the purified proteins to the radiolabeled probe was done as described for probe p97. The unlabeled probe was added as a specific competitor in an excess of up to 66×. The negative control was performed as described for probe p97 (Fig. 7). The experiments showed that only the constructs containing the pD1:1 C-terminal extension interacted with the DNA probe p518 (NCBI designation Synpcc7942_0422; bp 452 to 970). The strongest retardation was shown by protein B3 (Fig. 8), followed by pD1:1 (Fig. 7) and fragment B5 (Fig. 8).
FIG. 7.
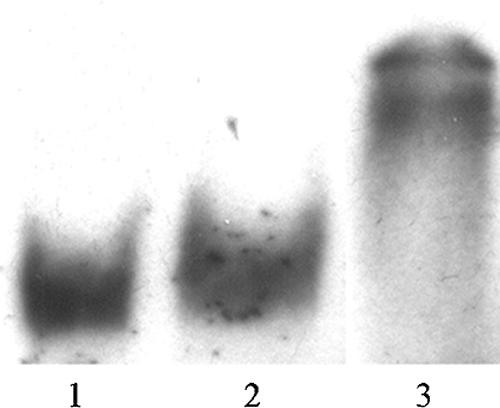
Electrophoretic mobility shift assay as a negative control for the probe p518. Lane 1 contains the assay mix without protein. Lane 2 contains 1.615 μg of overexpressed tags. Lane 3 contains 840 ng of pD1 protein.
FIG. 8.
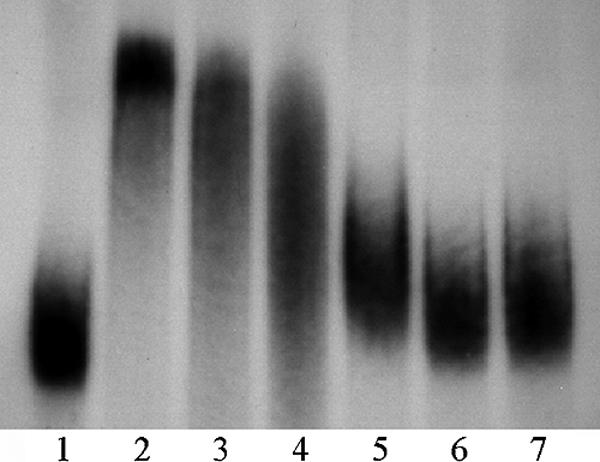
Gel retardation assay of probe p518 with recombinant proteins B3 and B5. Lane 1 displays the assay mix without protein. Lanes 2 to 4 contain 290 ng B3, and lanes 5 to 7 contain 845 ng of fragment B5. In lanes 3 and 6, a 16× excess of unlabeled probe was added. Lanes 4 and 7 contain a 66× excess of unlabeled p518.
DISCUSSION
It is well established that in cyanobacteria, psbA transcription and translation are under specific regulatory control (28). The regulation of D1 synthesis, in which the protein itself regulates its own transcription like a conveyor belt, was discussed earlier (5, 29). Specific binding of an unidentified protein to the 5′ region of the psbAI gene was shown for Synechococcus strain PCC 7942 (18). In the present paper, specific protein-DNA interactions of the recombinant pD1:1 protein and its potentially occurring proteolytic cleavage products from Synechococcus strain PCC 7942 were demonstrated. The observed multiple binding activities of the proteins led to the assumption that the D1:1 protein is involved in the regulation of psbAI transcription. The experiments showed that the D1:1 protein binds in its precursor form to the psbAI promoter region. This interaction could induce enhanced transcription of the psbAI gene under low-light conditions. In addition, the five potential D1:1 cleavage products (Fig. 1) bind to the upstream region of the gene. After ROS-mediated conformational changes in the D1 protein and proteolytic degradation, the breakdown products are possibly made accessible to DNA or released by the FtsH protease into the cytoplasm (14, 19, 23), where they contact the DNA attached to the thylakoid membrane. The occurrence of a 25-kDa soluble D1 degradation product, possibly fragment B4, has been demonstrated in the present paper (Fig. 2). The fraction did not contain membranes, as demonstrated by the absence of a signal for the mature protein of 32 kDa. The 83-kDa band cannot be assigned.
The highest binding affinity for the DNA probe p97 (Fig. 9) was shown by protein fragment B2 (Fig. 1), the cleavage product which occurs after a two-step degradation by FtsH (24). In the case of the N-terminal product B4 (Fig. 1), which results from proteolytic cleavage in the DE loop solely, the protein binds equally to the psbAI promoter region, but with a lower affinity, as concluded from the protein concentration necessary for signal detection. The absence of a signal from protein fragment B5, which consists of B2 and B3, can possibly be attributed to protein folding in E. coli.
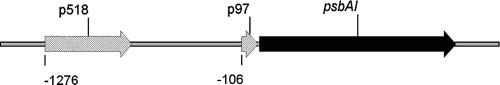
FIG. 9.
Positions of probes p97 and p518 in the 5′ upstream region of psbAI. Numbers indicate the 5′ sites of the probes relative to the start codon of psbAI.
