Abstract
The type III secretion system (T3SS) of Pseudomonas aeruginosa plays a significant role in pathogenesis. We have previously identified type III secretion factor (TSF), which is required for effective secretion of the type III effector molecules, in addition to the low calcium signal. TSF includes many low-affinity high-capacity calcium binding proteins, such as serum albumin and casein. A search for the TSF binding targets on the bacterial outer membrane resulted in identification of PopN, a component of the T3SS that is readily detectable on the bacterial cell surface. PopN specifically interacts with Pcr1, and both popN and pcr1 mutants have a constitutive type III secretion phenotype, suggesting that the two proteins form a complex that functions as a T3SS repressor. Further analysis of the popN operon genes resulted in identification of protein-protein interactions between Pcr1 and Pcr4 and between Pcr4 and Pcr3, as well as between PopN and Pcr2 in the presence of PscB. Unlike popN and pcr1 mutants, pcr3 and pcr4 mutants are totally defective in type III secretion, while a pcr2 mutant exhibits reduced type III secretion. Interestingly, PopN, Pcr1, Pcr2, and Pcr4 are all secreted in a type III secretion machinery-dependent manner, while Pcr3 is not. These findings imply that these components have important regulatory roles in controlling type III secretion.
A type III secretion system (T3SS) is present in many gram-negative bacterial pathogens and plays an important role in bacterial pathogenesis (8, 17, 24). T3SS use a needle-like structure to translocate effectors into animal or plant host cells, causing cytotoxicity. In Pseudomonas aeruginosa, nearly 30 genes are involved in the regulation and biogenesis of the T3SS needle structure. These genes are clustered in a locus and form five operons, including the pscN, popN, pcrG, exsC, and exsD operons (55). The T3SS needle is about 60 nm long, has a 7-nm hollow center, and is composed mainly of the PscF protein (40).
The T3SS of P. aeruginosa responds to various environmental signals, such as a low concentration of calcium and type III secretion factor (TSF) or direct contact with host cells (14, 31, 52). TSF consists of abundant host proteins that are low-affinity, high-capacity calcium binding proteins, such as albumin and casein (31). ExsA, ExsC, ExsD, and ExsE form a complex regulatory network regulating the expression of T3SS genes in response to specific environmental signals (57). Upon activation, the type III secretion apparatus translocates effector molecules into the cytoplasm of the host cell, resulting in cell rounding, lifting, and death by necrosis or apoptosis (11, 16, 26, 29, 32, 41). There are four known effector molecules, including ExoS and ExoT, both of which have ADP ribosyltransferase activity and a GTPase-activating protein activity, the acute cytotoxin ExoU, and the adenylate cyclase ExoY (11, 21, 54, 56). ExoS preferentially ADP ribosylates several Ras families of GTP-binding proteins required for the regulation of intracellular vesicle transport, cell proliferation, and differentiation (7, 18). The ADP ribosyltransferase activity of ExoS causes programmed cell death in various types of host cells (26, 27, 29). ExoT preferentially ADP ribosylates Crk-I and Crk-II proteins in vivo (48) and inhibits host cell division by targeting cytokinesis (46). ExoU has been shown to have a lipase activity that causes rapid host cell membrane disruption (42).
In addition to the effector proteins, PopB, PopD, PopN, and PcrV are also secreted by the T3SS (49, 55). PopB and PopD are translocases that are necessary for the delivery of T3SS effector proteins into host cells. During infection, these two proteins form pores on the host cell membrane to facilitate effector translocation into the host cytosol (44). Without PopB or PopD, the expression and level of secretion of the T3SS are normal, as they are in the wild-type strain, but the capacity to translocate effectors into host cells is lost (49). PcrV is a surface-localized protein with homology to the LcrV protein of Yersinia (43). Although it is not part of the pore formed by PopB and PopD, it affects the size of the pore (19, 23). A pcrV mutant can secret effectors constitutively but does not deliver effectors into host cells (43, 49). The PopN gene is the first gene in an operon and is followed by four small open reading frames, whose products are designated Pcr1, Pcr2, Pcr3, and Pcr4. A mutant with a mutation in the popN gene has previously been shown to have a constitutive type III secretion phenotype (49). The functions of the four small pcr genes are not well understood.
In this study, PopN was identified as one of the TSF binding targets, and this protein is readily detectable on the bacterial cell surface. Further analysis of the proteins interacting with PopN resulted in identification of Pcr1, and a mutation in either gene resulted in constitutive type III secretion, suggesting that a PopN-Pcr1 complex functions as a type III repressor. Pcr1 also binds Pcr4, while Pcr4 interacts with Pcr3. A mutation in pcr3 or pcr4 completely knocked out the type III secretion function. Interestingly, the members of an interacting triplex, PopN, Pcr1, and Pcr4, are all secreted under type III inducing conditions. Implications of these findings for our understanding of the mechanism of type III secretion are discussed below.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in L broth (LB). P. aeruginosa strains were grown in LB or Dulbecco modified Eagle medium (DMEM). When needed, antibiotics were used at the following concentrations: for E. coli, 100 μg/ml ampicillin, 50 μg/ml kanamycin, 10 μg/ml tetracycline, 10 μg/ml gentamicin, 40 μg/ml chloramphenicol, 25 μg/ml spectinomycin, and 25 μg/ml streptomycin; and for P. aeruginosa, 150 μg/ml carbenicillin, 100 μg/ml tetracycline, 100 μg/ml gentamicin, 200 μg/ml spectinomycin, and 200 μg/ml streptomycin.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli RS | BacterioMatch two-hybrid system reporter strain, Kmr | Stratagene |
| P. aeruginosa strains | ||
| PAK | Wild-type P. aeruginosa strain | 4 |
| PAKexsA::Ω | PAK with exsA disrupted by insertion of Ω cassette, Spr Smr | 15 |
| PAKΔpopN | PAK with the popN gene deleted | This study |
| PAKpcr1 | Point mutation of start codon in pcr1 gene of PAK | This study |
| PAKpcr2 | Point mutation of start codon in pcr2 gene of PAK | This study |
| PAKpcr3 | Point mutation of start codon in pcr3 gene of PAK | This study |
| PAKpcr4 | Point mutation of 13th codon in pcr4 gene of PAK | This study |
| PAKΔpcrV | PAK with the pcrV gene deleted | This study |
| PAKΔpopB | PAK with the popB gene deleted | This study |
| Plasmids | ||
| pCR2.1-TOPO | Cloning vector for the PCR products | Invitrogen |
| pFLAG-CTC | Fusion vector for cytoplasmic expression of C-terminal Flag tag, Apr | Sigma |
| pGEX-4T-2 | Fusion vector for N-terminal GST, Apr | Pharmacia |
| pQE30 | Fusion vector for N-terminal His tag, Apr | QIAGEN |
| pQE31 | Fusion vector for N-terminal His tag, Apr | QIAGEN |
