Abstract
In the opportunistic human pathogen Pseudomonas aeruginosa, quorum sensing (QS) is crucial for virulence. The RsaL protein directly represses the transcription of lasI, the synthase gene of the main QS signal molecule. On the basis of sequence homology, RsaL cannot be predicted to belong to any class of characterized DNA-binding proteins. In this study, an in silico model of the RsaL structure was inferred showing that RsaL belongs to the tetrahelical superclass of helix-turn-helix proteins. The overall structure of RsaL is very similar to the N-terminal domain of the lambda cI repressor and to the POU-specific domain of the mammalian transcription factor Oct-1 (Oct-1 POUs). Moreover, residues of Oct-1 POUs important for structural stability and/or DNA binding are conserved in the same positions in RsaL and in its homologs found in GenBank. These residues were independently replaced with Ala, and the activities of the mutated variants of RsaL were compared to that of the wild-type counterpart in vivo by complementation assays and in vitro by electrophoretic mobility shift assays. The results validated the RsaL in silico model and showed that residues Arg 20, Gln 38, Ser 42, Arg 43, and Glu 45 are important for RsaL function. Our data indicate that RsaL could be the founding member of a new protein family within the tetrahelical superclass of helix-turn-helix proteins. Finally, the minimum DNA sequence required for RsaL binding on the lasI promoter was determined, and our data support the hypothesis that RsaL binds DNA as a dimer.
Quorum sensing (QS) is a widespread bacterial communication system based on the production of signal molecules. In a bacterial population, when the concentration of the signal molecule reaches a threshold corresponding to a certain cell density, individual cells of the population coordinately respond by reprogramming the expression levels of several genes (16).
Pseudomonas aeruginosa is an important opportunistic human pathogen able to establish severe and hard-to-eradicate infections in immunocompromised hosts, such as burned, AIDS, and cystic fibrosis patients. In the last case, a persistent P. aeruginosa lung infection is the major cause of death (14). QS plays a key role in the P. aeruginosa infection process because it regulates the production of many virulence factors, as well as biofilm formation (24). Moreover, the QS signal molecules themselves play important roles in the cross talk between host and parasite (27). In animal models, P. aeruginosa mutants impaired in QS display reduced virulence, and QS-inhibitory compounds are effective in clearing the infecting bacteria and in reducing mortality (9, 19).
The QS response in P. aeruginosa is controlled by many regulators that are involved in the modulation of signal molecule production. The structures and modes of action of these regulators and their real impacts on the production of the QS signal molecules, as well as the stimuli to which they respond, in most cases are unknown (21, 26).
The RsaL protein is a pivotal negative regulator of QS, since the production of the main QS signal molecule of P. aeruginosa, i.e., N-3-oxo-dodecanoyl homoserine lactone (3OC12-HSL), is dramatically enhanced in an rsaL mutant strain. RsaL directly represses lasI, the 3OC12-HSL synthase gene, by binding its promoter, PlasI (7, 18). Importantly, besides the 3OC12-HSL-dependent activator LasR, RsaL is the only QS regulator known to bind PlasI (21, 22). The demonstration that RsaL is a DNA-binding protein has been of particular interest, considering that it shows homology only with the orthologous RsaL proteins of Pseudomonas putida strains IsoF, WCS358, and PCL1445 and with putative proteins present in some of the sequenced microbial genomes, whose functions have not yet been determined or predicted (3, 8, 18, 25). In addition, conventional in silico methods for the identification of domain-specific signature patterns (CDD [http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml] and SMART [http://smart.embl-heidelberg.de]) did not highlight any known domain for DNA binding in RsaL, suggesting that the protein could belong to a new class of transcriptional regulators.
In this study, an in silico model of RsaL structure has been inferred and experimentally validated, showing that RsaL belongs to the tetrahelical superclass of helix-turn-helix (H-T-H)-containing proteins (1). Many transcriptional factors from the three superkingdoms of life contain H-T-H domains belonging to this wide structural class, such as the N-terminal domain of the lambda cI repressor (λcI-NTD) and the POU-specific domain of the mammalian transcription factor Oct-1 (Oct-1 POUs). The POU domain is the DNA-binding domain of an important class of eukaryotic transcription factors involved in developmental regulation and cellular housekeeping. It is a bipartite structure composed of two autonomous DNA-binding subdomains, the POU-specific domain and the POU homeodomain, that are covalently connected by a flexible peptide linker (17). Interestingly, we have found that residues of the Oct-1 POUs important for structural stability and/or DNA binding are conserved in the same positions in RsaL and its homologs.
The characterization of the minimum consensus sequence for RsaL binding to PlasI and data suggesting that the protein binds DNA as a homodimer are also reported.
MATERIALS AND METHODS
In silico modeling.
The three-dimensional structures of RsaL from P. aeruginosa PAO1 (7) and from P. putida WCS358 (3) were modeled using the molecular-modeling software Rosetta (4) as implemented on the Robetta web server (http://robetta.bakerlab.org/).
Recombinant DNA techniques.
Preparation of plasmid DNA, purification of DNA fragments, ligation, and transformation of Escherichia coli were carried out by using standard procedures (20). PCR amplifications were performed using Red-Taq polymerase (Sigma). The sequences of oligonucleotides used for PCR are shown in Table 1. Automated sequencing was performed to check the DNA inserts of all the plasmids constructed in this study (MWG Biotech).
TABLE 1.
