Abstract
The activity of a fourth conserved tyrosine site-specific recombinase (Tsr) of Bacteroides fragilis was characterized. Its gene, tsr19, is adjacent to mpi, encoding the global DNA invertase regulating capsular polysaccharide biosynthesis. Unlike the other described Tsrs of B. fragilis, Tsr19 brings about inversion of two DNA regions, one local and one located distantly.
Bacteroides fragilis is a member of the intestinal microbiota and can become an opportunistic pathogen if released into extraintestinal sites. We have found that DNA inversions regulate expression of key virulence factors and may also modulate expression of surface factors involved in intestinal symbiosis. The B. fragilis NCTC 9343 genome encodes a large number of site-specific recombinases: three members of the serine site-specific recombinase (Ssr) family and 28 members of the tyrosine site-specific recombinase (Tsr) family. We have previously shown that two of the ssr genes and nine of the tsr genes are conserved in the species (3). We have demonstrated that Mpi is a global DNA invertase of the Ssr family that is involved in inverting 13 distinct DNA regions that are flanked by inverted repeats (IRs), including the promoters of seven of the capsular polysaccharide biosynthesis regions (3). All of the regions that Mpi inverts are located at regions remote from mpi. We have also shown that three of the conserved tsr genes (tsr15, tsr25, and tsr26) encode DNA invertases that act locally to invert promoter regions immediately downstream of their gene (6).
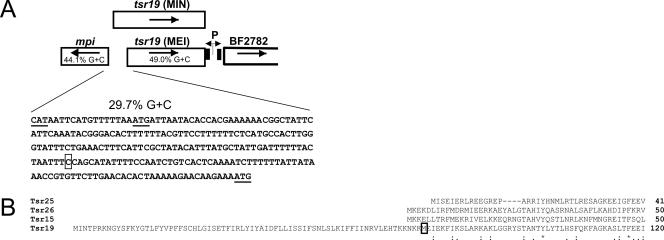
Another conserved tsr, tsr19, is present on the B. fragilis genome adjacent to mpi and is transcribed divergently (Fig. 1A). The genomes of the two sequenced B. fragilis strains have been annotated such that the start codon of mpi is separated from the start codon of the divergently transcribed tsr19 by only 15 bp (2, 4). The close clustering of these genes does not allow for the presence of intergenic promoter regions for either mpi or tsr19. Downstream of tsr19 is a 315-bp region that is flanked by inverted repeats and undergoes inversion (4). It has been suggested that a promoter within this invertible region may be involved in transcription of mpi (4). We sought to determine whether Tsr19 is a DNA invertase and, if so, whether it inverts this downstream IR region. We also sought to determine whether there is a functional promoter in this invertible region and, if so, whether it is necessary for the transcription of mpi.
FIG. 1.
Open reading frame map of the mpi-tsr19 genetic region. A. mpi and tsr19 are divergently transcribed. The predicted start of the tsr19 gene based on the genome annotation (beginning with MIN) and an alternative predicted start site (beginning with MEI) are shown. Inverted repeat regions downstream of tsr19 are indicated as black boxes with an intervening consensus promoter designated as P. The G+C content of the 234-bp intergenic region resulting from the MEI start of Tsr19 as well as the G+C contents of the adjacent genes are shown. The mpi start codon and the annotated and putative alternate tsr19 start codons are underlined. The first base of the insert contained in pLEC84 is boxed. B. Alignment of Tsr19 with the three other functionally characterized tyrosine family site-specific recombinases of B. fragilis. A methionine representing an alternative putative start site for Tsr19 is boxed.
Alignment of the three characterized Tsr family DNA invertases of B. fragilis with Tsr19 revealed that Tsr19 has a large N-terminal region of approximately 70 amino acids that the other three products lack (Fig. 1B). Furthermore, the second methionine of Tsr19 aligns within 4 amino acids of the initial methionines of the other three Tsrs. If it is assumed that the ATG corresponding to this second methionine of Tsr19 is the actual start codon, the percent G+C content of the resulting 234-bp intergenic region would be 29.7%, very dissimilar to the G+C contents of the surrounding genes (44.1% and 49.0% for mpi and tsr19, respectively). The low G+C content of this region suggests that it may be an intergenic region rather than part of the coding region of tsr19.