| pDN191acΩ | Promoterless lacZ fusion vector, Spr Smr Tcr | 51 |
| pDN19 | Broad-host-range shuttle vector, Tcr | 38 |
| pUCP19 | Broad-host-range shuttle vector, Apr | 45 |
| pEX18Gm | Gene replacement vector, GmroriT+sacB+ | 22 |
| pEX18Tc | Gene replacement vector, TcroriT+sacB+ | 22 |
| pPcrV19 | pcrV gene clone in pUCP19, Apr | This study |
| pBT | Bait vector encoding full-length bacterial phage λcI protein, Chlr | Stratagene |
| pTRG | Target vector encoding RNAP α-subunit protein, Tcr | Stratagene |
| pBT-LGF2 | Dimerization domain of Gal4 in bait vector, Chlr | Stratagene |
| pTRG-Gal 11p | Gal11 on target vector, Tcr | Stratagene |
| pHW0006 | exoT-lacZ fusion reporter in pDN19lacZΩ, Spr Smr Tcr | 20 |
| pPppB-Flag | PppB-Flag fusion reporter in pDN19, Apr Tcr | 1 |
| pYAN0515 | Fragment containing pcr1 gene in pEX18Gm, Gmr | This study |
| pYAN0512 | Fragment containing pcr2 and pcr3 genes in pEX18Gm, Gmr | This study |
| pYAN0625 | Fragment containing pcr4 gene in pEX18Gm, Gmr | This study |
| pPopNExT | Fragment containing popN gene in pEX18Tc, Tcr | This study |
| pPopNDT | popN gene with ScaI/HincII deletion in pPopNExT, Tcr | This study |
| pPcrVDG | pcrV with HincII fragment deletion cloned in pEX18Gm, Gmr | This study |
| pPopBDA | popB with EcoRV/NotI fragment deletion cloned in pEX18Ap, Apr | This study |
| pYAN0522 | pcr1 with start codon changed to TTC in pYAN0515, Gmr | This study |
| pYAN0525 | pcr2 with start codon changed to ATC in pYAN0512, Gmr | This study |
| pYAN0523 | pcr3 with start codon changed to CTC in pYAN0512, Gmr | This study |
| pYAN0626 | pcr4 with 13th codon changed to TAG in pYAN0625, Gmr | This study |
| pPopN-F | popN-Flag fusion in pUCP19, Apr | This study |
| pYAN0628 | pcr1-Flag fusion gene in pFLAG-CTC, Apr | This study |
| pYAN0629 | pcr2-Flag fusion gene in pFLAG-CTC, Apr | This study |
| pYAN0630 | pcr3-Flag fusion gene in pFLAG-CTC, Apr | This study |
| pYAN0631 | pcr4-Flag fusion gene in pFLAG-CTC, Apr | This study |
| pYAN0632 | pYAN0628 fused with pDN19, Apr Tcr | This study |
| pYAN0633 | pYAN0629 fused with pDN19, Apr Tcr | This study |
| pYAN0634 | pYAN0630 fused with pDN19, Apr Tcr | This study |
| pYAN0635 | pYAN0631 fused with pDN19, Apr Tcr | This study |
| pYAN0636 | pFLAG-CTC fused with pDN19, Tcr Apr | This study |
| pYAN0601 | GST-pcr1 fusion in pGEX-4T-2 vector, Apr | This study |
| pYAN0602 | GST-pcr3 fusion in pGEX-4T-2 vector, Apr | This study |
| pPopN-His | His-popN fusion in pQE31 vector, Apr | This study |
| pPcrV-His | His-pcrV fusion in pQE31 vector, Apr | This study |
| pPcr4-His | His-pcr4 fusion in pQE30 vector, Apr | This study |
| pBT-popN | popN gene in pBT vector, Chlr | This study |
| pBT-pcr1 | pcr1 gene in pBT vector, Chlr | This study |
| pBT-pcr2 | pcr2 gene in pBT vector, Chlr | This study |
| pBT-pcr3 | pcr3 gene in pBT vector, Chlr | This study |
| pBT-pcr4 | pcr4 gene in pBT vector, Chlr | This study |
| pBT-pcrD | pcrD gene in pBT vector, Chlr | This study |
| pBT-pcrR | pcrR gene in pBT vector, Chlr | This study |
| pTRG-popN | popN gene in pTRG vector, Tcr | This study |
| pTRG-pcr1 | pcr1 gene in pTRG vector, Tcr | This study |
| pTRG-pcr2 | pcr2 gene in pTRG vector, Tcr | This study |
| pTRG-pcr3 | pcr3 gene in pTRG vector, Tcr | This study |
| pTRG-pcr4 | pcr4 gene in pTRG vector, Tcr | This study |
| pTRG-pcrD | pcrD gene in pTRG vector, Tcr | This study |
| pTRG-pcrR | pcrR gene in pTRG vector, Tcr | This study |
| pYAN0671 | pscB gene cloned into pTRG-pcr2, Tcr | This study |
Casein-agarose-mediated affinity purification.
Wild-type P. aeruginosa strain PAK was grown under type III inducing conditions (LB containing 5 mM EGTA) or noninducing conditions (LB). Bacterial outer membrane fractions were isolated by sucrose gradient centrifugation as described previously (39). The outer membrane proteins were solubilized by sonication in a buffer containing Triton X-100 (50 mM Tris Cl [pH 8.0], 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.1% Triton X-100) and then ultracentrifuged to remove membrane debris. Solubilized proteins were mixed with casein-agarose conjugate (Sigma) and incubated overnight at 4°C with gentle agitation. The agarose beads were washed thoroughly with the same buffer, and bound proteins were eluted with 1× protein loading buffer. Eluted proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining. Protein bands were excised from the gel and subjected to mass spectrometry analysis.
Whole-cell ELISA.
A whole-cell enzyme-linked immunosorbent assay (ELISA) was carried out as described previously (53), with minor modifications. The bacterial cells were grown under type III inducing conditions (LB containing 5 mM EGTA) or noninducing conditions (LB) for 4 h at 37°C. Cells were collected and resuspended in 50 mM sodium carbonate buffer (pH 9.6). Each well of a 96-well ELISA plate was coated with 108 bacterial cells overnight at 4°C. The wells were washed once with phosphate-buffered saline (PBS) containing 0.1% Tween 20 and blocked with PBS containing 5% nonfat milk for 2 h at room temperature. Anti-Flag monoclonal antibody (1:1,000 dilution) was added and incubated at room temperature for 2 h. The plates were washed three times with PBS containing 0.1% Tween 20. Horseradish peroxidase-conjugated secondary antibody (1:1,000 dilution) was added and incubated for another 2 h at room temperature. Following three washes, a substrate solution containing 10 mg O-phenylenediamaine in 10 ml citrate buffer (0.2 M Na2PO4, 0.1 M citric acid; pH 5.0) supplemented with 8 μl 30% H2O2 was added. The reaction was stopped by addition of 0.1 ml of 2 N sulfuric acid, and the absorbance at 492 nm was determined.
Generation of P. aeruginosa mutants.
Primers were designed to amplify fragments containing target genes with flanking regions (see Table S1 in the supplemental material). The PCR products were first cloned into the pCR2.1-TOPO vector (Invitrogen) and sequenced to confirm that the clones were the correct clones. The fragments were then subcloned into suicide vector pEx18G or pEx18Tc (22). For the popN, pcrV, and popB genes, ScaI/HincII (207 bp), HincII (250 bp), and EcoRV/NotI (520 bp) internal fragments, respectively, were deleted. For the pcr1, pcr2, pcr3, and pcr4 genes, the start codons were changed to TTC, ATC, CTC, and TAG, respectively, by site-directed mutagenesis. The oligonucleotides used for mutagenesis are also listed in Table S1 in the supplemental material. New restriction enzyme sites were introduced at the mutation sites for ease of screening after crosses into the chromosomes of P. aeruginosa. The mutated plasmid constructs were electroporated into P. aeruginosa, and double-cross mutants were isolated as described previously (22). A complementing plasmid for the pcrV mutant (pPcrV19) was constructed by PCR cloning of the gene into pUCP19 (the primers are shown in Table S1 in the supplemental material), while C-terminally Flag-tagged gene clones were used to complement popN, pcr1, pcr2, pcr3, and pcr4 mutants (see below).
Overexpression of tagged fusion proteins.
