Abstract
Staphylococcus epidermidis is both a human skin commensal and an opportunistic pathogen, causing infections linked to implanted medical devices. This paper describes localized tufts of fibrillar appendages on a subpopulation (25%) of wild-type (WT) S. epidermidis NCTC 11047 cells. The fibrils (122.2 ± 10.8 nm long) are usually in a lateral position on the cells. Fibrillar (Fib+) and nonfibrillar (Fib−) subpopulations were separated (enriched) by 34 sequential partitions of WT cells between a buffer phase and a hexadecane phase. Following enrichment, hydrophobic cells from the hexadecane phase comprised 70% Fib+ cells and the less hydrophobic cells from the buffer phase entirely comprised Fib− cells. The Fib+ and Fib− subpopulations did not revert on subculture (34 times) on solid medium. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of cell surface proteins from WT, Fib+, and Fib− cells revealed two high-molecular-mass proteins (280 kDa and 230 kDa) on the WT and Fib+ cells that were absent from the Fib− cells. Amino acid sequencing revealed that fragments of both the 280- and 230-kDa proteins had 100% identity to the accumulation-associated protein (Aap). Aap is known to cause biofilm formation if it is truncated by loss of the terminal A domain. Immunogold staining with anti-Aap antibodies labeled tuft fibrils of the WT and Fib+ cells but not the cell surface of Fib− cells. The tufts were labeled with N-terminally directed antibodies (anti-A domain), showing that the fibrillar Aap was not truncated on the cell surface. Thus, the presence of full-length Aap correlated with the low biofilm-forming abilities of both WT and Fib+ S. epidermidis NCTC 11047 populations. Reverse transcription-PCR showed that aap was transcribed in both Fib+ and Fib− cells. We therefore propose that full-length Aap is expressed on cells of S. epidermidis NCTC 11047 as tufts of short fibrils and that fibril expression is regulated at a posttranscriptional level.
Staphylococcus epidermidis is an opportunistic pathogen causing serious nosocomial infections associated with implanted medical devices such as intravascular catheters, prosthetic heart valves, artificial hips and knees, continuous ambulatory peritoneal dialysis catheters, and cardiac pacemakers (9, 32). S. epidermidis cells form biofilms on the implant materials, resulting in infections, and multilayered clusters of cells are known to be recalcitrant to antibiotics (13), so surgical removal of the device is the only means of combating the infection (25). Biofilm formation is considered to be the main virulence factor of S. epidermidis (43) and can be divided into two main stages: adhesion and accumulation (41). Very early events of adhesion to uncoated device materials mainly involve nonspecific physicochemical forces such as hydrophobic interactions and van der Waals forces (24) and may be mediated by the capsular polysaccharide adhesin (35, 38), the autolysin AtlE (20), or two staphylococcal surface proteins, SSP-1 and SSP-2 (55). However, once inserted into the body the device materials are very quickly covered by a film of extracellular matrix proteins, adhesion to which is mediated mainly by specific protein adhesins. These include the vitronectin binding autolysin AtlE (20); the fibrinogen, fibronectin, and vitronectin binding autolysin Aae (22); the fibrinogen binding protein Fbe (19, 39); and the extracellular matrix binding protein (Embp), which binds fibronectin as well as other extracellular matrix proteins (56). In addition cell wall teichoic acid can augment adhesion of S. epidermidis to fibronectin (26).
The accumulation phase completes biofilm formation and involves cell multiplication and the formation of multilayered cell clusters (33). Accumulation requires intercellular adhesion, mediated by a polysaccharide intercellular adhesin (PIA) (21, 34) synthesized by the products of the ica operon. In addition, a 140-kDa cell surface protein known as the accumulation-associated protein (Aap) is involved in vitro in accumulation of strain RP62A on polymer surfaces (27) and has been linked to virulence of S. epidermidis (53). Rohde et al. (47) proposed that a 140-kDa Aap species found in S. epidermidis 5179 is the truncated isoform of the larger 220-kDa Aap and that the smaller protein is functional in biofilm accumulation independently of PIA. This truncation was shown to be necessary for biofilm formation, as the Aap-negative, biofilm-negative strain S. epidermidis 1585 was able to form a biofilm when it expressed the truncated form of Aap but remained biofilm negative when full-length Aap was expressed (47). Aap is prevalent in clinical isolates of S. epidermidis (46). The structure and location of Aap on the cell surface are so far unknown.