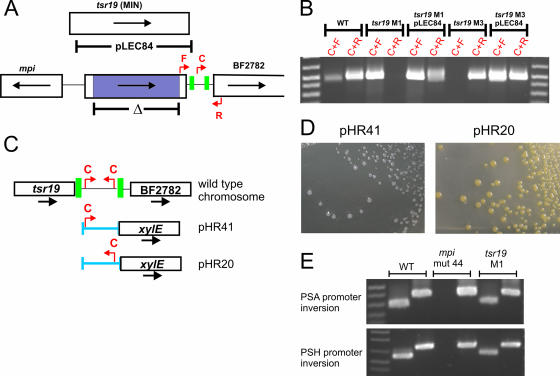
Based on the finding that Tsr15, Tsr25, and Tsr26 act on inverted repeat regions in their immediate downstream region, we predicted that Tsr19 has DNA invertase activity and mediates the inversion of its downstream IR region. Deletion mutants of tsr19 where a 733-bp internal region was removed by allelic replacement were created (primers used in this study are listed in Table 1). The resulting mutants were tested for the inversion of the tsr19 downstream region by using forward and central or central and reverse primer pairs. As shown in Fig. 2B, compared to that in the wild type, the DNA region between the IRs does not invert in the tsr19 mutants. Mutants where this region was locked in each orientation were obtained (mutants M1 and M3), demonstrating that Tsr19 is necessary for the inversion of this region in both directions. For complementation studies, tsr19 was cloned into the BamHI site of the B. fragilis expression vector pFD340 (5), creating pLEC84. Based on the possibility that tsr19 may be shorter than predicted, pLEC84 was created so that the first 141 bp of the larger gene was omitted (Fig. 2A), and this shorter clone was tested for invertase activity. As shown in Fig. 2B, pLEC84 is able to mediate the inversion of this region in both Δtsr19 M1 and Δtsr19 M3. These data demonstrate that Tsr19 is a DNA invertase involved in the inversion of its downstream IR region and that the N terminus of the annotated gene is not necessary for its activity.
TABLE 1.
Primers used in this study
| Purpose | Primer or plasmid | Primer sequencea |
|---|---|---|
| Creation of tsr19 deletion mutants | tsr19 flank 1 forward | TACCATGGTGCTTCAGAGAAGGTGACATACAT |
| tsr19 flank 1 reverse | TCGGATCCATTTGCCATAGTTCGTGAGTCTCT | |
| tsr19 flank 2 forward | CTGGATCCACTTTCTGTTCCTTGAAACCATTC | |
| tsr19 flank 2 reverse | ATCCATGGTATTGGCTGTGCTGTAACG | |
| Demonstration of inversion of tsr19 downstream region | tsr19 forward | TGCTTCAGAGAAGGTGACATACAT |
| tsr19 central | ATTCATTTGTTTTATCGCTG | |
| tsr19 reverse | ATACAATCCTTTTAATGACT | |
| Cloning of tsr19 for complementation (pLEC84) | tsr19 forward | CTCGGATCCAGCATATTTTCCAATCTGTCACTC |
| tsr19 reverse | TCTGGATCCGCAGCGATAAAACAAATGAATCT | |
| Demonstration of inversion of PSA and PSH promoter regions | PSA forward | TGTGTAAATGATAGGAGGCTAGGG |
| PSA central | ACCTTTTTTCGACCTTTTCTAAAAATC | |
| PSA reverse | AGAAAACTCCTGGTCCTTCTTTG | |
| PSH forward | CTTTGCCAGTTCCCGTATGT | |