Oligonucleotides (see Table S1 in the supplemental material) were designed to fuse target genes with various tags in fusion vectors, including the Flag tag vector pFlag-CTC (Sigma), the His tag vector pQE30 (QIAGEN), and the glutathione S-transferase (GST) tag vector pGEX-4T-2 (Pharmacia). PCR products were first cloned into pCR2.1-TOPO and then subcloned into the fusion vectors. The resulting clones were sequenced to confirm in-frame fusions.
The Flag-tagged genes described above were also used for complementation experiments. Most of the genes were driven by a tac promoter on the vector; the only exception was popN, which contained its own promoter. The pCTC-Flag-based plasmids were fused with the pDN19 vector (38) at a BamHI site, while the EcoRI/XmnI fragment containing popN::flag was ligated into the EcoRI/SmaI sites of pUCP19 (45).
Bacterial two-hybrid system.
A BacterioMatch II two-hybrid vector kit (Stratagene) was used to detect protein-protein interactions. First, all genes were PCR cloned into pCR2.1-TOPO and subcloned into the bait vector pBT, as well as the target vector pTRG. The PCR primers were designed to keep every gene in frame in both vectors (see Table S1 in the supplemental material). In-frame fusions were confirmed by DNA sequencing. The members of each pair of plasmid constructs to be tested were electroporated simultaneously into a reporter strain, E. coli RS. The resulting transformants were grown overnight in L broth, diluted fivefold into fresh L broth containing 0.01 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and shaken at 30°C for 5 h. Cells were then collected, and a beta-galactosidase activity assay was performed as described previously (34). Each assay was repeated at least three times independently. The assay results were compared with the results for a Gal4-Gal11 positive control (provided by the manufacturer), as well as an empty vector negative control.
Protein pull-down assay.
Target proteins with either a His tag or a GST tag were purified with Ni-agarose or glutathione-agarose. In the case of His-Pcr4, the protein was insoluble and thus subjected to denaturation and renaturation as described previously (28). For a pull-down assay, purified His-tagged proteins were incubated with E. coli cell extract expressing GST-tagged proteins at 4°C overnight. The mixtures were subjected to affinity purification using glutathione-agarose for GST. The beads were washed three times with PBS, and bound proteins were eluted with 10 mM reduced glutathione in PBS buffer. Samples were separated by SDS-PAGE, stained with Coomassie blue, and blotted with monoclonal antibody against the His tag.
T3SS induction conditions.
Bacterial cells were grown overnight in LB with the proper antibiotics. Unless indicated otherwise, 1% of an overnight culture was inoculated into fresh LB containing 5 mM EGTA to induce the T3SS. Cells were incubated at 37°C for 3 h with shaking. When DMEM was used, 5% fetal bovine serum was also added, and the preparation was kept in a tissue culture incubator at 37°C for 3 h without shaking. For the Western blot analysis, the sample loading volumes on the SDS-PAGE gel were adjusted based on the optical density at 600 nm of each sample.
HeLa cell infection experiments.
HeLa cells were seeded into six-well plates to obtain 50% confluence after 24 h of growth. Log-phase bacterial cells were used to infect the HeLa cells. The cell densities were estimated based on the optical density at 600 nm. The multiplicity of infection (MOI) was 20. Two hours postinfection, HeLa cells were scraped and centrifuged at 500 × g for 5 min to pellet them. The supernatant was centrifuged again at top speed for 5 min to remove the bacterial cells in the pellet. To the HeLa cell pellets, 50 μl PBS containing 0.25% Triton X-100 was added, and then the preparation was kept on ice for 5 min and centrifuged at the top speed at 4°C for 15 min. The supernatant containing HeLa cell cytosolic proteins was mixed with an equal volume of 2× loading buffer and boiled for 10 min. Proteins in the culture supernatant were precipitated with 15% trichloroacetic acid, washed with cold acetone, and resuspended in 1× loading buffer. The protein samples were separated by 10% SDS-PAGE and subjected to Western blot analysis.
Cell lifting assay.
HeLa cells (5 × 104 cells) were seeded into a 24-well plate. The cells were cultured in DMEM with 5% fetal bovine serum at 37°C in the presence of 5% CO2 for 24 h. HeLa cells were infected with log-phase bacterial cells at an MOI of 20. Cell lifting assays were performed after 4 h of infection. Culture medium was aspirated, washed twice with PBS, and stained with 0.05% crystal violet for 5 min. The stain solution was discarded, and the plates were washed twice with water. Then 0.25 ml of 95% ethanol was added to each well and incubated at room temperature for 30 min with gentle shaking. The ethanol solution with dissolved crystal violet dye was used to determine the absorbance at 590 nm.
RESULTS
Casein and serum albumin specifically bind PopN of P. aeruginosa.
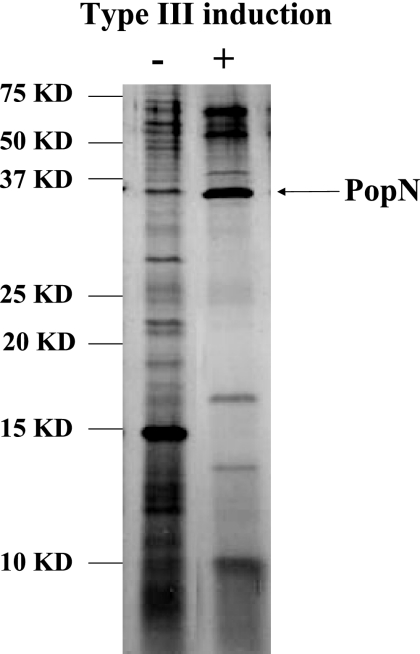
We have previously reported that activation of type III secretion in P. aeruginosa requires not only a low calcium concentration but also a protein factor called TSF which contains casein and serum albumin (31). To understand the role of TSF in the activation of type III secretion, we analyzed its interaction with the bacterial cell membrane components. Bacterial outer membrane fractions were prepared from type III induced or noninduced cells of wild-type strain PAK, and the outer membrane proteins were solubilized by sonication in a buffer containing Triton X-100 (see Materials and Methods). Solubilized proteins were subjected to affinity purification through casein-agarose conjugate as the affinity matrix. The affinity beads were washed, and proteins bound to the casein were directly eluted with 1× protein loading buffer. The eluted samples were subjected to separation by 12% SDS-PAGE and were visualized by silver staining. As Fig. 1 shows, numerous protein bands from both induced and noninduced membrane fractions bound to the casein-agarose. Interestingly, a 30-kDa protein band was dominant in the induced bacterial cell membrane. Mass spectrometry analysis of the protein band identified it as the PopN protein, a component of the T3SS.
FIG. 1.
Casein binding proteins on the P. aeruginosa outer membrane. Outer membrane fractions were isolated from strain PAK grown under type III inducing conditions (lane +) or noninducing conditions (lane −), and Triton X-100-solubilized proteins were subjected to affinity purification using casein-agarose. Specific bound proteins were subjected to 12% SDS—PAGE, followed by silver staining. KD, kilodaltons.
To further confirm the results described above, the PopN protein was overexpressed in the form of an N-terminally His-tagged fusion protein in E. coli. Purified His-PopN protein was tested for binding to both casein and bovine serum albumin (BSA). As a control, another key T3SS component, PcrV, was also overexpressed and purified (Fig. 2A). Purified His-PopN and His-PcrV proteins were incubated with either BSA-agarose or casein-agarose, and bound proteins were subjected to separation by 12% SDS-PAGE, followed by Western blotting using antibody against the His tag. As shown in Fig. 2B and C, both BSA-agarose and casein-agarose specifically pulled down His-PopN but not His-PcrV, proving that there was a specific interaction between TSF and PopN. The binding was further validated by an ELISA. First, a 96-well plate was coated with 1 μg/ml BSA overnight, washed with PBS, and incubated with serial dilutions of purified His-PopN or cell lysate of E. coli expressing His-PopN. As controls, purified His-PcrV and cell lysates of E. coli expressing His-PcrV were used. The plates were washed with PBS, and bound His-PopN or His-PcrV was detected by anti-His tag antibody conjugated with horseradish peroxidase. As shown in Fig. 2D, wells coated with BSA specifically bound His-PopN (purified or in cell lysates) but not His-PcrV. When the results described above were combined, it was clear that TSF specifically bound to the PopN protein.