Aap is a cell wall-anchored protein with an N-terminal signal sequence and a C-terminal sorting signal including an LPXTG motif followed by a hydrophobic transmembrane region and a positively charged cytoplasmic tail (3, 45). It has a mosaic structure with an N-terminal region of short, 16-amino-acid (aa) repeats, an all-β A region, and a B-repeat region consisting of 12 repeats of 128 aa (in S. epidermidis RP62A) and 19 repeats of 6 aa, with a sortase recognition sequence of LPDTG, used for linking the polypeptide to peptidoglycan, at the C terminus (3). Aap is also named SesF (S. epidermidis surface protein) in the RP62A amino acid sequence posted by The Institute for Genomic Research (SERP2398) (14).
This paper reports on the presence of localized tufts of novel fibrillar appendages on a subpopulation of cells of S. epidermidis NCTC 11047 and reports that the fibrils are comprised of Aap.
MATERIALS AND METHODS
Bacterial strains and culturing conditions.
Bacterial strains used in this study were the nasal isolate S. epidermidis NCTC 11047 (ATCC 14990); S. epidermidis RP62A (ATCC 35984), a clinical, biofilm-forming strain (5), the genome sequence of which has recently been determined (14); and S. epidermidis ATCC 12228, a biofilm-negative strain (57). Bacteria were grown on tryptone soy agar (Oxoid) or in tryptone soy broth (TSB) (Oxoid) for 18 h under static conditions at 37°C unless otherwise stated. Colony morphology on Congo red agar was used as a surrogate marker of PIA production according to the method of Ziebuhr et al. (58).
Escherichia coli DH5α and M15 pREP4 (QIAGEN) were used for expression of recombinant hexahistidine (His6)-tagged proteins. Both strains were grown in LB broth, and medium was supplemented with ampicillin (100 μg ml−1), kanamycin (25 μg ml−1), or IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) as appropriate.
Transmission electron microscopy.
Cell surface structures of S. epidermidis NCTC 11047 were visualized by negative staining in the transmission electron microscope (TEM) using published methods (17). Formvar-coated copper grids (600 mesh; Agar Scientific) were carbon coated (Bio-Rad E6200 Carbon Coater) and plasma glowed (Plasma Barrel Etcher PT7150; Fisons) prior to use to render the surface hydrophilic. Cells from 18-h stationary-phase cultures were harvested by centrifugation (16,000 × g for 1 min), washed three times, and resuspended in distilled water (dH2O). A drop of cell suspension was applied to the grids, and the bacteria were stained with 2% (wt/vol) methylamine tungstate (pH 6.5) (Agar Scientific). The lengths of fibrils were measured from the base to the tips of the longest fibrils on about 20 to 30 cells from a batch of cells. To investigate the effects of protease and lysostaphin on the fibrils, cells were either incubated in phosphate-buffered saline (PBS) (140 mM NaCl, 270 μM KCl, 430 μM Na2HPO4, 147 μM KH2PO4, pH 7.4) containing 0.5 mg ml−1 protease XIV (Sigma) for 1 h at 37°C or incubated in PBS containing 100 μg ml−1 lysostaphin (Sigma) and 1 mM phenylmethylsulfonyl fluoride (Sigma) for 4 h at 37°C prior to being washed in water and applied to the grids for negative staining. No stabilization of protoplasts was needed as neither enzyme lysed the cells in the conditions used.