Oligonucleotides utilized for site-directed mutagenesis of RsaLPAO
| Name | Sequence (5′-3′)a | Positionb | Introduced mutation |
|---|---|---|---|
| FW-RsaL | ATGCCATGGCTTCACACGAGAGAA | +1 | |
| RV-RsaL | CCCAAGCTTCTCTCTGATCTTGCCTCT | +240 | |
| FW R20A | AAGGCCACCGCCACCGCCC | +49 | R20A: CGC→GCC |
| RV-R20A | GGGCGGTGGCGGTGGCCTT | +67 | R20A: CGC→GCC |
| FW-Q38A | GGGATAAGCGCCTCCTGCGGC | +103 | Q38A: CAA→GCC |
| RV-Q38A | GCCGCAGGAGGCGCTTATCCC | +123 | Q38A: TTG→GGC |
| FW-C40A | GCCAATCCGCCGGCAGTCGT | +110 | C40A: TGC→GCC |
| RV-C40A | ACGACTGCCGGCGGATTGGC | +129 | C40A: GCA→GGC |
| FW-S42A | TCCTGCGGCGCCCGTTTCGAG | +115 | S42A: AGT→GCC |
| RV-S42A | CTCGAAACGGGCGCCGCAGGA | +135 | S42A: ACT→GGC |
| FW-R43A | CTGCGGCAGTGCCTTCGAGAATG | +117 | R43A: CGT→GCC |
| RV-R43A | CATTCTCGAAGGCACTGCCGCAG | +139 | R43A: ACG→GGC |
| FW-E45A | CAGTCGTTTCGCGAATGGCGAG | +123 | E45A: GAG→GCG |
| RV-E45A | CTCGCCATTCGCGCAACGACTG | +144 | E45A: CTC→CGC |
| FW-R20E | AAGGCCACCGAGACCGCCCG | +49 | R20E: CGC→GAG |
| RV-R20E | CGGGCGGTCTCGGTGGCCTT | +68 | R20E: GCG→CTC |
| FW-E45R | CAGTCGTTTCCGCAATGGCGAGA | +123 | E45R: GAG→CGC |
| RV-E45R | TCTCGCCATTGCGGAAACGACTG | +145 | E45R: CTC→GCG |
| P0FW | GGGTACCAATAATTTTGTTTAACTTTA | 329 | |
| P0RV | GGGATCCATTGCTCAGCGGTGGCAGC | 71 |
Introduced restriction sites (NcoI, HindIII, KpnI, and BamHI) are underlined. Substituted nucleotides are in boldface.
Distance in base pairs from the rsaL ATG (for P0FW and P0RV, the position with respect to the pET28b(+) sequence is given).
Site-directed mutagenesis and protein purification.
Site-directed mutations were generated in rsaL using the splicing by overlap extension PCR method (10) with the primers described in Table 1. In the first step, two distinct PCRs (PCR-1 and PCR-2) were carried out for each rsaL derivative mutant, using chromosomal P. aeruginosa DNA as a template. Each PCR-1 was performed with the forward (FW) primer FW-RsaL and with a mutagenic reverse (RV) primer carrying the mutation in the desired codon. Each PCR-2 was performed with a mutagenic FW primer complementary to the RV primer utilized in the corresponding PCR-1 and with primer RV-RsaL. In the second PCR step, products obtained from PCR-1 and PCR-2 for each mutation were spliced together using the FW-RsaL and RV-RsaL primers. To generate rsaLR20E-E45R, a first round of two-step PCR mutagenesis was performed to introduce the Arg 20 Glu substitution in the RsaL coding sequence. Afterwards, the PCR product of this first round was used as a template for a second round of two-step PCR mutagenesis, in order to introduce the second mutation, Glu 45 Arg.
The different PCR products were digested by NcoI and HindIII, ligated into the expression vector pET28b(+) (Novagen), and introduced into the expression host E. coli BL21(DE3)pLysS (Novagen) for overexpression. Purification by Ni2+ affinity of mutant proteins, expressed by the different pET28b(+) derivatives and six-His tagged at the C terminus, was carried out as previously described for RsaLPAO (18). Proteins were further purified by gel permeation chromatography on a Superdex 200 HR column (Amersham Pharmacia Biotech).
Complementation assay and measurement of 3OC12-HSL.
In order to express the wild-type and mutated rsaL variants fused to six-His coding sequences in P. aeruginosa PAO1 (wild type; University of Washington Genome Center) and in its rsaL derivative (University of Washington Genome Center), the differently mutated variants of rsaL were cloned into the broad-host-range expression vector pBBR1MCS-5 under the control of the constitutive Plac promoter (13). For this purpose, the different pET28b(+) derivatives constructed to purify the wild-type and mutated RsaL proteins (see above) were used as DNA templates for PCR, together with primers P0FW and P0RV (Table 1). The different PCR products were digested by KpnI and BamHI, ligated into pBBR1MCS-5, and conjugated into P. aeruginosa PAO1 and its rsaL-mutated derivative as previously described (18). 3OC12-HSL was extracted from spent-growth medium and quantified by the use of a P. putida SM17(prsaL220) HSL biosensor strain. The P. putida SM17(prsaL220) sensor strain was constructed based on the PpuI/R quorum-sensing system of P. putida WCS358, which produces and responds to the 3OC12-HSL signal molecule (3). In this strain, 3OC12-HSL is produced by the PpuI synthase, encoded by the ppuI gene that is contiguous to rsaL (3). The 3OC12-HSL response regulator PpuR activates transcription of both ppuI and rsaL (3). A kanamycin resistance cassette was inserted within a BamHI-KpnI fragment between the SalI-XhoI sites of ppuI and rsaL, respectively, yielding pMPQS::Km. This plasmid was then used to inactivate the chromosomal ppuI and rsaL genes of P. putida by a marker exchange technique. Plasmid pPH1JI was used as the incoming IncP1-incompatible plasmid (6). The resultant P. putida SM17 strain contained a deletion in both ppuI and rsaL and consequently did not produce 3OC12-HSL and the RsaL repressor. 3OC12-HSL could then be quantified by measuring β-galactosidase activity using P. putida SM17 carrying plasmid prsaL220. This plasmid contains the PpuR-3OC12-HSL-regulated rsaL promoter transcriptionally fused to lacZ (3).