FIG. 2.
PopN specifically binds casein and albumin. (A) Both PopN and PcrV were overexpressed in the form of N-terminal His-tagged proteins and were affinity purified from E. coli using Ni-agarose. (B) His-PopN was specifically pulled down by casein-agarose. (C) His-PopN was specifically pulled down by albumin-agarose. (D) High-level anti-His tag signal was detected in a BSA-coated 96-well plate following incubation with either purified His-PopN or a crude E. coli cell lysate containing His-PopN.
PopN functions as an inhibitor of the T3SS.
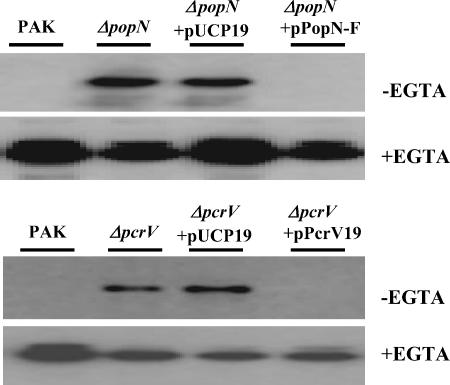
To understand the role of PopN in the type III secretion of P. aeruginosa, a popN gene knockout mutant was generated. Since the popN gene is the first gene in a type III operon, a deletion mutant was generated to avoid polar effects on downstream genes. Similarly, a pcrV deletion mutant was also generated. As shown in Fig. 3, secretion of ExoS in L broth was partially constitutive in both popN and pcrV mutants, and addition of EGTA further stimulated type III secretion. The altered secretion phenotypes of the mutants were effectively corrected to the wild-type phenotype by introduction of corresponding wild-type gene clones (Fig. 3). Taken together, the results described above indicated that PopN and PcrV function as inhibitors of the type III secretion apparatus, and the observed interaction between TSF and PopN suggests that TSF may trigger type III secretion by disrupting the inhibitor function of PopN or PcrV.
FIG. 3.
Constitutive ExoS secretion by a popN mutant. ExoS secretion by wild-type strain PAK and isogenic popN and pcrV mutant strains, as well as mutants harboring either vector plasmid pUCP19 or vector plasmid pUCP19 containing the corresponding wild-type genes, was examined. Bacterial cells were grown in L broth in the presence or absence of 5 mM EGTA, and secreted ExoS in the culture supernatants was detected by Western blotting using anti-ExoS antibody.
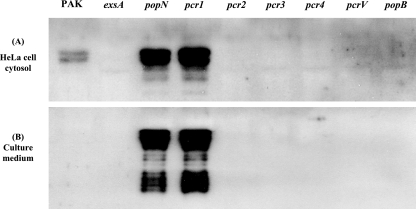
Previously, the pcrV mutant was shown to be defective in type III injection into host cells (polarized translocation) (43, 50). To determine if the popN mutant was also defective in targeted injection of the effector molecules, HeLa cells were infected with strain PAK or a popN or pcrV mutant at an MOI of 20 for 2 h. As negative controls, HeLa cells were infected with an exsA or popB mutant. The HeLa cell cytosolic proteins were then subjected to SDS-PAGE, followed by Western blotting using antibody against ExoS. Surprisingly, a larger amount of ExoS was injected into HeLa cells by the popN mutant than by PAK, while the pcrV mutant injected no detectable ExoS (Fig. 4A). Consistent with this observation, the popN mutant caused faster HeLa cell lifting than PAK caused, while pcrV had no effect on the HeLa cells. Moreover, a large amount of ExoS was also detected in the culture medium of HeLa cells infected by the popN mutant, while no ExoS was detected in the culture medium of HeLa cells infected by PAK (Fig. 4B). These results indicated that the PopN protein controls type III secretion but plays no role in polarized translocation. In contrast, the PcrV protein controls type III secretion, as well as polarized translocation.
FIG. 4.
Mutational effects of different genes on ExoS secretion. Type III secretion system-mediated injection of the ExoS protein into the HeLa cell cytosol (A) and secretion into the culture supernatant (B) by different mutant strains were examined. HeLa cells were infected with the P. aeruginosa strains at an MOI of 20 for 2 h.
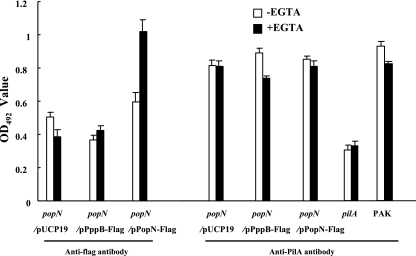
PopN is exposed on the bacterial cell surface. The PopN-TSF interaction data suggested that PopN is an outer membrane-associated protein, possibly binding to the TSF on the bacterial cell surface. To test this possibility further, whole-cell ELISA experiments were conducted. We generated a PopN-Flag fusion construct which was capable of complementing a popN mutant strain (Fig. 5A) and thus encoded a functional PopN protein. The popN mutant complemented by the pPopN-Flag construct (popN/pPopN-Flag) was grown under either type III inducing or noninducing conditions and coated onto a 96-well plate (see Materials and Methods). Surface exposure of the PopN-Flag fusion protein was detected by a whole-cell ELISA using antibody against the Flag tag. As a negative control, strain PAK harboring a cytoplasmic Flag fusion protein, PppB-Flag (1), was utilized. The tests detected significantly more Flag tag signal on the surface of popN/pPopN-Flag cells grown under type III inducing conditions than on the surface of noninduced cells or cells of the negative control strain PAK/pPppB-Flag grown under either inducing or noninducing conditions (Fig. 6). Using the same cells, surface localization of pili was also detected with antibody against the pilin subunit (PilA). As Fig. 6 shows, high levels of PilA were detected on all strains, while the signal for the pilA mutant was significantly lower. Similar levels of PilA on the test strains also served as controls for the equal numbers of bacterial cells adhering to the 96-well plates. As shown by the results described above, PopN-Flag was readily detected on the surface of bacterial cells, and the amount was influenced by the type III inducing signal.
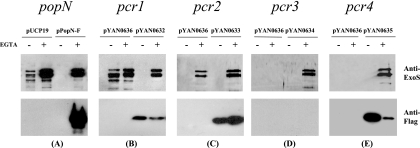
FIG. 5.
Secretion status of PopN, Pcr1, Pcr2, Pcr3, and Pcr4. popN (A), pcr1 (B), pcr2 (C), pcr3 (D), and pcr4 (E) mutant strains harboring an empty vector (pUCP19 or pYAN0636) or a vector containing the corresponding genes fused to a Flag tag at the C terminus were grown in L broth with (+) or without (−) EGTA. Culture supernatants were subjected to Western blotting to detect ExoS (upper panels) or Flag-tagged proteins (lower panels). Vector control plasmid pYAN0636 is a product of fusion of pDN19 and pCTC-Flag. Plasmids pYAN0632, pYAN0633, pYAN0634, and pYAN0635 contained Pcr1-Flag, Pcr2-Flag, Pcr3-Flag, and Pcr4-Flag, respectively.
FIG. 6.