| PSH central | TGATGAAATTCAGAACCGGATA | |
| PSH reverse | CGCTCGTTCTTGACGATGTA | |
| Construction of xylE transcriptional fusion clones in pLEC23 | pHR20 and pHR41 | ACGGATCCATTCATTTGTTTTATCGCTG |
| ACGGATCCTGCGGAATAGCGAAAAGATA | ||
| pLEC55 | TAGGATCCGTACTTACTCTCAAATAAGC | |
| TAGGATCCTAAACATAAAACAGGGGCTGG | ||
| pMCL25 | ACGGATCCATTCATTTGTTTTATCGCTG | |
| GTTGGGATCCCTTTTAGAAATGTTTGCGATAGTTG | ||
| pMCL26 | ACGGATCCTGCGGAATAGCGAAAAGATA | |
| GTTGGGATCCCTTTTAGAAATGTTTGCGATAGTTG | ||
| pMCL21 | CGTTGGATCCTGATAAATCAGTCCTTTTGTTGC | |
| TCAGGATCCGCTAATGGTTTAATGTTCATAGTTACTT | ||
| pMCL22 | ACGTGGATCCAATTTTAATGAATATATGGCTATGCA | |
| TCAGGATCCGCTAATGGTTTAATGTTCATAGTTACTT | ||
| pMCL23 | CGTTGGATCCTGATAAATCAGTCCTTTTGTTGC | |
| GTGAGGATCCAATGTTAAGTATTTGTTGTTTCTTTAGTG | ||
| pMCL24 | ACGTGGATCCAATTTTAATGAATATATGGCTATGCA | |
| GTGAGGATCCAATGTTAAGTATTTGTTGTTTCTTTAGTG | ||
| Demonstration of inversion of distant inverted repeat region | Forward | AACAGTATCGCCTGCCTTTAATAC |
| Central | GTTGCTGCAAATGTATGGATTCT | |
| Reverse | TCAATGACACTTGTTTCATGTTTG | |
| PCR amplification of groEL probe for Northern blot analysis | groEL probe forward | GTGCAAGCCCTATGGATATTAAAC |
| groEL probe reverse | AATACCGGTAGTCTCGTCATCATT |
Restriction sites are underlined.
FIG. 2.
Tsr19 is necessary for inversion of the downstream promoter region. A. Open reading frame map of the mpi-tsr19 region showing the region that was deleted in the tsr19 mutant (purple) and the extent of DNA used to complement this mutant (pLEC84). The red arrows show the primers used to assay for inversion. Inverted repeats are shown as green boxes. B. Ethidium bromide-stained agarose gel of the inversion event from the wild type (WT), tsr19 mutants, and complemented mutants. Primers are those shown in panel A. C. Genetic regions (blue) cloned upstream of xylE in relation to the orientation of primer C, which undergoes inversion in wild-type bacteria and therefore is present in each orientation from a population of wild type bacteria. pHR20 and pHR41 contain identical DNA cloned in opposite orientations upstream of xylE. D. Analysis of functional promoter activity from B. fragilis 9343 containing pHR20 or pHR41 in trans. The transconjugants were sprayed with catechol, which is the substrate for XylE and convert the colonies to yellow if a functional promoter has been cloned. E. Ethidium bromide-stained agarose gels showing the inversion of the PSA and PSH promoter regions from the wild type, mpi deletion mutant 44, and Δtsr19 M3.