N-Acyl-homoserine lactone extracts of different P. aeruginosa strains were prepared as previously described (18), and the quantity of 3OC12-HSL was determined using P. putida SM17(prsal220). Overnight cultures of P. putida SM17(prsaL220) were diluted in LB medium to an A660 of 0.1. The N-acyl-homoserine lactone extract was then added, and after 6 h of growth at 30°C, β-galactosidase activity was determined (15). The amounts for a minimal and for a maximal saturated response were determined using synthetic 3OC12-HSL, and the quantifications corresponded in the linear dose response between these two values. This sensor is very specific for detection and quantification of 3OC12-HSL, as several synthetic N-acyl-homoserine lactones were tested, and apart from the cognate signal molecule, only 3OC10-HSL was an agonist (data not shown). The linear dose responses for 3OC12-HSL were from 0.1 μM to 5 μM, making this a well-suited biosensor for quantification of this molecule.
EMSA.
Different DNA probes were obtained by annealing in vitro complementary oligonucleotides corresponding either to nucleotides −4 to −28 of the wild-type lasI promoter (the numbering refers to the lasI transcription start point [23]) or to the same sequence carrying substitutions (see Fig. 5A). The annealed oligonucleotides were cloned into pDRIVE (QIAGEN), excised by EcoRI digestion, and 5′ end labeled. The procedures for [α-32P]dATP labeling of the probes, as well as the electrophoretic mobility shift assay (EMSA), have been previously described (18). The concentrations either of the different proteins (see the EMSA experiment shown in Fig. 3) or of the different DNA probes (see the EMSA experiment shown in Fig. 5) were previously equilibrated, and the experiments were carried out simultaneously, with identical polyacrylamide gels run in parallel.
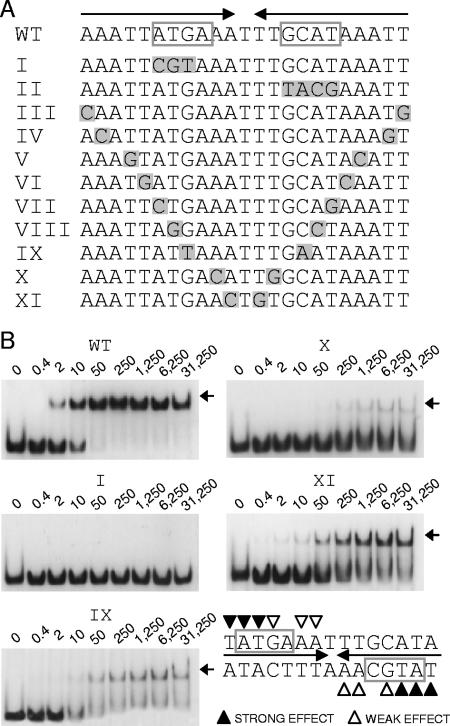
FIG. 5.
Characterization of the RsaLPAO DNA-binding site. (A) Sequences of different DNA probes used for EMSA. WT, wild-type probe encompassing the RsaLPAO-binding site on PlasI. The repeats forming a palindrome are indicated by arrows. The ATG(C/A) sequences are boxed. Probes I to XI correspond to different mutated variants of the WT probe. Mutated nucleotides are shaded gray. (B) EMSA carried out with RsaLPAO and with the DNA probes indicated above each gel. The concentrations of the different DNA probes were previously equilibrated, and the experiments were carried out simultaneously, with identical polyacrylamide gels run in parallel. The patterns of EMSA carried out with probes III, IV, and V (not shown) were identical to that of probe WT. The patterns of EMSA carried out with probes II, VI, VII, and VIII (not shown) were identical to that of probe I. The RsaLPAO-DNA complexes are indicated by arrows. The protein concentration (nM) for each sample is indicated above its lane. At the lower right is indicated the minimum sequence required for RsaLPAO binding. The triangles indicate the nucleotides whose replacement affects binding.
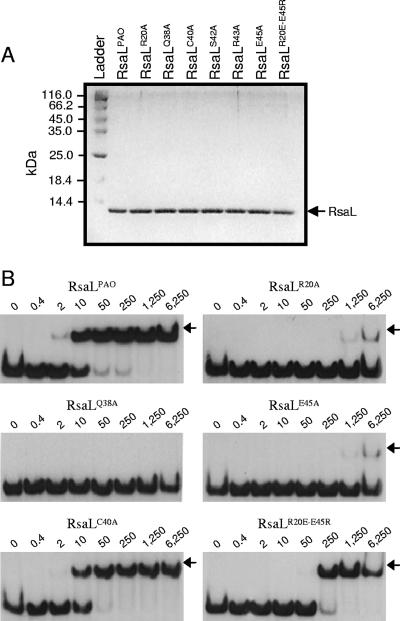
FIG. 3.
In vitro validation of the RsaLPAO model. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis showing that purified RsaLPAO and its mutated derivatives (indicated above each lane) are very pure and that their concentrations are equilibrated. Two microliters of each protein was loaded on the gel. Ladder, PageRuler protein ladder (Fermentas); the molecular mass of each band is indicated on the left. (B) EMSA carried out with a DNA probe encompassing the RsaLPAO-binding site on PlasI and either RsaLPAO or its mutated derivatives (indicated above each gel). The EMSAs were carried out simultaneously, starting from the same mixture containing the DNA probe, with identical polyacrylamide gels run in parallel. RsaLS42A and RsaLR43A showed an EMSA pattern (not shown) identical to that of RsaLQ38A. The RsaL-DNA complexes are indicated by arrows. The protein concentration (nM) for each sample is indicated above its lane.
RESULTS
RsaL is predicted to belong to the tetrahelical superclass of H-T-H proteins.