Immunogold negative staining was carried out using a modification of the method of McNab et al. (37). Cells from 18-h stationary-phase cultures were harvested by centrifugation (16,000 × g for 1 min), washed three times, and resuspended in dH2O. Formvar-coated copper grids (400 mesh; Agar Scientific) were carbon coated (Bio-Rad E6200 Carbon Coater), plasma glowed (Plasma Barrel Etcher PT7150; Fisons), and then placed for 5 min onto drops of cell suspension that had been washed three times in water. The grids were then inverted onto drops of anti-Aap antisera raised against the recombinant A domain (anti-Aap53-608) or the recombinant B domain (anti-Aap609-752) of Aap. The antisera were diluted in buffer (0.05 M Tris [pH 8.0] containing 1% [wt/vol] ovalbumin, 0.1% [wt/vol] gelatin, and 0.05% [vol/vol] Tween 20) and incubated at room temperature for 30 min. The grid was then washed five times in drops of dH2O and incubated for 30 min with 10-nm-diameter gold-anti-rabbit immunoglobulin G at room temperature. Grids were washed five times in drops of dH2O again and then stained with 2% (wt/vol) methylamine tungstate (pH 6.5) (Agar Scientific). Control grids were prepared using either preimmune serum instead of anti-Aap antisera or no primary antibody, to test for nonspecific binding of the gold particles.
All cells were photographed using an FEI Tecnai 12 electron microscope (FEI Company, Eindhoven, The Netherlands) at 100 kV.
Hexadecane enrichment assay.
Strain NCTC 11047 was separated using a hexadecane enrichment procedure into subpopulations that either carried (Fib+) or lacked (Fib−) fibrillar surface appendages (18, 49). Cells from a stationary-phase culture of S. epidermidis NCTC 11047 were harvested by centrifugation, washed in Sorenson's phosphate buffer (SPB; 66.6 mM Na2HPO4, 66.6 mM KH2PO4, pH 7.2), and suspended to an optical density at 440 nm (OD440) of 0.5. A 3-ml aliquot of cell suspension was partitioned with 200 μl hexadecane (Sigma). Vortex and partition times varied depending on whether hydrophobic or hydrophilic cells were being enriched. For enrichment of hydrophobic cells, the tube was vortexed for 10 s; after a 1-min partition time a 20-μl aliquot was removed from the hexadecane and inoculated into 20 ml TSB. This culture was grown for 18 h at 37°C and was then used for the next hydrophobic enrichment. For enrichment of hydrophilic cells the tube was vortexed for 1 min and partitioned for 15 min, after which a 20-μl aliquot was removed from the buffer phase and inoculated into 20 ml TSB and then grown for 18 h at 37°C. This culture was then used for the next hydrophilic enrichment. Hydrophilic and hydrophobic enrichments of cells were repeated, and the cells were examined by negative staining in the TEM at intervals during the 34 enrichments for the presence of fibrils.
Cell surface hydrophobicity assays.
A comparison of the cell surface hydrophobicities of the S. epidermidis subpopulations was performed by two procedures. Firstly, the same hexadecane affinity partition assay previously used for the enrichment process (49) was employed. Briefly, stationary-phase cells (18-h cultures) were harvested by washing cells three times and resuspending them to an OD440 of 0.5 in SPB, dH2O, or PBS. A 3-ml aliquot of bacterial cell suspension was vortexed for 1 min with 200 μl n-hexadecane (Sigma). The suspension was allowed to stand for 15 min for partitioning, after which 1 ml was removed from the aqueous layer. Cell surface hydrophobicity was calculated as the percent decrease in OD due to cells removed from the aqueous phase after vortexing with hexadecane.
Secondly, hydrophobic interaction chromatography (HIC) was used to further quantify the relative differences in cell surface hydrophobicity of the subpopulations. In this method affinity for the hydrophobic octyl-Sepharose in columns was measured using a modification of the technique developed by Smyth et al. (50). Columns were prepared from short glass pipettes (internal diameter, 5 mm; length, 150 mm) (Poulten & Graf Ltd., Barking, United Kingdom), with the necks plugged with a small amount of glass wool and with clamped Teflon tubing attached to control the flow. Columns were packed with 0.7 ml octyl-Sepharose CL-4B or Sepharose CL-4B (both from Sigma, United Kingdom), as a control to test for retention by the nonhydrophobic agarose gel. The gel columns had a bed height of ∼15 mm and were washed through with 5 ml of 1 M NaCl prior to the experiment. An overnight culture of cells was washed three times in 1 M NaCl and set to an OD470 of 0.2, and 2 ml of cells (∼2.6 × 107 CFU·ml−1) was applied to the columns. The cells were washed through with 5 ml of 1 M NaCl, and the eluates were collected. The viable counts of the cell suspension applied to the top of the column and of the cells eluted from the bottom of the column were quantified by the Miles and Misra technique. The percent retention of cells on the columns was calculated as follows: % retention = [(numbers of cells applied to the column − numbers of cells washed through)/numbers of cells applied] × 100. Three separate batches of cells were tested, each in triplicate.