Like the other three characterized Tsr-regulated IR regions of B. fragilis, the region between the IRs of the tsr19 downstream region has a perfect match with the described B. fragilis consensus promoter sequence (1). As it was predicted that mpi may be transcribed from this invertible promoter, it was necessary to determine if this region in fact contained an active promoter. A 284-bp DNA region between the IRs was PCR amplified and cloned in each orientation into the BamHI site of the xylE reporter plasmid pLEC23 (3) (Fig. 2C). The resulting clones, pHR20 and pHR41, were conjugally transferred into B. fragilis, and the presence of a promoter was qualitatively assessed by spraying the plates with the substrate catechol and by performing quantitative XylE assays as described previously (3). As shown in Fig. 2D, when this putative promoter is oriented to drive transcription of xylE (pHR20), 356 ± 23 milliunits XylE/mg protein is produced, versus 0 milliunits XylE/mg protein when the promoter is oriented in the opposite direction in clone pHR41 (mean ± standard deviation for two assays). Therefore, in the Δtsr19 M3 mutant, the promoter is oriented so that it could constitutively transcribe mpi if this was indeed the promoter for mpi, whereas in the tsr19 M1 mutant, the promoter is locked in the opposite orientation and thus could not drive transcription of mpi.
If the invertible promoter downstream of tsr19 is necessary for the transcription of mpi, Δtsr19 M1 should not express Mpi. To determine whether Mpi is synthesized in Δtsr19 M1, we screened two polysaccharide promoter regions (PSA and PSH) known to invert in an Mpi-dependent manner (3). As a control, we used an mpi deletion mutant (mutant 44) which is unable to invert its polysaccharide promoters and has the PSA promoter locked ON and the PSH promoter locked OFF (3). As shown in Fig. 2E, inversion of these two polysaccharide promoter regions in Δtsr19 M1 was similar to that in the wild type. Therefore, the promoter between the IRs of the tsr19 downstream region is not necessary for expression of Mpi.
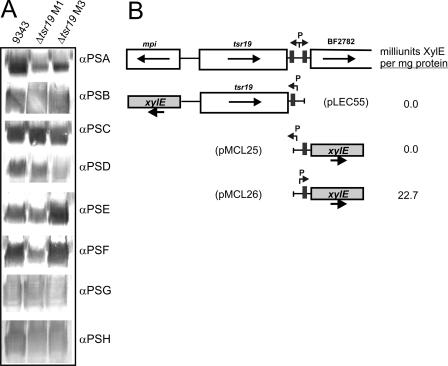
We next performed Western immunoblot analysis to determine if expression of any of the eight characterized capsular polysaccharides was affected by the deletion of tsr19. As shown in Fig. 3A, each of the eight capsular polysaccharides was synthesized in both Δtsr19 M1 and Δtsr19 M3, demonstrating that the invertible promoter downstream of tsr19 does not affect capsular polysaccharide biosynthesis. To determine if this invertible promoter region may instead drive transcription of the downstream gene BF2782, xylE transcriptional fusion clones were constructed as shown in Fig. 3B. For both constructs, the upstream IR was omitted so that the promoter was locked either ON (pMCL26) or OFF (pMCL25) by PCR amplifying the region to be cloned from Δtsr19 M1 or Δtsr19 M3, respectively (Fig. 3B). No XylE activity was detected from pMCL25, in which the invertible promoter is oriented away from BF2782; however, when the promoter is oriented toward the gene (pMCL26), 22.7 milliunits XylE/mg protein is produced. Therefore, under the conditions of this assay, transcription of BF2782 is dependent on the invertible promoter in its upstream region.
FIG. 3.
The Tsr19-regulated invertible promoter does not affect capsular polysaccharide expression. A. Western immunoblot analysis of whole-cell lysates of the wild type, Δtsr19 M1, and Δtsr19 M3 probed with antisera to each of the eight capsular polysaccharides. B. Analysis of the transcription of mpi and BF2782 by the Tsr19-regulated invertible promoter, using xylE transcriptional fusion clones. pLEC55 detects transcription into mpi when the invertible promoter is oriented toward mpi, and pMCL25 and pMCL26 detect transcription into BF2782 when the invertible promoter is cloned in either the OFF or ON orientation, respectively. The resulting milliunits of XylE activity per milligram protein are reported for each of these constructs when in trans in B. fragilis 9343.