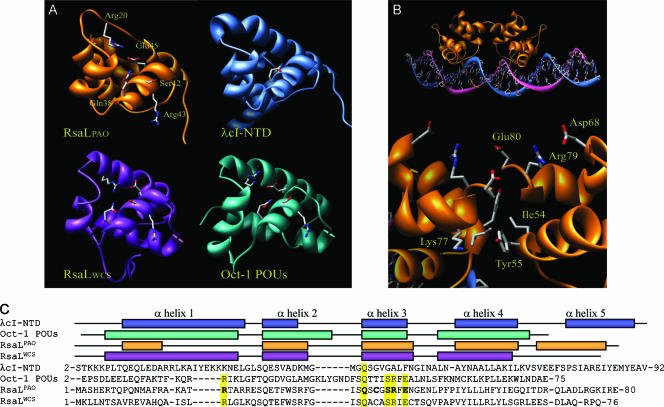
The orthologous RsaL proteins of P. aeruginosa PAO1 and P. putida WCS358 (RsaLPAO and RsaLWCS, respectively) are 42% identical. Their three-dimensional structures were independently modeled using the Rosetta software (4), which selected the structures of λcI-NTD (11) and of Oct-1 POUs (5) as templates for comparative modeling for RsaLPAO and RsaLWCS, respectively (Fig. 1A). Despite the lack of sequence homology, λcI-NTD and Oct-1 POUs share a common fold consisting of four alpha helices, in which helices 2 and 3 form an H-T-H motif for DNA binding, with helix 3 directly contacting the DNA major groove (2, 5, 11, 12). This bundle characterizes the tetrahelical H-T-H superclass, one of the major monophyletic groups in which the H-T-H proteins are clustered (1). Both RsaL models display this four-helix H-T-H fold, while differences are observed in the N- and C-terminal regions between the two models. In addition, despite the low overall sequence homology between Oct-1 POUs, RsaLPAO, and RsaLWCS, a striking conservation of several residues (Gln 38, Ser 42, Arg 43, and Glu 45; the numbering refers to RsaLPAO) is observed in the region corresponding to helix 3 (residues 38 to 46). Also, Arg 20 in helix 1, Glu 27 at the beginning of helix 2, and Ser 37 just before the beginning of helix 3 are conserved. With the exception of Gln 27 and Gln 38, the above-mentioned residues are not conserved in λcI-NTD (Fig. 1C). It has been determined that Gln 38, Ser 42, and Arg 43 are important for the specific interaction of Oct-1 POUs with its target DNA (5). We also observed in DNase I protection experiments that the ATGC DNA motif typically bound by Oct-1 POUs is present within the DNA region protected by RsaLPAO (5, 17, 18).
FIG. 1.
In silico model of RsaL. (A) Schematic representation of the molecular models of RsaLPAO and RsaLWCS and of the experimentally determined structures of λcI-NTD (11) (protein data base [PDB] code, 1RIO) and Oct-1 POUs (5) (PDB code, 1CQT). Selected residues are shown in stick representation and labeled in the RsaLPAO structure. Corresponding residues in the other three proteins are also shown. (B) Global and enlarged views of the complex between the putative RsaLPAO dimer and DNA, modeled on the basis of the λcI-NTD-DNA complex. Residues involved in the interactions between the two monomers are shown in stick representation and are labeled on only one of the two monomers. (C) Structure-based sequence alignment of RsaLPAO, RsaLWCS, λcI-NTD, and Oct-1 POUs. The secondary structure of each protein is shown above the alignment. Residues shown in stick representation in panel A are highlighted in yellow. Residues selected for site-directed mutagenesis are in boldface.
Another interesting feature that is predicted to be shared by Oct-1 POUs, RsaLPAO, and RsaLWCS is the presence of a salt bridge between Arg 20 and Glu 45 (Fig. 1A). In λcI-NTD, this salt bridge is lacking, while in Oct-1 POUs, it links helices 1 and 3, contributing to the overall stability of the protein (5).
Validation of the model: key functional residues are conserved in RsaLPAO and Oct-1 POUs.
In order to experimentally confirm the proposed structural model for RsaLPAO, conserved residues predicted to contact DNA (Gln 38, Ser 42, and Arg 43) and to form the salt bridge (Arg 20 and Glu 45) were mutated to Ala. As a control, the nonconserved Cys 40 residue that, although located in helix 3, is not predicted to interact with DNA in our model was also replaced with Ala. In addition, the residues predicted to form the salt bridge were inverted by introducing the double substitution Arg 20 Glu/Glu 45 Arg.
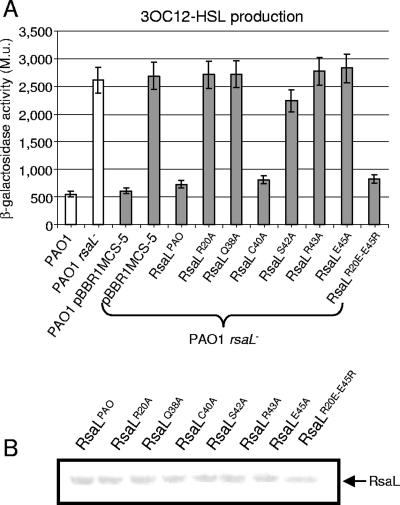
A recent study showed that 3OC12-HSL production was enhanced in an rsaL mutant of P. aeruginosa PAO1 (18). This unambiguous phenotype makes it possible to compare in vivo the activities of the wild-type and mutated variants of RsaLPAO by a complementation assay. Therefore, the rsaL wild-type and mutated coding sequences, fused with a six-His tag at their C termini to allow the detection of the expressed proteins by Western assay, were cloned into the broad-host-range expression plasmid pBBR1MCS-5 and independently introduced into the rsaL mutant of P. aeruginosa PAO1. 3OC12-HSLs were extracted from the growth media of the strains carrying the different plasmids, and their levels were compared using the P. putida SM17(prsaL220) sensor strain that was constructed here for the purpose (see Materials and Methods). Briefly, in this strain, the expression of lacZ is under the control of a 3OC12-HSL-responsive promoter, so that β-galactosidase activity is proportional to the amount of 3OC12-HSL present in the growth medium. The results of this experiment (Fig. 2A) showed that the 3OC12-HSL level produced by the P. aeruginosa wild type was enhanced about fivefold in the P. aeruginosa rsaL mutant. Complementation of this mutant with rsaL or with its mutated variants rsaLC40A and rsaLR20E-E45R restored the 3OC12-HSL production to wild-type levels. Conversely, 3OC12-HSL levels were not restored upon complementation of the rsaL mutant strain with plasmids expressing the mutated proteins RsaLR20A, RsaLQ38A, RsaLS42A, RsaLR43A, and RsaLE45A (Fig. 2A). The Western analysis carried out on the soluble cellular fractions of the same cultures in which 3OC12-HSL was measured showed that the levels of wild-type RsaLPAO and its mutated derivatives were comparable, demonstrating that the lack of complementation ability was not due to decreased expression of the recombinant proteins (Fig. 2B). Therefore, RsaLPAO residues Arg 20, Gln 38, Ser 42, Arg 43, and Glu 45 are essential for the activity of RsaLPAO in the cell.
