Abstract
The thioquinolobactin siderophore from Pseudomonas fluorescens ATCC 17400 utilizes a variation of the sulfur transfer chemistry found in thiamine and molydobterin biosynthesis. A JAMM motif protein cleaves the C-terminal amino acid residues following a diglycine moiety on a small sulfur carrier protein, and the modified C terminus is activated and sulfurylated, forming a thiocarboxylate.
Thioquinolobactin (8-hydroxy-4-methoxy-thioquinaldic acid; structure 1) is a siderophore produced by the fluorescent pseudomonad Psuedomonas fluorescens ATCC 17400 (10-12). The genes responsible for the biosynthesis of thioquinolobactin have been cloned, and putative functions have been assigned based on homology searches. The biosynthesis of thioquinolobactin appears to be a combination of two pathways; the first pathway involves tryptophan catabolism to hydroxykynurenine (7), and the second involves formation of thioquinolobactin (structure 1) from quinolobactin by a mechanism likely to be similar to that found in thiamine and molybdopterin biosynthetic pathways and a recently discovered cysteine biosynthetic pathway (2, 3, 13, 16). Here we describe the formation of an advanced sulfur donor (QbsE with a C-terminal thiocarboxylate) and identification of an unanticipated protease activity.
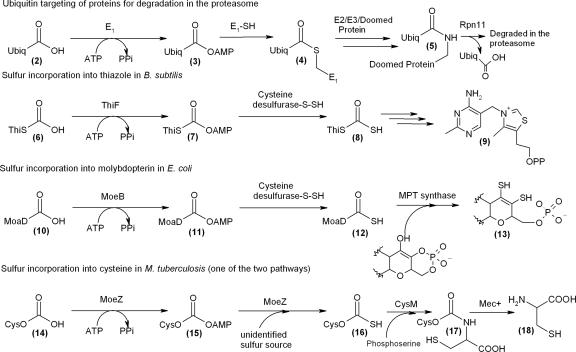
There are a number of naturally occurring molecules with diverse structures that contain sulfur, including thiamine, molybdopterin, and cysteine. In the biosynthesis of these molecules, small sulfur carrier proteins (ThiS structure 6, MoaD structure 10, and CysO structure 14) are converted to carboxy-terminal thiocarboxylates (structures 8, 12, and 16), which are responsible for the delivery of sulfur to the small target molecules (Fig. 1). This chemistry is analogous to the transformations that occur in the ubiquitin (structure 2) targeting of doomed proteins to the proteosome (5). In the thiamine and molybdopterin biosynthetic pathways, the small sulfur carrier protein (ThiS or MoaD) clusters with an adenylating protein (ThiF or MoeB) within the biosynthetic operon (2, 3, 8). The C terminus of each of the sulfur carrier proteins has a highly conserved -GG-COOH motif that functions as a flexible linker, picking up sulfide from one active site and delivering it to another. The similarity of these sulfur transfer systems suggested that a small sulfur carrier protein may also be utilized in the biosynthesis of other sulfur-containing molecules, such as thioquinolobactin (Fig. 2).
FIG. 1.
This-related proteins. Ubiquitin targeting of proteins for degradation in the proteasome, sulfur incorporation into thiazole in Bacillus subtilis, sulfur incorporation into molybdopterin in E. coli, and sulfur incorporation into cysteine in M. tuberculosis (one of two pathways) are shown. Ubiq, ubiquitin; MPT, molybdopterins.
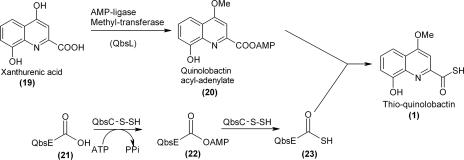
FIG. 2.
Proposed biosynthesis of the thioquinolobactin siderophore (structure 1) involving the use of a small sulfur carrier protein.
The thioquinolobactin gene cluster contains a gene encoding a putative small sulfur carrier protein, QbsE, which is clustered with an adenylating protein (QbsC) similar to ThiF and MoeB. Although QbsE has a diglycine sequence, this sequence is followed by two additional amino acids (cysteine-phenylalanine) that are not found in ThiS and MoaD (Fig. 3a). Also clustered with QbsC and QbsE is a putative metal-dependent hydrolase (QbsD) with a JAMM motif (2). This motif is found in some metalloproteases that cleave isopeptide bonds and consists of a conserved putative metal-binding domain (15). Since QbsD clusters with QbsC and QbsE, we predicted that QbsD might hydrolyze the two C-terminal residues of QbsE, thus generating the flexible diglycine moiety at its C terminus.