Detection of PopN-Flag and pilin on the bacterial cell surface by whole-cell ELISA. Microplates were coated with 108 bacterial cells grown under type III inducing conditions (+EGTA) or noninducing conditions (−EGTA). PopN-Flag and pili were detected with anti-Flag monoclonal antibody and rabbit anti-PilA polyclonal antibody, respectively. PAK, wild-type P. aeruginosa strain; popN−, popN mutant of PAK; pilA−, pilA mutant of PAK; pUCP19, vector control; pPppB-Flag, fusion construct with Flag tag at the C terminus of cytoplasmic protein PppB; pPopN-Flag, popN::flag fusion construct. OD492, optical density at 492 nm.
Interactions among the popN operon gene products.
To understand the possible interactions among the popN operon gene products, a BacterioMatch two-hybrid system (Stratagene) was utilized. This system is based on the principle that the binding of RNA polymerase (RNAP) to a promoter can be stabilized through strong protein-protein binding between a DNA-binding activator and the α subunit of RNA polymerase. The bait plasmid contains a λcI gene, while the prey plasmid contains an RNAP α-subunit gene; both genes are driven by lacUV5 promoters. In the reporter strain, a λcI binding sequence is located upstream of two reporter genes, bla and lacZ. Interaction of the bait and prey stabilizes the binding of λcI and the RNAP α subunit in the promoter areas of the bla and lacZ genes and activates the expression of these two genes, giving rise to ampicillin-resistant colonies. Expression of the β-galactosidase gene is then used as a secondary marker.
Each of the seven popN operon genes was fused to the λcI gene in the bait, as well as to the RNAP α-subunit gene in the prey constructs. Pairs of plasmids were cotransformed into the E. coli reporter strain to conduct β-galactosidase assays. As shown in Table 2, the presence of pcr3 and pcr4 plasmids together resulted in β-galactosidase activity that was as high as the activity of the Gal4-Gal11 positive control provided by the kit manufacturer. Also, the β-galactosidase activities when the popN and pcr1 plasmids or the pcr1 and pcr4 plasmids were present together were significantly higher than the activities observed with the corresponding plasmids cotransformed with empty vector pBT or pTRG, suggesting that there were specific protein-protein interactions between PopN and Pcr1, between Pcr1 and Pcr4, and between Pcr3 and Pcr4.
TABLE 2.
Interaction study using BacterioMatch two-hybrid system
| pBT plasmid | Interaction witha:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vector | pTRG-Gal11p | pTRG-popN | pTRG-pcr1 | pTRG-pcr2 | pTRG-pcr3 | pTRG-pcr4 | pTRG-pcrD | pTRG-pcrR | pTRG-pcr2 + pscB | |
| Vector | − | − | − | − | − | − | − | − | − | − |
| pBT-LGF2 | − | +++b | NT | NT | NT | NT | NT | NT | NT | NT |
| pBT-popN | − | NT | − | ++ | − | − | − | − | − | ++ |
| pBT-pcr1 | − | NT | ++ | − | − | − | + | − | − | NT |
| pBT-pcr2 | − | NT | − | − | − | − | − | − | − | NT |
| pBT-pcr3 | − | NT | − | − | − | − | +++ | − | − | NT |
| pBT-pcr4 | − | NT | − | + | − | ++ | − | − | − | NT |
| pBT-pcrD | − | NT | − | − | − | − | − | − | − | NT |
| pBT-pcrR | − | NT | − | − | − | − | − | − | − | NT |
−, no interaction (β-galactosidase activity, <180); +, positive interaction with β-galactosidase activity of 250 to 350; ++, positive interaction with β-galactosidase activity of 350 to 450; +++, positive interaction with β-galactosidase activity of >450; NT, not tested.
Positive control provided with the kit.
To confirm the results described above, a protein pull-down assay was performed. The PopN and Pcr4 proteins were overexpressed as N-terminal His tag fusions, while Pcr1 and Pcr3 were overexpressed as GST fusions (Fig. 7A and C). E. coli cells expressing the fusions were lysed by sonication, and soluble proteins were recovered by high-speed centrifugation. To examine the interaction between PopN and Pcr1, cell lysate containing His-PopN was mixed with cell lysate containing GST-Pcr1. As a negative control, GST was used in place of GST-Pcr1. Glutathione-agarose beads were added to the mixtures and allowed to bind GST-Pcr1 or GST. The beads were recovered from the mixture by centrifugation. Following washes, the bead-associated proteins were eluted with reduced glutathione, separated by 12% SDS—PAGE, and visualized by Coomassie blue staining or Western blotting using antibody against the His tag. As shown in Fig. 7B, both GST and GST-Pcr1 were effectively pulled down from the mixture by the affinity beads. However, His-PopN was detected only with the pull-down sample of GST-Pcr1 and not with the pull-down sample of GST, demonstrating that there was a specific interaction between Pcr1 and PopN. Using a similar approach, specific interactions between Pcr1 and Pcr4, as well as between Pcr3 and Pcr4, were also observed (Fig. 7D).
FIG. 7.
Pull-down assay of interacting proteins. Proteins were separated by SDS-PAGE and stained with Coomassie blue (A and C) or subjected to Western blotting with anti-His tag monoclonal antibody (B and D). Lanes 1 and 6, GST; lanes 2 and 7, GST-Pcr1; lane 3, lysate of His-PopN-expressing cells; lane 4, GST pull-down of His-PopN; lane 5, GST-Pcr1 pull-down of His-PopN; lane 8, GST-Pcr3; lane 9, purified His-Pcr4; lane 10, GST pull-down of His-Pcr4; lane 11, GST-Pcr1 pull-down of His-Pcr4; lane 12, GST-Pcr3 pull-down of His-Pcr4.
According to a previous study with Yersinia, SycN (a homolog of Pcr2) and YscB (a homolog of PscB) form a complex, and the complex functions as a chaperone for the YopN protein. Neither SycN alone nor YscB alone can interact with the YopN protein (9). Thus, it is possible that the interaction between PopN and Pcr2 requires the presence of PscB at the same time. Indeed, simultaneous introduction of an intact pscB gene in the two-hybrid assay for the PopN-Pcr2 interaction resulted in higher β-galactosidase activity (Table 2).
Mutational analysis of the pcr1, pcr2, pcr3, and pcr4 genes.
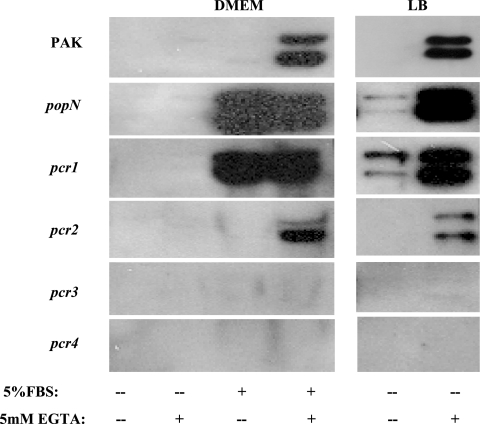
Using the results described above, the phenotypes of the mutants defective in the pcr1, pcr2, pcr3, and pcr4 genes were investigated. Each of the genes was mutated by replacing the start codon (ATG) with a nonstart codon (TTC, ATC, CTC, and TAG, respectively). First, the levels of secretion of the ExoS and ExoT proteins in the mutants were determined by Western blotting using antibody against ExoS. As Fig. 8 shows, the pcr1 mutant secreted ExoS constitutively, similar to the popN mutant, while a mutation in the pcr3 or pcr4 gene totally abolished ExoS secretion. A pcr2 mutation had a minor effect on type III secretion, and the mutant secreted about 75% of the ExoS secreted by wild-type strain PAK. The altered ExoS secretion phenotypes were readily corrected by introduction of corresponding wild-type gene clones in the form of C-terminally Flag-tagged fusions (Fig. 5).