FIG. 2.
In vivo validation of the RsaLPAO model. (A) Measurement of 3OC12-HSL produced by P. aeruginosa PAO1 and its rsaL mutant derivative (white bars) and by the same strains carrying either the pBBR1-MCS5 plasmid, as a control, or its derivatives expressing RsaLPAO or the indicated mutant proteins (gray bars). 3OC12-HSL was extracted from the spent-growth medium of each strain and quantitatively measured as described in Materials and Methods, using the P. putida SM17(prsaL220) strain, in which β-galactosidase activity (given in Miller units [M.u.]) is proportional to the levels of 3OC12-HSL. Standard deviations (error bars) are based on the mean values for three parallel cultures. (B) Western hybridization performed with anti-six-His tag antibody (Sigma) on the cellular soluble fractions derived from the same cultures in which 3OC12-HSL production was measured. The protein expressed in each sample is indicated above its lane.
At the molecular level, RsaLPAO acts as a transcriptional repressor of the 3OC12-HSL synthase gene by direct binding to its promoter, PlasI (18). In order to assess whether the introduced mutations affect the DNA-binding activity of RsaLPAO, we performed EMSAs comparing the ability of purified wild-type RsaLPAO to bind a DNA probe encompassing the RsaLPAO-binding site of PlasI with those of its mutated derivatives. All of the proteins were purified to homogeneity, and their concentrations were equilibrated (Fig. 3A). Incubation of the DNA probe with wild-type RsaLPAO led to the formation of a complex having lower electrophoretic mobility than the probe alone (Fig. 3B). This result was expected and was consistent with that of a previously published EMSA experiment (18). RsaLPAO was able to shift about 50% of the probe at a concentration of 10 nM, while RsaLQ38A, RsaLS42A, and RsaLR43A were unable to shift the same DNA probe even at a concentration 625-fold higher (Fig. 3B; only EMSA carried out with RsaLQ38A is shown). On the other hand, the DNA-binding ability of RsaLC40A was comparable to that of the wild-type protein (Fig. 3B). Moreover, the substitution Arg 20 Ala or Glu 45 Ala had a strong negative influence on the RsaL DNA-binding ability that was partially restored by inverting the positions of these residues in RsaLR20E-E45R (Fig. 3B). The above-mentioned results validated our proposed in silico model, in which the helix 3 residues Gln 38, Ser 42, and Arg 43, but not Cys 40, are important to establish direct interactions with DNA, while Arg 20 and Glu 45 stabilize the RsaLPAO structure by forming a salt bridge.
RsaLPAO binds to an eptameric inverted repeat as a dimer.
The RsaLPAO-binding site on PlasI was previously characterized by DNase I protection experiments and contains two inverted repeats forming a palindrome, suggesting that RsaLPAO binds DNA as a dimer (18). Accordingly, the modeling of the RsaLPAO dimer, based on the λcI-NTD dimer, revealed the presence of several interactions that could stabilize the formation of the RsaLPAO dimer (Fig. 1B). In particular, Lys 77 on one monomer could form a favorable electrostatic interaction with Tyr 55 on the other monomer, and Asp 68, Arg 79, and Glu 80 could form an extended charge network with the corresponding residues of the second monomer. In addition, hydrophobic interactions could take place between Ile 54 and Tyr 55 of both monomers, potentially providing additional stability to the dimer.
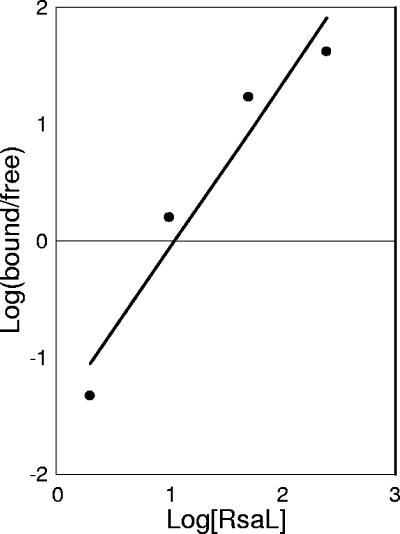
To assess the oligomeric state of RsaLPAO, we performed gel permeation chromatography showing that RsaL is a monomer in solution (data not shown). However, the Hill plot inferred from the EMSA performed with RsaLPAO indicated that binding of this protein to the target DNA is cooperative (Hill coefficient, 1.4) (Fig. 4), suggesting that DNA could play a role in RsaL dimerization. Moreover, the absence of intermediate complexes in the EMSA indicates that only the complex between an RsaLPAO dimer and DNA could be stable enough to be detectable. If this is the case, disrupting the palindromic structure of the DNA probe bound by RsaLPAO would lead to the abolition of the complex formation in an EMSA assay. To validate this hypothesis, we performed EMSA experiments using mutated DNA probes in which the palindromic structure of the RsaLPAO-binding site was disrupted. To select the nucleotides to be mutagenized, we considered that Oct-1 POUs specifically contact the ATGC motif, mainly through the conserved residues Gln 38, Ser 42, and Arg 43 (5, 12). This DNA motif is also present in the core of the right repeat of the RsaLPAO-binding site (complementary strand) and is partially conserved in the left repeat (Fig. 5A). Thus, we constructed two DNA probes in which the ATG sequence of the left repeat or the ATGC sequence of the right repeat were alternatively substituted (Fig. 5A, probes I and II, respectively), and the abilities of RsaLPAO to bind the mutated and wild-type DNA fragments were compared by EMSA. The results showed that the substitution of each of these sequences abrogated RsaLPAO binding even at protein concentrations 625-fold higher than that sufficient to completely shift the same amount of wild-type probe. These results strongly support our hypothesis that the formation of a dimer of RsaLPAO on the target DNA is necessary for the formation of a stable complex (Fig. 5B; EMSA carried out with probe II, identical to that with probe I, is not shown).