Quantitative determination of biofilm formation.
Biofilm formation of wild-type (WT) S. epidermidis NCTC 11047 and those of its two subpopulations were compared using the crystal violet biofilm assay (58), which also quantified biofilm formation by S. epidermidis strains. The assay was first described by Christensen et al. (6) and has been subsequently used extensively to quantify biofilm formation by many genera (8). Cells were grown to stationary phase (18 h) in TSB, each culture was adjusted to an OD490 of 1.0, and the culture was then diluted 1:100 in fresh TSB. Individual wells of a sterile 96-well, flat-bottomed tissue culture plate (Corning Inc.) were inoculated with 200-μl aliquots of diluted culture. Every other row was not inoculated and contained 200 μl TSB as a negative control. Plates were incubated at 37°C for 24 h. After incubation the OD490 of each well was measured to ensure that each culture had reached the same density, and then each well was washed three times with PBS (pH 7.4). Biofilms were stained with 200 μl of 0.4% (vol/vol) crystal violet (Sigma) for 10 min. Plates were rinsed with PBS and air dried; a 200-μl aliquot of 100% ethanol was added to each well to redissolve the crystal violet. Plates were incubated at 4°C for 4 h, after which the OD490 was read in a microtiter plate reader (Synergy HT; Bio-Tek), using the mean of the negative-control wells as a blank. S. epidermidis RP62A (ATCC 35984) and S. epidermidis ACTC 12228 were included as positive and negative controls, respectively.
Quantitative determination of initial adhesion to polystyrene.
Initial adhesion of S. epidermidis NCTC 11047 and the Fib− and Fib+ subpopulations to polystyrene was quantified by quantifying adhesion in a 96-well plate. Overnight cultures were resuspended in fresh TSB to an OD490 of 1.0; 200 μl of culture was then added to each well, and the plates were incubated at 37°C for 2 h. Plates were then washed three times with PBS, and each well was stained with 200 μl 4% (vol/vol) crystal violet for 10 min. Cells were then washed five times with PBS and dried. Aliquots (200 μl) of 100% ethanol were then added to each well, the plate was incubated for 4 h at 4°C, and the OD490 of each well was then read. The assay was repeated three times.
Preparation of cell wall proteins and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
Cell surface polypeptides were prepared according to the method of Timmerman et al. (52) with slight modifications. Stationary-phase cells (20 ml) were adjusted to an OD450 of 1.3. Cells were washed in PBS and harvested by centrifugation at 3,500 × g for 10 min. The resulting pellet was resuspended in 150 μl of PBS containing 100 μg ml−1 lysostaphin (Sigma) and 1 mM phenylmethylsulfonyl fluoride (Sigma). The suspension was then incubated with shaking at 37°C for 4 h, after which the supernatant, containing cell surface proteins (CSPs), was collected by centrifugation for 30 min at 16,000 × g. To investigate the effects of protease on CSPs, the supernatant containing CSPs extracted with lysostaphin was incubated with 0.5 mg ml−1 protease XIV (Sigma) for 1 h at 37°C.
Aliquots (10 μl) of the supernatant were dissolved in an equal amount of 2× SDS loading buffer (4% [wt/vol] SDS, 200 mM dithiothreitol, 20% [wt/vol] glycerol, 0.2% [wt/vol] bromophenol blue in 100 mM Tris-HCl buffer, pH 6.8), and the solution was boiled for 5 min. Samples were subjected to electrophoresis through a 10% (wt/vol) acrylamide resolving gel and a 5% (wt/vol) acrylamide stacking gel according to the method of Laemmli and Favre (29). Gels were stained with Coomassie brilliant blue R-250 (Fluka). Molecular masses of proteins were estimated by comparing distance migrated to a plot of migration distances against log molecular mass of marker proteins (Bio-Rad prestained SDS-PAGE standards, broad range).