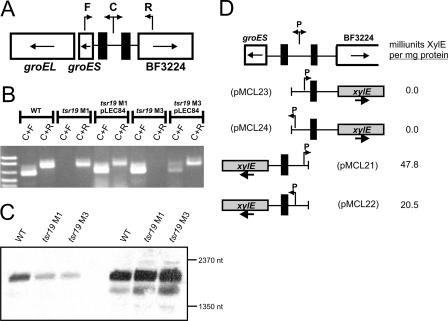
Inverted repeats similar to those downstream of tsr19 were detected in one other location of the B. fragilis genome at a distance of approximately 550 kb from tsr19 (4). These IRs are separated by 261 bp of DNA that undergoes inversion (2, 4). To determine if Tsr19 is responsible for this inversion, PCR was performed using DNAs from the tsr19 mutants and complemented mutants with forward and central or central and reverse primer pairs (Table 1; Fig. 4A). As shown in Fig. 4B, this distal region is unable to invert in tsr19 mutants, and inversion is restored when tsr19 is added in trans (pLEC84). In addition, in Δtsr19 M1, this region is locked in the orientation opposite to that of Δtsr19 M3.
FIG. 4.
Tsr19 is necessary for inversion of a distant chromosomal region. A. Schematic showing the genetic regions flanking the Tsr19-like inverted repeats (filled boxes). The direction of transcription of each gene is shown with arrows inside the boxes, and primers used to demonstrate inversion of DNA between the IRs are indicated as F, C, and R. B. Ethidium bromide-stained agarose gel of the resulting PCR products from the wild type (WT), tsr19 mutants, and complemented mutants, using the F, C, and R primers shown in panel A. C. Northern blot analysis of wild type and tsr19 mutants grown to early log phase (optical density at 600 nm = 0.3) (first three lanes) and mid-log phase (optical density at 600 nm = 0.8) (last three lanes), using 30 μg of RNA per well probed with an internal portion of groEL. D. Analysis of the transcription of groES and BF3224 by the adjacent Tsr19-regulated invertible promoter, using xylE transcriptional fusion clones. pMCL23 and pMCL24 detect transcription into BF3224 when the invertible promoter is cloned in either the ON or OFF orientation, respectively. pMCL21 and pMCL22 detect transcription into groES when the invertible promoter is cloned in either the OFF or ON orientation, respectively. The resulting milliunits of XylE activity per milligram of protein are reported for each of these constructs when in trans in B. fragilis 9343.
This second Tsr19-regulated invertible region contains a perfect B. fragilis consensus promoter sequence which may be involved in the transcription of either of two divergently transcribed operons. The groESL operon is located 163 bp from the invertible region, and a divergently transcribed gene (BF3224) is located 223 bp from the invertible region. The genetic organization and close proximity of the groES start codon to the invertible promoter suggested that transcription of the groESL operon might be regulated or enhanced by this invertible promoter. To determine if groESL transcription is dependent on this invertible promoter, Northern blot analysis was employed using RNA from the wild type, Δtsr19 M1, and Δtsr19 M3. Using an internal portion of groEL as a probe, we found that there were similar amounts of groEL transcript produced by Δtsr19 M3 (where the promoter is locked OFF for groESL transcription) and Δtsr19 M1 (where the promoter would constitutively drive transcription) from both early- and mid-log-phase cultures (Fig. 4C). These data demonstrate that this invertible promoter is not necessary for transcription of groEL. The hybridizing transcript size of ∼2,000 nucleotides is the expected size for the groESL operon. To determine if this invertible promoter is involved in the transcription of the divergently transcribed gene BF3224, xylE transcriptional fusions similar to those of the other Tsr19-regulated invertible promoter region were constructed (Fig. 4C). No XylE activity was detected when this promoter was locked in either the OFF or ON orientation for transcription into BF3224. In addition, when the invertible promoter was locked to constitutively transcribe groES, no augmentation of XylE activity was detected compared to when this promoter was locked OFF for groES transcription. These data demonstrate that under the conditions in which the bacteria were grown, this invertible promoter does not contribute to the transcription either of the groESL operon or of BF3224. The biological significance of this invertible promoter, therefore, is yet to be determined.