FIG. 4.
Hill plot inferred from the EMSA performed with RsaLPAO and a DNA probe encompassing the RsaLPAO-binding site on the lasI promoter (shown in Fig. 3B). The amounts of bound and free probe for each lane were measured by a densitometric analysis carried out with the software Kodak 1D 3.5. The protein concentration is in molar excess with respect to the probe concentration. The Hill coefficient (n50 = 1.4) indicates cooperative binding.
To better define the minimum sequence requirements for RsaLPAO binding to PlasI, we tested the abilities of a set of DNA probes (mutated in each pair of specular nucleotides contributing to form the palindrome) to bind RsaLPAO (Fig. 5A, probes III to XI). We found that the EMSA carried out with probes III to V had a pattern identical to that obtained with the wild-type probe, indicating that the mutagenized nucleotides are not involved in the formation of the RsaLPAO-DNA complex (In Fig. 5B, only the EMSA with the wild-type probe is shown). On the other hand, the DNA probes VI to VIII had a pattern identical to that of probe I and therefore could not form stable complexes with RsaLPAO (in Fig. 5B, only the EMSA carried out with probe I is shown). Therefore, the specular nucleotides mutagenized in these probes are essential for the formation of the RsaLPAO-DNA complex. Finally, RsaLPAO was able to form unstable, smeared complexes with probes IX to XI, indicating that the specular nucleotides mutagenized in these probes also play roles in RsaLPAO binding. In summary, the above data showed that the palindromic structure of the RsaLPAO-binding site is required for the formation of a stable complex between RsaLPAO and the target DNA and allowed us to define the minimum half-site for RsaLPAO binding on PlasI as TATG(A/C)AA (Fig. 5B).
The RsaL-like proteins.
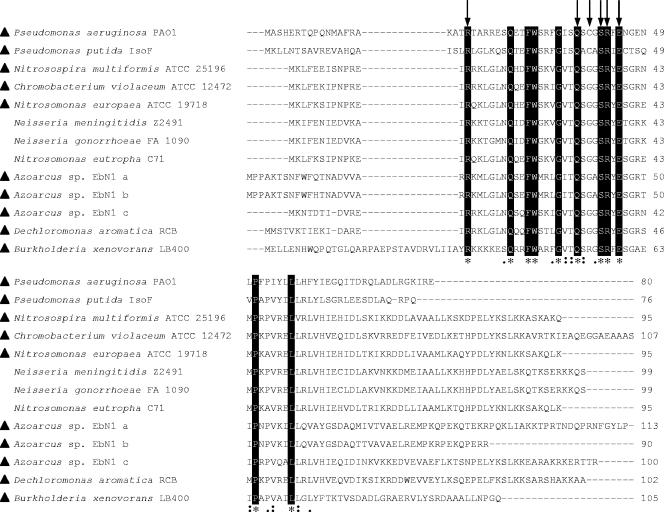
A PSI-BLASTP analysis of the RsaLPAO sequence was performed, retrieving 12 bacterial proteins showing significant levels of homology (Fig. 6) (http://www.ncbi.nlm.nih.gov/BLAST). With the exception of RsaL from P. putida strains IsoF, WCS358, and PCL1445 (100% identical), the RsaL homologs are classified as hypothetical proteins. The rsaL genes in the above-mentioned P. putida strains are located in the intergenic region between lasR-like and lasI-like genes and have been shown to regulate QS (3, 8, 25). Since acyl-HSL-based QS systems are widespread in bacteria, we asked whether the other rsaL-like genes could be involved in QS regulation. Several of the sequenced genomes of bacteria containing RsaL-like proteins contain homologs of the acyl-HSL synthases LasI and/or LuxI (Fig. 6). However, only in the case of Burkholderia xenovorans LB400 was the rsaL-like gene located in the intergenic region between lasR and lasI homologs (GenBank accession no. YP_554691 and YP_554693, respectively), suggesting that at least in this strain, the rsaL-like gene could behave as a regulator of QS.
FIG. 6.
Alignment of putative bacterial proteins showing significant levels of homology with RsaLPAO in a PSI-BLASTP analysis. The alignment is presented as in the ClustalW ALN output (http://www.ebi.ac.uk/clustalw/) (asterisks, identical residues; colons, conserved substitutions; periods, semiconserved substitutions) with the following modifications: identical residues are also black boxed, residues mutagenized in RsaLPAO are indicated by arrows, and genomes containing lasI-like genes are indicated by triangles. GenBank accession numbers are as follows: Pseudomonas aeruginosa PAO1, NP_250122; Pseudomonas putida IsoF, AAM75412; Nitrosospira multiformis ATCC 25196, YP_412629; Chromobacterium violaceum ATCC 12472, NP_902047; Nitrosomonas europaea ATCC 19718, NP_841560; Neisseria meningitidis Z2491, NP_275082; Neisseria gonorrhoeae FA 1090, YP_209002; Nitrosomonas eutropha C71, ZP_00669043; Azoarcus sp. strain EbN1 a, YP_157206; Azoarcus sp. strain EbN1 b, YP_195632; Azoarcus sp. strain EbN1 c, YP_157355; Dechloromonas aromatica RCB, YP_284082; Burkholderia xenovorans LB400, YP_554692. Note that the amino acid sequences of RsaL proteins from P. putida strains IsoF, WCS358, and PCL1445 are 100% identical.
DISCUSSION
The regulation of the QS circuitry of P. aeruginosa is very complex, since many regulators modulate the production of the QS signal molecules. However, the modes of action of these regulators have been investigated at the molecular level in only a few cases (reviewed in references 22 and 27). To our knowledge, no structural data on any P. aeruginosa QS transcriptional regulator have been published. In this study, new insights into the structure and function of the lasI transcriptional repressor RsaLPAO are provided.