In order to detect glycoproteins, the cell surface proteins were run on a 12% SDS-polyacrylamide gel and stained using a glycoprotein detection kit (Sigma), containing periodic-Schiff reagents. Horseradish peroxidase was included as a positive control, and any bands containing glycoprotein stained pink.
Amino acid sequence determination by tandem mass spectrometry analysis of protein bands on SDS-polyacrylamide gels.
Protein fragments from polypeptide bands of interest were analyzed using nanoelectrospray on a quadrupole time of flight mass spectrometer (Q-Tof; Waters, Manchester, United Kingdom). Tandem mass spectrometry was used to fragment the tryptic peptides to be able to elucidate the amino acid sequence. The amino acid sequence was then used for database searching using the BLAST function on the NCBI database (www.ncbi.nlm.nih.gov) to identify the protein.
Isolation of genomic DNA.
Genomic DNA was prepared from S. epidermidis NCTC 11047 WT, Fib+, and Fib− subpopulations and S. epidermidis RP62A using a QIAmp DNA minikit (QIAGEN) as directed by the manufacturer, with the modification that cells were incubated at 37°C for 1 h with 200 μg ml−1 lysostaphin prior to the lysis step; after 1 h 20 μl proteinase K was added to the cells, and the culture was incubated at 56°C for 30 min and then 95°C for 15 min in order to lyse the cells.
Construction of recombinant His6-tagged fusion proteins.
A single B-repeat unit of aap of S. epidermidis NCTC 11047 was amplified using primer 9 and primer 10 (Table 1). PCR was carried out using Expand High Fidelity PCR system according to the manufacturer's instructions (Table 1). Products were run on an agarose gel, and a band at ∼400 bp (corresponding to a single 128-aa repeat) was excised from the gel and purified using a gel extraction kit (QIAGEN). The purified DNA fragment was cleaved with the restriction endonucleases BamHI (Roche) and HindIII (Roche) and ligated into pQE30 Xa vector (QIAGEN). Ligation products were then transformed into E. coli DH5α. Plasmids with the correct insert (pQE30 Xa::rAap609-752 and pQE30::rAap53-608) were transformed into E. coli M15 pREP4 for overexpression.
TABLE 1.
Oligonucleotide primers used for PCR and RT-PCRs
| Primer no. | Primer namea | Sequence (5′-3′)b | Reference |
|---|---|---|---|
| 1 | aap 1 F | TAC ATG GGA GGT ATA ATA TGG | This paper |
| 2 | aap 2 R | CCA AAT ATG AAC AAT GAT CCG | This paper |
| 3 | rAAP1357-1756 F | ATA CAA CTG GTG CAG ATG GTT G | 53 |
| 4 | rAAP1357-1756 R | GTA GCC GTC CAA GTT TTA CCA G | 53 |
| 5 | aap rev 2 | TGG TTC TGC TTT TGT TGG ACC ATA C | 47 |
| 6 | rAAP53-608 F | CCG GGATCC GCA GAA GAA AAA CAA GTT GAT C | 44 |
| 7 | rAAP53-608 R | CGG AAGCTT TTA TGC TTT AGG AGT GTA TGT CAA TG | 44 |
| 8 | aap repeat_end | GGA CCA TAT TTT STC ACA TC | This paper |
| 9 | aap repeat 1_BamHI | CCG GGATCC GTC GAT GGA GAT CCA ATT ATA TC | This paper |
| 10 | aap repeats_end_HindIII | CGG AAGCTT GGA CCA TAT TTT STC ACA TC | This paper |
| 11 | aap US 1 F | AGC ACA AGC AGA AGA TGG | This paper |
| 12 | icaR F | TTA TCA ATG CCG CAG TTG TC | 1 |
| 13 | icaR R | GTT TAA CGC GAG TGC GCT AT | 1 |
| 14 | icaA F | TAA TCC CGA ATT TTT GTG AA | 1 |
| 15 | icaA R | AAC GCA ATA ACC TTA TTT TCC | 1 |
| 16 | icaD F | ACA GTC GCT ACG AAA AGA AA | 1 |
| 17 | icaD R | GGA AAT GCC ATA ATG ACA AC | 1 |
| 18 | icaB F | ATG GTC AAG CCC AGA CAG AG | 1 |
| 19 | icaB R | CGT GTT TTC AAC ATT TAA TGC AA | 1 |
| 20 | icaC F | CTG ATC AAG AAT TTA AAT CAC AAA | 1 |
| 21 | icaC R | AAA GTC CCA TAA GCC TGT TT | 1 |
| 22 | icaAB-F | TAA CTT TAG GCG CAT ATG TTT T | 10 |
| 23 | icaAB-R | TTC CAG TTA GGC TGG TAT TG | 10 |
F, forward primer; R, reverse primer.