FIG. 8.
ExoS secretion by various mutant strains. Secretion of ExoS by mutants grown under different conditions (DMEM or L broth with or without EGTA) was detected by Western blotting. FBS, fetal bovine serum.
Next, the mutational effects of these genes on HeLa cell infection were investigated. Wild-type strain PAK was used as a positive control, while an exsA mutant and a pcrV mutant were used as negative controls. Two hours postinfection (MOI, 20), both popN and pcr1 mutant strains caused 100% cell rounding and lifting, whereas wild-type strain PAK and the pcr2 mutant caused around 90% cell rounding but no lifting. Mutants with mutations in pcr3 and pcr4 caused no cell rounding or lifting. Infected HeLa cells were collected after 2 h of infection, and translocated ExoS was detected in the cytosolic protein samples by Western blot analysis (see Materials and Methods). As Fig. 4 shows, both popN and pcr1 mutants translocated much more effector proteins into HeLa cell cytoplasm than wild-type strain PAK translocated. Furthermore, the popN and pcr1 mutants also secreted ExoS and ExoT into the culture medium, while no such secretion by wild-type strain PAK was observed (Fig. 4). Consistent with the cell rounding assay results, the pcr3 and pcr4 mutants translocated no detectable ExoS into the HeLa cells, while the pcr2 mutant translocated a reduced amount of the ExoS.
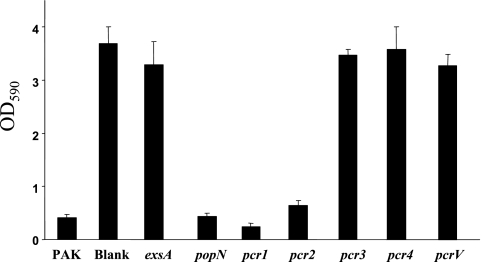
Finally, cell lifting was quantified after 4 h of infection by measuring the crystal violet staining associated with the adhering HeLa cells (see Materials and Methods). As Fig. 9 shows, the popN and pcr1 mutants caused amounts of cell lifting similar to the amounts caused by PAK and the pcr2 mutant, whereas the pcr3 and pcr4 mutants did not cause any lifting, similar to the results obtained with the negative controls (exsA and pcrV mutants).
FIG. 9.
HeLa cell lifting assay. HeLa cells were infected with different strains at an MOI of 20 for 4 h, and the cells that adhered to the culture plates were stained with crystal violet. The dye was dissolved in 95% ethanol, and the absorbance at 590 nm (OD590) was determined.
In summary, mutations in the popN and pcr1 genes resulted in identical constitutive type III secretion phenotypes. Considering the fact that these two components specifically bind to each other, the two components are likely to form a functional repressor complex. Disruption of pcr3 or pcr4 resulted in no detectable secretion or translocation of the type III effector proteins; thus, both the pcr3 and pcr4 genes are indispensable for type III secretion. Pcr2, a component implicated as a PopN chaperone together with PscB, has a minimal effect on type III secretion.
Secretion status of the Pcr1, Pcr2, Pcr3, and Pcr4 proteins.
Since PopN is known to be secreted under type III inducing conditions, we were interested in determining if any of the PopN binding components are also secreted. Each gene was fused to a Flag tag at the C terminus, and the resulting constructs were tested to determine their abilities to complement corresponding gene knockout mutants. An ExoS secretion assay (Fig. 5, upper panels) indicated that all of the Flag-tagged proteins were able to complement corresponding gene mutants, demonstrating that the Flag tags did not interfere with the functions of the target genes. Under the same conditions, PopN-Flag, Pcr1-Flag, Pcr2-Flag, and Pcr4-Flag were readily detectable by Western blotting in the culture supernatant (Fig. 5, lower panels), while Pcr3-Flag was not detectable. A negative control (PppB-Flag fusion) was also not secreted under the assay conditions (data not shown). Interestingly, secretion of PopN-Flag required EGTA, while secretion of Pcr1, Pcr2, and Pcr4 seemed to be relatively independent of EGTA; in fact, EGTA inhibited the secretion of Pcr1 and Pcr4. Nonetheless, secretion of these components was dependent on the T3SS, as secretion did not occur with an exsA mutant background (data not shown). The difference in the secretion patterns might reflect ordered secretion during T3SS needle biogenesis.
DISCUSSION
Type III secretion in P. aeruginosa has previously been shown to require both a low calcium signal (14, 52) and a protein factor called TSF, which includes caseins and serum albumin (31). Since TSFs are large protein molecules, their effects on type III secretion are most likely through direct interactions with the bacterial surface components. In this study, we isolated TSF binding proteins from the outer membranes of type III induced and noninduced cells of P. aeruginosa using casein-agarose. Although multiple proteins were bound to the TSF, PopN was the dominant protein from the type III induced cells (Fig. 1). Also, using a whole-cell ELISA approach, surface-exposed PopN-Flag was readily detectable in type III induced cells, suggesting that there may be a TSF-PopN interaction during type III activation. Elimination of the PopN protein (popN mutant) resulted in constitutive ExoS secretion, further suggesting the importance of the observed TSF-PopN interaction, although possible involvement of additional TSF binding proteins, as shown in Fig. 1 for type III secretion, cannot be ruled out at this time. Efforts are under way to identify and test the roles of these proteins in type III secretion.
Using a bacterial two-hybrid system, several protein-protein interactions among the popN operon gene products were found. The β-galactosidase activity assay indicated that there were strong interactions between PopN and Pcr1, as well as between Pcr3 and Pcr4, consistent with previous reports of Pcr3-Pcr4 interactions, as well as similar interactions in Yersinia (YopN-TyeA and YscX-YscY interactions) (5, 25). Most interestingly, we found a novel protein-protein interaction between Pcr1 and Pcr4 linking a chain of interactions, namely PopN-Pcr1-Pcr4-Pcr3. All of these interactions were readily observable in protein pull-down assays, further increasing the likelihood of in vivo interactions. The observed protein interaction chain may well represent part of a complex controlling type III secretion. Studies have suggested that in Yersinia the LcrH-YscY-YscX and LcrH-YopD-LcrQ regulatory loops are involved in regulating gene expression and the ordered secretion of type III substrates (2, 5, 6, 13).
There are a number of important differences between the T3SS of Yersinia and P. aeruginosa. The first difference is the secretion status of the Pcr3 and Pcr4 proteins. In contrast to the Yersinia pestis system, where YscX is secreted into the culture supernatant while YscY is not (10), in P. aeruginosa Pcr4, instead of Pcr3, is actually secreted through the type III secretion machinery. It has been reported previously that Pcr4 is unable to complement a yscY null mutant in Yersinia, despite the ability of YscY to function in P. aeruginosa (5). Second, a mutation in the popN gene of P. aeruginosa results in constitutive type III secretion and increased effector injection into the host cells, while a mutation in yopN of Yersinia results in a defect in effector injection. Third, mutation of the pcrV gene of P. aeruginosa results in constitutive effector secretion (43, 50), while an lcrV mutant of Yersinia is down-regulated for Yop expression and secretion (3, 36, 47). These differences may highlight the organism-specific regulatory mechanisms of the T3SS.