From the obtained results in the present investigation, it can be concluded that the pD1:1 protein and its degradation products B2, B3, and B4 may influence D1:1 transcription by binding in or upstream of the promoter region under low-light conditions. The protein binding region is represented by probe p97 (Fig. 9). The importance of sequences upstream of the −35 element for promoter activity was demonstrated earlier (2, 6, 11, 18). The present investigation shows binding of the recombinant proteins between base pairs −106 and −10 upstream of the psbAI gene from Synechococcus. The resulting hypothesis is that the D1 protein regulates its own resynthesis during light-dependent turnover by its cleavage products. In the present paper, a model for the transcriptional regulation of the psbAI gene is proposed, in which the D1 protein adjusts its own transcription. After the accessibility of the D1 degradation products, fragments bind specifically to the upstream region of psbAI and thus, possibly, play the role of transcription factors.
In contrast to the N-terminal cleavage products, the C-terminal cleavage products B3 and B5 from Synechococcus (Fig. 1) show high binding affinities for the DNA probe called p518 (Fig. 9). The same is true for the entire D1 protein in its precursor form. The sequence p518, which is 1.3 to 0.8 kb upstream of psbAI (NCBI designation Synpcc7942_0422; bp 452 to 970), shows similarity to isiB (1), a gene coding for flavodoxin, a soluble electron acceptor, which substitutes for ferredoxin under iron-deficient conditions in cyanobacteria. The derepression of isiB is also triggered under iron-replete conditions by oxidative and high-light stress (10, 12). It was shown for Synechocystis that flavodoxin may be involved in cyclic electron transport around PS I in addition to ferredoxin (9). In the present paper, a specific protein-DNA interaction between the pD1:1 C terminus and the isiB-like sequence from Synechococcus is described.
One could expect that C-terminal D1:1 protein binding may inhibit the transcription of this isiB-like gene under low-light conditions. The transcription inhibition may be lifted when the cells switch from the D1:1 form to the D1:2 form of the protein upon transfer to high light. The resulting protein may then take part in cyclic electron transport, together with ferredoxin (9). At this point, it is important to take into consideration that the recombinant constructs pD1:1, B3, and B5 carry the C-terminal extension typical for the precursor protein (17), which consists of 16 amino acid residues in cyanobacteria (20). The potential C-terminal fragments B3 and B5 or the processed C-terminal extension of the D1:1 protein may be the protein-binding partner of the isiB-like DNA sequence p518 (Fig. 9) in vivo.
Acknowledgments
Susan S. Golden is thanked for sharing the information on the 5′ upstream region of psbAI in Synechococcus prior to publication, and Roberto Barbato is thanked for the kind gift of the antiserum to the D1 protein.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asayama, M., H. Kato, J. Shibato, M. Shirai, and T. Ohyama. 2002. The curved DNA structure in the 5′-upstream region of the light-responsive genes: its universality, binding factor and function for cyanobacterial psbA transcription. Nucleic Acids Res. 30:4658-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbato, R., C. A. Shipton, G. M. Giacometti, and J. Barber. 1991. New evidence suggests that the initial photoinduced cleavage of the D1-protein may not occur near the PEST sequence. FEBS Lett. 290:162-166. [DOI] [PubMed] [Google Scholar]
- 4.Chiba, S., Y. Akiyama, and K. Ito. 2002. Membrane protein degradation by FtsH can be initiated from either end. J. Bacteriol. 184:4775-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Critchley, C. 1988. The chloroplast thylakoid membrane system is a molecular conveyor belt. Photosynth. Res. 19:265-276. [DOI] [PubMed] [Google Scholar]
- 6.Eriksson, J., G. F. Salih, H. Ghebramedhin, and C. Jansson. 2000. Deletion mutagenesis of the 5′ psbA2 region in Synechocystis 6803: identification of a putative cis element involved in photoregulation. Mol. Cell Biol. Res. Commun. 3:292-298. [DOI] [PubMed] [Google Scholar]
- 7.Golden, S. S. 1995. Light-responsive gene expression in cyanobacteria. J. Bacteriol. 177:1651-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golden, S. S., J. Brusslan, and R. Haselkorn. 1986. Expression of a family of psbA genes encoding a photosystem II polypeptide in the cyanobacterium Anacystis nidulans R2. EMBO J. 5:2789-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagemann, M., R. Jeanjean, S. Fulda, M. Havaux, F. Joset, and N. Erdmann. 1999. Flavodoxin accumulation contributes to enhanced cyclic electron flow around photosystem I in salt-stressed cells of Synechocystis sp. strain PCC 6803. Physiol. Plant 105:670-678. [Google Scholar]