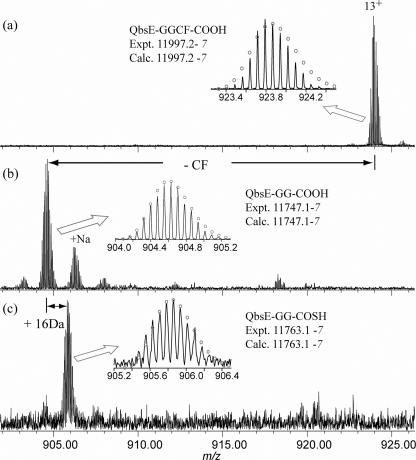
FIG. 3.
ESI-FTMS analysis of QbsE. (a) His-tagged QbsE coexpressed with QbsC showing no modification to QbsE; (b) His-tagged QbsE coexpressed with QbsD showing removal of the carboxy-terminal Cys-Phe dipeptide; (c) His-tagged QbsE coexpressed with QbsD and QbsC showing replacement of the carboxy-terminal Cys-Phe dipeptide with sulfide.
The qbsD and qbsE genes were PCR amplified from P. fluorescens ATCC 17400 genomic DNA and cloned into the pET28a and pACYC duet vectors, respectively, using standard methods (14). Proteins were overexpressed in Escherichia coli Tuner in Luria-Bertani medium with 1% sorbitol at 15°C for 13 h after induction with 0.5 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Lysis of the cell pellet in the presence of 0.1% Triton X-100 increased the yield of soluble protein, and the proteins were purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography. QbsD and QbsE showed only minimal solubility when they were heterologously overexpressed in E. coli. However, when His-tagged QbsD was coexpressed with His-tagged QbsE, the solubility of both proteins increased dramatically. These two proteins were copurified by Ni-NTA affinity chromatography, and a species whose molecular mass was 250 Da less than the expected molecular mass of unmodified QbsE was detected by electrospray ionization-Fourier transform mass spectrometry (ESI-FTMS) (Fig. 3b) (17). This difference corresponded to removal of the cysteine-phenylalanine carboxy terminus of QbsE. Mass spectrometry-mass spectrometry analysis confirmed that the modification occurred at the C terminus of QbsE. Assignments of the fragment masses and compositions were made with the computer program THRASH (6).
The reaction catalyzed by QbsD resembles the reaction catalyzed by Rpn11, a proteasomal cap protein containing a JAMM motif that is responsible for the deubiquitination of doomed proteins (Fig. 1) (15). Although the JAMM motif is found in prokaryotes, archaea, and eukaryotes, complete characterization of its activity and even functional assignment remain elusive (1). Only one other JAMM motif protein in prokaryotes has a functional assignment, the cysteine biosynthetic protein Mec+. Mec+ hydrolyzes cysteine from CysO in the alternative cysteine biosynthetic pathway found in Mycobacterium tuberculosis (2). QbsD and Mec+ exhibit significant sequence homology (47% identity and 67% similarity).
Generation of the flexible diglycine sequence at the carboxy terminus of QbsE allows subsequent sulfurylation of QbsE by QbsC. Before sulfurylation of QbsE can occur, the C terminus must be activated as the acyl adenylate. qbsC was cloned into a pET28a vector and expressed and purified as described above for qbsD and qbsE. Using [α-32P]ATP and taking advantage of the instability of the QbsE acyl adenylate, we were able to detect the QbsC- and QbsE-GG-COOH-dependent formation of [α-32P]AMP by thin-layer chromatography (4; unpublished results). Aliquots of the reaction mixture, which contained 8.5 μM QbsC, 11 μM QbsE, 100 μM ATP, and 1 mM MgCl2 in 250 μl of 50 mM Tris HCl (pH 7.8)-100 mM NaCl, were removed at various times and quenched by direct spotting on a silica thin-layer chromatography plate. The plates were developed in n-butanol—water-acetic acid (4:1:1) and exposed to a phorphorimaging screen.