The PopN and Pcr1 proteins seem to form a complex to block T3SS expression in P. aeruginosa. Their counterparts in Yersinia, YopN and TyeA, form a complex with the two other proteins, namely SycN and YscB, which are homologues of Pcr2 and PscB in P. aeruginosa. This complex was proposed to block T3SS expression in the absence of induction signals. In our bacterial two-hybrid experiment we did not observe a positive interaction between the popN and pcr2 genes, while the presence of pscB resulted in a positive interaction, consistent with similar interactions in Yersinia, where SycN and YscB form a complex and the complex functions as the chaperone for the YopN protein and neither SycN alone nor YscB alone can interact with the YopN protein (9). Furthermore, consistent with the proposed chaperone role of the Pcr2 protein, a pcr2 knockout mutant exhibited a slight decrease in type III secretion, secreting about 75% of the amount secreted by the wild type. Also, a pcr2 mutation does not affect popN gene expression; it affects only PopN secretion (H. Yang and S. Jin, unpublished results). An unexpected observation is that the Pcr2 protein is secreted in a T3SS-dependent manner. Since chaperone proteins are normally not secreted, this observation seems to indicate that the Pcr2 protein may have additional functions besides its role as a PopN chaperone. The fact that secretion of Pcr2 (and also Pcr2 and Pcr4) seems to occur before secretion of PopN (in the absence of EGTA) may imply that there is stage-specific secretion of these components during T3SS needle biogenesis. Efforts are under way to understand the molecular details of this process.
Secretion of the type III effectors was proposed to be controlled by a “Plug” or “CAP” on the tip of the type III needle in Yersinia and Shigella strains (12, 25, 30). In Yersinia, Yop secretion is blocked in the presence of calcium ions prior to contact with the host cell. The block in Yop secretion is dependent on the secreted YopN protein, TyeA, and LcrG (12, 25, 37). Mutational inactivation of the yopN, tyeA, or lcrG gene results in uncontrolled secretion prior to host cell contact and in a loss of polarized translocation after host cell contact. Surface-exposed YopN is thought to function as a regulatory plug that prevents Yop secretion prior to host cell contact (12). In Shigella flexneri, the secreted IpaB and IpaD proteins are required to prevent T3SS prior to cell contact (33). The membrane-associated IpaB-IpaD complex has been proposed to function as a CAP that prevents secretion prior to contact with the surface of eukaryotic cells. Recently, the hypothetical CAP structure has been visualized by electron microscopy on the tips of type III needles from Yersinia, demonstrating for the first time the presence of the putative CAP-like structure on the type III needles (35). However, the exact composition of the CAP structure, the mechanism by which CAP blocks secretion, and the physiological signals that release the block in secretion are unknown.
Previously published data, as well as our experimental data presented here, support the hypothesis that there is a similar mechanism of control in the T3SS of P. aeruginosa. First, PopN and PcrV are surface localized, and mutations in the genes result in constitutive type III secretion; second, the three interacting components of PopN-Pcr1-Pcr4, as well as PcrV, are all secreted under type III inducing conditions and thus are likely be part of the CAP complex; and third, a mutation in the pcr1 gene results in a constitutive phenotype identical to that of the popN mutant and thus PopN and Pcr1 likely form a functional repressor complex. The exact composition of the CAP structure needs to be elucidated in order to understand the mechanism by which type III inducing signals trigger the secretion of type III effector molecules.
Although activation of type III secretion requires both a low calcium concentration and TSF, the binding of the PopN protein by either albumin or casein was independent of calcium (data not shown), suggesting that a low calcium concentration is not required to increase the binding affinity between TSF and PopN; rather, calcium affects some other aspects. Based on the proposed CAP model, it is possible that a low calcium level may weaken the association of CAP with the type III needle, enabling the TSF-PopN interaction to displace the CAP from the type III needle. Alternatively, another calcium-dependent TSF binding protein(s) might play a key role(s) in the activation of type III secretion. Further studies are needed to understand how TSF molecules without obvious amino acid sequence similarities can bind the same target. Two likely possibilities involve binding different epitopes of the same protein or binding pockets involving specific three-dimensional structures and/or specific amino acids. Efforts are under way to address these possibilities.
Supplementary Material
Acknowledgments
We thank Daniel J. Wozniak of Wake Forest University for the gift of anti-PilA antibody and Jinghua Jia and Xiaoling Wang for technical assistance.
This work was supported by a research scholarship award from the American Cancer Society to S.J.
Footnotes
Published ahead of print on 19 January 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahn, K.-S., U. Ha, J. Jia, D. Wu, and S. Jin. 2004. The truA gene of Pseudomonas aeruginosa is required for the expression of type III secretory genes. Microbiology 150:539-547. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, D. M., K. S. Ramamurthi, C. Tam, and O. Schneewind. 2002. YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J. Bacteriol. 184:1287-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergman, T., S. Hakansson, A. Forsberg, L. Norlander, A. Macellaro, A. Backman, I. Bolin, and H. Wolf-Watz. 1991. Analysis of the V antigen lcrGVH-yopBD operon of Yersinia pseudotuberculosis: evidence for a regulatory role of LcrH and LcrV. J. Bacteriol. 173:1607-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley, T. J., and N. H. Khan. 1974. The production of extracellular lipids by Pseudomonas aeruginosa NCTC 2000 in stationary liquid media containing macrogols. J. Pharm. Pharmacol. 26:900-902. [DOI] [PubMed] [Google Scholar]
- 5.Broms, J. E., P. J. Edqvist, K. E. Carlsson, A. Forsberg, and M. S. Francis. 2005. Mapping of a YscY binding domain within the LcrH chaperone that is required for regulation of Yersinia type III secretion. J. Bacteriol. 187:7738-7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cambronne, E. D., and O. Schneewind. 2002. Yersinia enterocolitica type III secretion: yscM1 and yscM2 regulate yop gene expression by a posttranscriptional mechanism that targets the 5′ untranslated region of yop mRNA. J. Bacteriol. 184:5880-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coburn, J., and D. M. Gill. 1991. ADP-ribosylation of p21ras and related proteins by Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 59:4259-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coombes, B. K., and B. B. Finlay. 2005. Insertion of the bacterial type III translocon: not your average needle stick. Trends Microbiol. 13:92-95. [DOI] [PubMed] [Google Scholar]
- 9.Day, J. B., and G. V. Plano. 1998. A complex composed of SycN and YscB functions as a specific chaperone for YopN in Yersinia pestis. Mol. Microbiol. 30:777-788. [DOI] [PubMed] [Google Scholar]
- 10.Day, J. B., and G. V. Plano. 2000. The Yersinia pestis YscY protein directly binds YscX, a secreted component of the type III secretion machinery. J. Bacteriol. 182:1834-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finck-Barbancon, V., J. Goranson, L. Zhu, T. Sawa, J. P. Wiener-Kronish, S. M. Fleiszig, C. Wu, L. Mende-Mueller, and D. W. Frank. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25:547-557. [DOI] [PubMed] [Google Scholar]
- 12.Forsberg, A., A. M. Viitanen, M. Skurnik, and H. Wolf-Watz. 1991. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol. Microbiol. 5:977-986. [DOI] [PubMed] [Google Scholar]
- 13.Francis, M. S., S. A. Lloyd, and H. Wolf-Watz. 2001. The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 42:1075-1093. [DOI] [PubMed] [Google Scholar]
- 14.Frank, D. W. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26:621-629. [DOI] [PubMed] [Google Scholar]