- 10.Havaux, M., G. Guedeney, M. Hagemann, N. Yeremenko, H. C. Matthijs, and R. Jeanjean. 2005. The chlorophyll-binding protein IsiA is inducible by high light and protects the cyanobacterium Synechocystis PCC6803 from photooxidative stress. FEBS Lett. 579:2289-2293. [DOI] [PubMed] [Google Scholar]
- 11.Ito, Y., M. Asayama, and M. Shirai. 2003. Light-responsive psbA transcription requires the −35 hexamer in the promoter and its proximal upstream element, UPE, in cyanobacteria. Biosci. Biotechnol. Biochem. 67:1382-1390. [DOI] [PubMed] [Google Scholar]
- 12.Jeanjean, R., E. Zuther, N. Yeremenko, M. Havaux, H. C. Matthijs, and M. Hagemann. 2003. A photosystem 1 psaFJ-null mutant of the cyanobacterium Synechocystis PCC 6803 expresses the isiAB operon under iron replete conditions. FEBS Lett. 549:52-56. [DOI] [PubMed] [Google Scholar]
- 13.Kanervo, E., N. Murata, and E.-M. Aro. 1998. Massive breakdown of the photosystem II polypeptides in a mutant of the cyanobacterium Synechocystis sp. PCC 6803. Photosynth. Res. 57:81-91. [Google Scholar]
- 14.Kihara, A., Y. Akiyama, and K. Ito. 1999. Dislocation of membrane proteins in FtsH-mediated proteolysis. EMBO J. 18:2970-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulkarni, R. D., M. R. Schaefer, and S. S. Golden. 1992. Transcriptional and posttranscriptional components of psbA response to high light intensity in Synechococcus sp. strain PCC 7942. J. Bacteriol. 174:3775-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Marder, J. B., P. Goloubinoff, and M. Edelman. 1984. Molecular architecture of the rapidly metabolized 32-kilodalton protein of photosystem II. Indications for COOH-terminal processing of a chloroplast membrane polypeptide. J. Biol. Chem. 259:3900-3908. [PubMed] [Google Scholar]
- 18.Nair, U., C. Thomas, and S. S. Golden. 2001. Functional elements of the strong psbAI promoter of Synechococcus elongatus PCC 7942. J. Bacteriol. 183:1740-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nixon, P. J., M. Barker, M. Boehm, R. De Vries, and J. Komenda. 2005. FtsH-mediated repair of the photosystem II complex in response to light stress. J. Exp. Bot. 56:357-363. [DOI] [PubMed] [Google Scholar]
- 20.Nixon, P. J., J. T. Trost, and B. A. Diner. 1992. Role of the carboxy terminus of polypeptide D1 in the assembly of a functional water-oxidizing manganese cluster in photosystem II of the cyanobacterium Synechocystis sp. PCC 6803: assembly requires a free carboxyl group at C-terminal position 344. Biochemistry 31:10859-10871. [DOI] [PubMed] [Google Scholar]
- 21.Nyström, T. 2005. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 24:1311-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma, J., M. Panico, C. A. Shipton, F. Nilsson, H. R. Morris, and J. Barber. 1997. Primary structure characterization of the photosystem II D1 and D2 subunits. J. Biol. Chem. 272:33158-33166. [DOI] [PubMed] [Google Scholar]
- 23.Silva, P., Y. J. Choi, H. A. Hassan, and P. J. Nixon. 2002. Involvement of the HtrA family of proteases in the protection of the cyanobacterium Synechocystis PCC 6803 from light stress and in the repair of photosystem II. Philos. Trans. R. Soc. Lond. B 357:1461-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva, P., E. Thompson, S. Bailey, O. Kruse, C. W. Mullineaux, C. Robinson, N. H. Mann, and P. J. Nixon. 2003. FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp. PCC 6803. Plant Cell 15:2152-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stadtman, E. R. 1993. Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu. Rev. Biochem. 62:797-821. [DOI] [PubMed] [Google Scholar]
- 26.Tambussi, E. A., G. C. Bartoli, J. Beltrano, J. J. Guiamet, and J. L. Araus. 2000. Oxidative damage to thylakoid proteins in water-stressed leaves of wheat (Triticum aestivum). Physiol. Plant 108:398-404. [Google Scholar]
- 27.Therien, A. G., M. Glibowicka, and C. M. Deber. 2002. Expression and purification of two hydrophobic double-spanning membrane proteins derived from the cystic fibrosis transmembrane conductance regulator. Protein Expr. Purif. 25:81-86. [DOI] [PubMed] [Google Scholar]
- 28.Tyystjärvi, T., M. Herranen, and E.-M. Aro. 2001. Regulation of translation elongation in cyanobacteria: membrane targeting of the ribosome nascent-chain complexes controls the synthesis of D1 protein. Mol. Microbiol. 40:476-484. [DOI] [PubMed] [Google Scholar]
- 29.Tyystjärvi, T., P. Mulo, P. Mäenpää, and E.-M. Aro. 1996. D1 polypeptide degradation may regulate psbA gene expression at transcriptional and translational levels in Synechocystis sp. PCC 6803. Photosynth. Res. 47:111-120. [DOI] [PubMed] [Google Scholar]
- 30.Wiklund, R., G. F. Salih, P. Mäenpää, and C. Jansson. 2001. Engineering of the protein environment around the redox-active TyrZ in photosystem II. The role of F186 and P162 in the D1 protein of Synechocystis 6803. Eur. J. Biochem. 268:5356-5364. [DOI] [PubMed] [Google Scholar]