In addition, QbsC contains a rhodanese domain that could be involved in the sulfur transfer to the activated C terminus of modified QbsE. We reconstituted the rhodanese activity of this domain by demonstrating that it catalyzes the transfer of sulfur from thiosulfate to cyanide via a protein-bound persulfide. QbsC was assayed for rhodanese activity by detecting the formation of thiocyanate (9). QbsC has a Km for thiosulfate of 2.3 mM and a kcat of 362 min−1 (4; unpublished results). Although thiosulfate acts as the in vitro sulfur donor, the ultimate in vivo sulfur donor is unknown. The persulfide that is generated on QbsC could then add to the C-terminal QbsE-acyl adenylate to form a QbsC-QbsE acyl disulfide. This compound could then be reduced to QbsE thiocarboxylate (structure 28).
When the genes encoding the sulfur transfer proteins, QbsCDE, were cloned as a contiguous unit into a pET28a vector and overexpressed in E. coli as described above for qbsD and qbsE, the solubility of each of the proteins increased compared with the solubility observed for overexpression of the proteins individually. In this construct, only QbsC possessed a His tag. Ni-NTA affinity chromatography resulted in copurification of QbsC and QbsE, indicating that these two proteins form a complex (4; unpublished results). Analysis of the complex by ESI-FTMS revealed a species whose molecular mass was 234 Da less than the expected average molecular mass of unmodified QbsE (Fig. 3c). This mass difference corresponded to replacement of the two C-terminal residues of QbsE with sulfide, resulting in a C-terminal diglycine thiocarboxylate on QbsE (structure 28). When QbsC was coexpressed and copurified with QbsE, no modification on QbsE could be detected, demonstrating that removal of the cysteine-phenylalanine dipeptide is essential for thiocarboxylate formation.
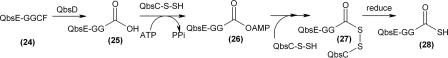
In this paper, we outline evidence pointing toward an interesting function for a JAMM motif protein (QbsD) in the modification of a sulfur carrier protein (QbsE) by cleavage of C-terminal amino acid residues. Based on this evidence, we propose a reaction sequence by which the modified QbsE thiocarboxylate is formed (Fig. 4). QbsD catalyzes removal of the carboxy-terminal dipeptide from QbsE (structure 24), exposing the flexible diglycine terminus (structure 25). The cleavage of the C-terminal residues of QbsE by QbsD is only the second characterized activity for a prokaryotic protein with a JAMM motif. This modification prepares QbsE for activation of its C terminus as the acyl adenylate (structure 26) by QbsC. A protein-bound persulfide on the rhodanese domain of QbsC attacks the QbsE-acyl adenylate, forming a QbsE-QbsC acyl persulfide (structure 27). The persulfide is reduced, releasing modified QbsE-thiocarboxylate (structure 28). QbsC is able to sulfurylate modified QbsE, QbsE-GG-COOH (structure 25), without QbsD present, and the role of these two proteins is almost certainly catalytic (4; unpublished results). The significance of the additional C-terminal amino acid residues is unknown. One possibility is that removal of the CF dipeptide by QbsD may play a regulatory role in the formation of thioquinolobactin (structure 1). The modification of the small sulfur carrier protein prior to formation of the carboxy-terminal thiocarboxylate represents an interesting deviation from the sulfur transfer systems found in thiamine, cysteine, and molybdopterin biosynthesis. A similar reaction sequence may be utilized in other sulfur transfer systems in which a protein with a JAMM motif clusters with a small sulfur carrier protein with additional amino acids after the diglycine at its C terminus.
FIG. 4.
Reaction sequence by which modified QbsE thiocarboxylate is formed. Structure 23 in Fig. 2 is equivalent to structure 28.
Acknowledgments
This research was supported by an NIH Chemical Biology Interface Traineeship to A.M.G. (grant GM008500) and by NIH grants to T.P.B. (grant GM069618) and F.W.M. (grant GM16609).
Footnotes
Published ahead of print on 5 January 2007.