- 15.Frank, D. W., G. Nair, and H. P. Schweizer. 1994. Construction and characterization of chromosomal insertional mutations of the Pseudomonas aeruginosa exoenzyme S trans-regulatory locus. Infect. Immun. 62:554-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frithz-Lindsten, E., Y. Du, R. Rosqvist, and A. Forsberg. 1997. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III-dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol. Microbiol. 25:1125-1139. [DOI] [PubMed] [Google Scholar]
- 17.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 18.Ganesan, A. K., L. Mende-Mueller, J. Selzer, and J. T. Barbieri. 1999. Pseudomonas aeruginosa exoenzyme S, a double ADP-ribosyltransferase, resembles vertebrate mono-ADP-ribosyltransferases. J. Biol. Chem. 274:9503-9508. [DOI] [PubMed] [Google Scholar]
- 19.Goure, J., A. Pastor, E. Faudry, J. Chabert, A. Dessen, and I. Attree. 2004. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect. Immun. 72:4741-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ha, U., and S. Jin. 2001. Growth phase-dependent invasion of Pseudomonas aeruginosa and its survival within HeLa cells. Infect. Immun. 69:4398-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauser, A. R., P. J. Kang, and J. N. Engel. 1998. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27:807-818. [DOI] [PubMed] [Google Scholar]
- 22.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 23.Holmstrom, A., J. Olsson, P. Cherepanov, E. Maier, R. Nordfelth, J. Pettersson, R. Benz, H. Wolf-Watz, and A. Forsberg. 2001. LcrV is a channel size-determining component of the Yop effector translocon of Yersinia. Mol. Microbiol. 39:620-632. [DOI] [PubMed] [Google Scholar]
- 24.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iriarte, M., M. P. Sory, A. Boland, A. P. Boyd, S. D. Mills, I. Lambermont, and G. R. Cornelis. 1998. TyeA, a protein involved in control of Yop release and in translocation of Yersinia Yop effectors. EMBO J. 17:1907-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia, J., M. Alaoui-El-Azher, M. Chow, T. C. Chambers, H. Baker, and S. Jin. 2003. c-Jun NH2-terminal kinase-mediated signaling is essential for Pseudomonas aeruginosa ExoS-induced apoptosis. Infect. Immun. 71:3361-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia, J., Y. Wang, L. Zhou, and S. Jin. 2006. Expression of Pseudomonas aeruginosa toxin ExoS effectively induces apoptosis in host cells. Infect. Immun. 74:6557-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin, S. G., T. Roitsch, P. J. Christie, and E. W. Nester. 1990. The regulatory VirG protein specifically binds to a cis-acting regulatory sequence involved in transcriptional activation of Agrobacterium tumefaciens virulence genes. J. Bacteriol. 172:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufman, M. R., J. Jia, L. Zeng, U. Ha, M. Chow, and S. Jin. 2000. Pseudomonas aeruginosa mediated apoptosis requires the ADP-ribosylating activity of ExoS. Microbiology 146:2531-2541. [DOI] [PubMed] [Google Scholar]
- 30.Kenjale, R., J. Wilson, S. F. Zenk, S. Saurya, W. L. Picking, W. D. Picking, and A. Blocker. 2005. The needle component of the type III secretion of Shigella regulates the activity of the secretion apparatus. J. Biol. Chem. 280:42929-42937. [DOI] [PubMed] [Google Scholar]
- 31.Kim, J., K. Ahn, S. Min, J. Jia, U. Ha, D. Wu, and S. Jin. 2005. Factors triggering type III secretion in Pseudomonas aeruginosa. Microbiology 151:3575-3587. [DOI] [PubMed] [Google Scholar]
- 32.Krall, R., G. Schmidt, K. Aktories, and J. T. Barbieri. 2000. Pseudomonas aeruginosa ExoT is a rho GTPase-activating protein. Infect. Immun. 68:6066-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menard, R., P. Sansonetti, and C. Parsot. 1994. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 13:5293-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 35.Mueller, C. A., P. Broz, S. A. Muller, P. Ringler, F. Erne-Brand, I. Sorg, M. Kuhn, A. Engel, and G. R. Cornelis. 2005. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310:674-676. [DOI] [PubMed] [Google Scholar]
- 36.Nilles, M. L., K. A. Fields, and S. C. Straley. 1998. The V antigen of Yersinia pestis regulates Yop vectorial targeting as well as Yop secretion through effects on YopB and LcrG. J. Bacteriol. 180:3410-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilles, M. L., A. W. Williams, E. Skrzypek, and S. C. Straley. 1997. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response. J. Bacteriol. 179:1307-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nunn, D., S. Bergman, and S. Lory. 1990. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 172:2911-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunn, D. N., and S. Lory. 1993. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J. Bacteriol. 175:4375-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pastor, A., J. Chabert, M. Louwagie, J. Garin, and I. Attree. 2005. PscF is a major component of the Pseudomonas aeruginosa type III secretion needle. FEMS Microbiol. Lett. 253:95-101. [DOI] [PubMed] [Google Scholar]
- 41.Pederson, K. J., A. J. Vallis, K. Aktories, D. W. Frank, and J. T. Barbieri. 1999. The amino-terminal domain of Pseudomonas aeruginosa ExoS disrupts actin filaments via small-molecular-weight GTP-binding proteins. Mol. Microbiol. 32:393-401. [DOI] [PubMed] [Google Scholar]
- 42.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawa, T., T. L. Yahr, M. Ohara, K. Kurahashi, M. A. Gropper, J. P. Wiener-Kronish, and D. W. Frank. 1999. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat. Med. 5:392-398. [DOI] [PubMed] [Google Scholar]
- 44.Schoehn, G., A. M. Di Guilmi, D. Lemaire, I. Attree, W. Weissenhorn, and A. Dessen. 2003. Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. EMBO J. 22:4957-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 46.Shafikhani, S. H., and J. Engel. 2006. Pseudomonas aeruginosa type III-secreted toxin ExoT inhibits host-cell division by targeting cytokinesis at multiple steps. Proc. Natl. Acad. Sci. USA 103:15605-15610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skrzypek, E., and S. C. Straley. 1995. Differential effects of deletions in lcrV on secretion of V antigen, regulation of the low-Ca2+ response, and virulence of Yersinia pestis. J. Bacteriol. 177:2530-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun, J., and J. T. Barbieri. 2003. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 regulator of kinase (Crk) proteins. J. Biol. Chem. 278:32794-32800. [DOI] [PubMed] [Google Scholar]
- 49.Sundin, C., J. Thelaus, J. E. Broms, and K. Forsberg. 2004. Polarisation of type III translocation by Pseudomonas aeruginosa requires PcrG, PcrV and PopN. Microb. Pathog. 37:313-322. [DOI] [PubMed] [Google Scholar]
- 50.Sundin, C., M. C. Wolfgang, S. Lory, A. Forsberg, and E. Frithz-Lindsten. 2002. Type IV pili are not specifically required for contact dependent translocation of exoenzymes by Pseudomonas aeruginosa. Microb. Pathog. 33:265-277. [DOI] [PubMed] [Google Scholar]
- 51.Totten, P. A., and S. Lory. 1990. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J. Bacteriol. 172:7188-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallis, A. J., T. L. Yahr, J. T. Barbieri, and D. W. Frank. 1999. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect. Immun. 67:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitchurch, C. B., A. J. Leech, M. D. Young, D. Kennedy, J. L. Sargent, J. J. Bertrand, A. B. Semmler, A. S. Mellick, P. R. Martin, R. A. Alm, M. Hobbs, S. A. Beatson, B. Huang, L. Nguyen, J. C. Commolli, J. N. Engel, A. Darzins, and J. S. Mattick. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52:873-893. [DOI] [PubMed] [Google Scholar]
- 54.Yahr, T. L., J. Goranson, and D. W. Frank. 1996. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol. Microbiol. 22:991-1003. [DOI] [PubMed] [Google Scholar]
- 55.Yahr, T. L., L. M. Mende-Mueller, M. B. Friese, and D. W. Frank. 1997. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 179:7165-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 95:13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yahr, T. L., and M. C. Wolfgang. 2006. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 62:631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.