Since RsaLPAO shows no homology with other DNA-binding proteins, an in silico approach was undertaken to predict the possible tertiary structure of RsaLPAO and its ortholog, RsaLWCS. The model prediction was strengthened by the fact that independent computational procedures selected two structures as templates, λcI-NTD for RsaLPAO and Oct-1 POUs for RsaLWCS, which are identical.
Although RsaLPAO was modeled on the λcI-NTD three-dimensional structure, key functional residues (Arg 20, Gln 38, Ser 42, Arg 43, and Glu 45) are shared with Oct-1 POUs. Both the RsaLPAO and RsaLWCS models show a salt bridge between residues Arg 20 and Glu 45. In Oct-1 POUs, this salt bridge is important for stabilizing the DNA-binding helix, while it is not present in the λcI-NTD three-dimensional structure. Thus, the presence of this salt bridge in the RsaLPAO model was not due to a bias imposed by the choice of the template used to build it. Our analysis confirmed the importance of this salt bridge, since the replacement of either Arg 20 or Glu 45 with Ala abrogated the DNA-binding ability of RsaLPAO, while the reconstruction of the salt bridge in the RsaLR20E-E45R double mutant partially restored DNA-binding ability. RsaLR20E-E45R is capable of a DNA-binding affinity about fivefold lower than that of RsaLPAO (compare the lanes corresponding to 250 nM and 50 nM protein, respectively, in Fig. 3B). This could be explained by considering that in Oct-1 POUs, Glu 45 hydrogen bonds to the conserved Gln 27 residue, in addition to Arg 20 (5). Moreover, the Arg 20 residue of Oct-1 POUs not only forms a salt bridge, but also contacts a phosphodiester oxygen of the DNA backbone. It is likely that the same interactions also occur in RsaLPAO, as indicated by the analysis of its structural model. However, the lower in vitro DNA-binding affinity of RsaLR20E-E45R proved to be insignificant in vivo, as shown by the complementation assay.
The residues Gln 38, Ser 42, and Arg 43, which in Oct-1 POUs directly contact an ATGC DNA motif, are also necessary for the DNA-binding activity of RsaLPAO. Our studies of the RsaLPAO-binding site on the lasI promoter defined the minimum half-site for RsaLPAO binding on PlasI as TATG(A/C)AA. This sequence contains the ATGC motif recognized by Oct-1 POUs and is also similar to the λcI-NTD half-consensus, TATCACCG (11, 12). It is known that the structural similarity between Oct-1 POUs and λcI-NTD corresponds to a striking similarity in their modes of binding to DNA. Indeed, despite the fact that the amino acids involved in DNA binding are not conserved, these two proteins establish similar networks of interactions with DNA (11, 12). The observation that the DNA sequence recognized by RsaLPAO is also similar to that of Oct-1 POUs and λcI-NTD adds further strength to the structural model proposed for RsaLPAO and indicates that RsaLPAO could dock similarly to DNA. At the quaternary structural level, there are some differences between Oct-1 POUs and λcI-NTD. The latter is a monomer in solution and binds to a pseudopalindromic DNA sequence as a homodimer. In particular, the fifth α helix of λcI-NTD constitutes the dimerization interface (11). Oct-1 POUs do not have a fifth helix and bind DNA as a pseudoheterodimer because they are covalently bound by a flexible linker to a second DNA-binding domain, the POU homeodomain. Therefore, in Oct-1 POUs the flexible linker functionally replaces the dimerization helix of λcI-NTD (5, 17). In our model, a fifth α helix is also present in RsaLPAO, though it is much shorter than that of λcI-NTD. Our analysis showed that RsaLPAO quaternary organization could be similar to that of λcI-NTD. Indeed, RsaLPAO exists as a monomer in solution but binds to DNA with high cooperativity, as shown by the Hill plot. Moreover a palindromic binding site is necessary for the formation of a stable complex between RsaLPAO and DNA, since disruption of the palindrome leads to lack of complex formation. Due to the high cooperativity, it is very likely that RsaL readily dimerizes in the presence of the palindromic binding site, and thus, intermediate complexes between monomeric RsaL and DNA cannot be detected. It is also possible that the complex between monomeric RsaL and DNA is a low-affinity complex that cannot be detected. Our in vitro results reported here strongly support the notion that the DNA-mediated RsaLPAO dimerization is essential to form a stable DNA-protein complex. This finding is further strengthened by the RsaLPAO dimer modeling (Fig. 5).
As far as RsaLWCS is concerned, in our model, the C-terminal region of the protein is not structured. This could be due to a bias imposed by the template Oct-1 POU structure. However, since the C-terminal region of RsaLWCS is shorter and shares no conserved residues with that of RsaLPAO, the possibility that RsaLWCS binds to DNA as a monomer cannot be excluded.
Proteins containing an H-T-H domain have recently been classified by Aravind and coworkers (1), taking into consideration structural features, the presence of additional domains, and sequence homology. The structural similarity between RsaL, λcI-NTD, and Oct-1 POUs indicates that RsaL belongs to the tetrahelical H-T-H superclass, which constitutes one of the major groups of H-T-H proteins, together with the winged-helix H-T-H proteins (1). On the basis of sequence homology and domain architectures, several distinct protein families have been identified within the tetrahelical H-T-H superclass, including the λcI, AraC, LuxR, LacI, DnaA, TrpR, and TetR families (1). In this analysis, RsaLPAO and three of its homologs (GenBank accession no. NP_902047, NP_275082, and NP_841560) have been considered, but they do not cluster with any of the above-mentioned families (ftp://ftp.ncbi.nih.gov/pub/aravind/HTH/). On this basis, the prokaryotic RsaL-like proteins could constitute a new family of the tetrahelical H-T-H superclass, sharing the following three distinctive features: (i) lack of significant homology with other proteins; (ii) very small size (spanning from 76 to 113 residues), consisting almost exclusively of the four-helix bundle core; and (iii) conservation of key residues. With respect to the last point, the Arg 20, Gln 38, Ser 42, Arg 43, and Glu 45 residues, conserved between RsaLPAO, RsaLWCS, and Oct-1 POUs, are also conserved in the other RsaLPAO homologs, in agreement with our data showing that these residues are important for RsaLPAO function (Fig. 6).