This study has demonstrated that a fourth conserved Tsr encoded by B. fragilis has DNA invertase activity. The three conserved Tsrs previously shown to have DNA invertase activity all localized to a single branch of a B. fragilis 9343 Tsr cladogram based on global protein alignment (3). Tsr19 also segregated to this same branch, allowing us to predict that this branch represents Tsr proteins with DNA invertase activity. Tsrs with other functions, such as transposition and integration, may segregate to other branches of this tree. Of the nine conserved Tsrs, five localize to this DNA invertase branch, and one other localizing to a distinct branch is predicted to be XerD (2), which is involved in chromosomal segregation. Therefore, few of the tsr genes that encode transposases or integrases are conserved in the species.
Tsr15, Tsr25, and Tsr26 each recognize almost identical inverted repeats; however, each is very specific in the inversion of its own downstream IR region (6). In contrast, Tsr19 recognizes two different, albeit related, IRs, one in its immediate downstream region, and one located distantly on the chromosome. This is the first report of a Tsr family DNA invertase of B. fragilis acting on a region distant from its gene, and Tsr19 shares this property with Mpi, the global Ssr of B. fragilis.
Studies designed to locate the mpi promoter by using the xylE promoter probing construct did not detect any promoter activity immediately upstream of mpi (data not shown) or in regions extending beyond the invertible promoter downstream of tsr19 (pLEC55) (Fig. 3B). Northern blot analysis also failed to detect an mpi transcript, suggesting that the gene is poorly transcribed from a very weak promoter. Due to this low level of mpi transcript, we used the functional assay of polysaccharide promoter inversion to assay for Mpi activity. The fact that Mpi activity was detected when the invertible promoter was locked in the OFF orientation for mpi transcription demonstrates that it is not necessary for its transcription.
We have shown that the invertible promoter downstream of tsr19 is involved in transcription of at least one of the genes in its downstream region, BF2782. The products of the three genes of this region are similar to products involved in various aspects of extracellular polysaccharide biosynthesis and secretion. As the data have shown that this invertible promoter is not involved in regulation of capsular polysaccharide biosynthesis, our continuing studies will examine the effects of this promoter on the synthesis of an as-yet-uncharacterized extracellular polysaccharide.
Acknowledgments
This work was supported by grant AI44193 from the National Institutes of Health (NIAID).
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. [DOI] [PubMed] [Google Scholar]
- 2.Cerdeno-Tarraga, A., S. Patrick, L. Crossman, G. Blakely, V. Abratt, N. Lennard, I. Poxton, B. Duerden, B. Harris, M. Quail, A. Barron, L. Clark, C. Corton, J. Doggett, M. Holden, N. Larke, A. Line, A. Lord, H. Norbertczak, D. Ormond, C. Price, E. Rabbinowitsch, J. Woodward, B. Barrell, and J. Parkhill. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463-1465. [DOI] [PubMed] [Google Scholar]
- 3.Coyne, M. J., K. G. Weinacht, C. M. Krinos, and L. E. Comstock. 2003. Mpi recombinase globally modulates the surface architecture of a human commensal bacterium. Proc. Natl. Acad. Sci. USA 100:10446-10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 101:14919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith, C. J., M. B. Rogers, and M. L. McKee. 1992. Heterologous gene expression in Bacteroides fragilis. Plasmid 27:141-154. [DOI] [PubMed] [Google Scholar]
- 6.Weinacht, K. G., H. Roche, C. M. Krinos, M. J. Coyne, J. Parkhill, and L. E. Comstock. 2004. Tyrosine site-specific recombinases mediate DNA inversions affecting the expression of outer surface proteins of Bacteroides fragilis. Mol. Microbiol. 53:1319-1330. [DOI] [PubMed] [Google Scholar]