REFERENCES
- 1.Ambroggio, X. I., D. C. Rees, and R. J. Deshales. 2004. JAMM: a mEtalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burns, K. E., S. Baumgart, P. C. Dorrestein, H. Zhai, F. W. McLafferty, and T. P. Begley. 2005. Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. J. Am. Chem. Soc. 127:11602-11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorrestein, P. C., H. Zhai, F. W. McLafferty, and T. P. Begley. 2004. The biosynthesis of the thiazole phosphate moiety of thiamin: the sulfur transfer mediated by the sulfur carrier protein ThiS. Chem. Biol. 11:1373-1381. [DOI] [PubMed] [Google Scholar]
- 4.Godert, A. M. 2006. Investigating the biosynthesis of thio-quinolobactin and the development of a proteomics probe for thiamin utilizing enzymes. Cornell University, Ithaca, NY.
- 5.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 6.Horn, D. M., R. A. Zubarev, and F. W. McLafferty. 2000. Automated reduction and interpretation of high-resolution electrospray mass spectra of large molecules. J. Am. Soc. Mass Spectrom. 11:320-332. [DOI] [PubMed] [Google Scholar]
- 7.Kurnasov, O., V. Goral, K. Colabroy, S. Gerdes, S. Anantha, A. Osterman, and T. P. Begley. 2003. NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 10:1195-1204. [DOI] [PubMed] [Google Scholar]
- 8.Leimkuhler, S., M. M. Wuebbens, and K. V. Rajagopalan. 2001. Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J. Biol. Chem. 276:34695-34701. [DOI] [PubMed] [Google Scholar]
- 9.Matthies, A., M. Nimtz, and S. Leimkuhler. 2005. Molybdenum cofactor biosynthesis in humans: identification of a persulfide group in the rhodanese-like domain of MOCS3 by mass spectrometry. Biochemistry 44:7912-7920. [DOI] [PubMed] [Google Scholar]
- 10.Matthijs, S., C. Baysse, N. Koedam, K. A. Tehrani, L. Verheyden, H. Budzikiewicz, M. Schaefer, B. Hoorelbeke, J.-M. Meyer, H. De Greve, and P. Cornelis. 2004. The Pseudomonas siderophore quinolobactin is synthesized from xanthurenic acid, an intermediate of the kynurenine pathway. Mol. Microbiol. 52:371-384. [DOI] [PubMed] [Google Scholar]
- 11.Mossialos, D., J.-M. Meyer, H. Budzikiewicz, U. Wolff, N. Koedam, C. Baysse, V. Anjaiah, and P. Cornelis. 2000. Quinolobactin, a new siderophore of Pseudomonas fluorescens ATCC 17400, the production of which is repressed by the cognate pyoverdine. Appl. Environ. Microbiol. 66:487-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neuenhaus, W., H. Budzikiewicz, H. Korth, and G. Pulverer. 1980. Bacterial components. Part IX. 8-Hydroxy-4-methoxymonothioquinaldic acid—a further thioacid from Pseudomonas. Z. Naturforsch. Teil B Anorg. Chem. Org. Chem. 35B:1569-1571. [Google Scholar]
- 13.Park, J.-H., P. C. Dorrestein, H. Zhai, C. Kinsland, F. W. McLafferty, and T. P. Begley. 2003. Biosynthesis of the thiazole moiety of thiamin pyrophosphate (vitamin B1). Biochemistry 42:12430-12438. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Plainview, NY.
- 15.Verma, R., L. Aravind, R. Oania, W. H. McDonald, J. R. I. Yates, E. V. Koonin, and R. J. Deshaies. 2002. Role of Rpn11 metalloproteases in deubiquitination and degredation by the 26S proteasome. Science 298:611-615. [DOI] [PubMed] [Google Scholar]
- 16.Wuebbens, M. M., and K. V. Rajagopalan. 2003. Mechanistic and mutational studies of Escherichia coli molybdopterin synthase clarify the final step of molybdopterin biosynthesis. J. Biol. Chem. 278:14523-14532. [DOI] [PubMed] [Google Scholar]
- 17.Zhai, H., X. Han, K. Breuker, and F. W. McLafferty. 2005. Consecutive ion activation for top down mass spectrometry: improved protein sequencing by nozzle-skimmer dissociation. Anal. Chem. 77:5777-5784. [DOI] [PubMed] [Google Scholar]