The four-helix bundle is a structure widely present in the three superkingdoms of life (1), suggesting that this fold was probably present in the last universal common ancestor. Given the huge evolutionary distance dividing bacteria and mammals, it is likely that the features shared by RsaLPAO and Oct-1 POUs, and by their cognate promoters, arose by convergent evolution from two distinct regulator-promoter pairs that independently evolved from a common ancestor. However, the possibility that these features have been maintained as ancestral characteristics cannot be ruled out.
Acknowledgments
We are grateful to P. Ascenzi, F. Imperi, and M. Milani for critical reading of the manuscript. We thank the University of Washington Genome Center for supplying the rsaL mutant of P. aeruginosa.
This work was supported by grants from ISPESL (B/98-1/DIPIA/03) and from University Roma Tre (CLAR 2005). I.B. was supported by a fellowship from the Italian Cystic Fibrosis Research Foundation, Verona, Italy. This work was also supported by a grant from the Italian Ministry of University and Research. PRIN-2006-entitled “Basi genetiche e molecolari della patogenicità batterica.”
Footnotes
Published ahead of print on 15 December 2006.
REFERENCES
- 1.Aravind, L., V. Anantharaman, S. Balaji, M. M. Babu, and L. M. Iyer. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231-262. [DOI] [PubMed] [Google Scholar]
- 2.Assa-Munt, N., R. J. Mortishire-Smith, R. Aurora, W. Herr, and P. E. Wright. 1993. The solution structure of the Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage l repressor DNA-binding domain. Cell 73:193-205. [DOI] [PubMed] [Google Scholar]
- 3.Bertani, I., and V. Venturi. 2004. Regulation of the N-acyl homoserine lactone-dependent quorum-sensing system in rhizosphere Pseudomonas putida WCS358 and cross-talk with the stationary-phase RpoS sigma factor and the global regulator GacA. Appl. Environ. Microbiol. 70:5493-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonneau, R., C. Strauss, C. Rohl, D. Chivian, P. Bradley, L. Malmstrom, T. Robertson, and D. Baker. 2002. De novo prediction of three-dimensional structures for major protein families. J. Mol. Biol. 322:65-78. [DOI] [PubMed] [Google Scholar]
- 5.Chasman, D., K. Cepek, P. A. Sharp, and C. O. Pabo. 1999. Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein-DNA interface. Genes Dev. 13:2650-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbin, D., G. Ditta, and D. R. Helinski. 1982. Clustering of nitrogen fixation (nif) genes in Rhizobium meliloti. J. Bacteriol. 149:221-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Kievit, T., P. C. Seed, J. Nezezon, L. Passador, and B. H. Iglewski. 1999. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 181:2175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubern, J. F., B. J. Lugtenberg, and G. V. Bloemberg. 2006. The ppuI-rsaL-ppuR quorum-sensing system regulates biofilm formation of Pseudomonas putida PCL1445 by controlling biosynthesis of the cyclic lipopeptides putisolvins I and II. J. Bacteriol. 188:2898-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heurlier, K., V. Denervaud, and D. Haas. 2006. Impact of quorum sensing on fitness of Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:93-102. [DOI] [PubMed] [Google Scholar]
- 10.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 11.Jain, D., B. E. Nickels, L. Sun, A. Hochshild, and S. A. Darst. 2004. Structure of a ternary transcription activation complex. Mol. Cell 12:45-53. [DOI] [PubMed] [Google Scholar]
- 12.Klemm, J. D., M. A. Rould, R. Aurora, W. Herr, and C. O. Pabo. 1994. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell 77:21-32. [DOI] [PubMed] [Google Scholar]
- 13.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 14.Lyczac, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051-1060. [DOI] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1972. Experiments in molecular genetics, p. 252-255. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Miller, M. B., and B. L. Bassler. 2001. Quorum-sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 17.Phillips, K., and B. Luisi. 2000. The virtuoso of versatility: POU proteins that flex to fit. J. Mol. Biol. 302:1023-1039. [DOI] [PubMed] [Google Scholar]
- 18.Rampioni, G., I. Bertani, E. Zennaro, F. Polticelli, V. Venturi, and L. Leoni. 2006. The quorum-sensing negative regulator RsaL of Pseudomonas aeruginosa binds to the lasI promoter. J. Bacteriol. 188:815-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen, T. B., and M. Givskov. 2006. Quorum sensing inhibitors: a bargain of effects. Microbiology 152:895-904. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Schuster, M., and E. P. Greenberg. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:73-81. [DOI] [PubMed] [Google Scholar]
- 22.Schuster, M., M. L. Urbanowski, and E. P. Greenberg. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 101:15833-15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seed, P. C., L. Passador, and B. H. Iglewski. 1995. Activation of the Pseudomonas aeruginosa lasI gene By LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, R. S., and B. H. Iglewski. 2003. Pseudomonas aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6:56-60. [DOI] [PubMed] [Google Scholar]
- 25.Steidle, A., M. Allesen-Holm, K. Riedel, G. Berg, M. Givskov, S. Molin, and L. Eberl. 2002. Identification and characterization of an N-acylhomoserine lactone-dependent quorum-sensing system in Pseudomonas putida strain IsoF. Appl. Environ. Microbiol. 68:6371-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venturi, V. 2006. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 30:274-291. [DOI] [PubMed] [Google Scholar]
- 27.Wagner, V. E., J. G. Frelinger, R. K. Barth, and B. H. Iglewski. 2006. Quorum sensing: dynamic response of Pseudomonas aeruginosa to external signals. Trends Microbiol. 14:55-58. [DOI] [PubMed] [Google Scholar]