Underlining indicates restriction sites.
Overexpression, purification, and concentration of recombinant proteins.
Cell pellets from 2 liters of overexpressed E. coli M15 pREP4 cultures with either pQE30::rAap53-608 (44) or pQE30 Xa::rAap609-752 were resuspended in 50 ml of 50 mM sodium phosphate, 0.3 M NaCl (pH 7). The cell suspension was then sonicated and centrifuged at 10,000 rpm for 30 min (Sorvall RC-5B Refrigerated Superspeed Centrifuge; DuPont Instruments). The His6-tagged protein was then purified from the cell lysate using a TALON histidine tag purification resin (BD Biosciences). Purified proteins were concentrated using a stirred ultrafiltration cell (Amicon). Aliquots of each protein were analyzed by SDS-PAGE to check purity.
Preparation of anti-Aap antibodies.
Antibodies to rAap53-608 (A region) and rAap609-752 (B region) were raised by Eurogentec (Seraing, Belgium) in New Zealand White rabbits according to the standard immunization protocol of the company.
PCR of aap and the ica operon.
PCR amplification of aap was carried out using the Expand Long Template PCR System (Roche) using primers 1 and 2 (Table 1). PCR was also carried out to detect the presence of the ica operon, which encodes polypeptides involved in the synthesis of capsular polysaccharide adhesin and PIA using the Expand High Fidelity PCR System (Roche). Primers used are detailed in Table 1. All reactions were carried out according to the manufacturer's instructions in a Bio-Rad i-Cycler. A positive control (genomic DNA of S. epidermidis RP62A) and a negative control (reaction mixture without DNA) were included in each PCR run.
Detection of the aap transcript by RT-PCR.
RNA was extracted from S. epidermidis NCTC 11047 WT, Fib−, and Fib+ cells and S. epidermidis RP62A. A 500-μl aliquot of an overnight culture was inoculated into 5 ml of fresh TSB and incubated at 37°C, 2 h (OD450, 0.6). Following incubation 400 μl of the culture was treated with RNAprotect bacterial reagent (QIAGEN) and RNA extraction was then carried out using an RNeasy minikit (QIAGEN). The S. epidermidis cells were lysed with TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) containing 200 μg ml−1 lysostaphin (Sigma), 400 U ml−1 mutanolysin (Sigma), and 40 μg ml−1 proteinase K (Roche). Contaminating DNA was eliminated by treating the RNA with DNase (QIAGEN). The amount of RNA contained in each sample was measured using an ND-100 spectrophotometer (Nanodrop), and a volume corresponding to 100 ng of RNA was used for first-strand synthesis. Reverse transcription of the RNA was carried out using a Reverse IT first-strand synthesis kit (ABgene) according to the manufacturer's instructions using the random decamers supplied. Following first-strand synthesis an amplification step was carried out. Two different fragments of aap were amplified using two primer pairs which amplified nucleotides 1357 to 1756 (primers 3 and 4) and 6657 to 7121 (primers 5 [reverse complement {r.c.}] and 2) (Table 1), using 5 μl of the first-strand reaction mix or 5 μl of RNA (negative control). Negative controls using RNA, without the first-strand synthesis stage, were included to ensure that positive results were not due to contaminating DNA. A negative control with dH2O was also included as well as a positive control with genomic DNA. Aliquots of the amplified products were separated on a 1% agarose gel and visualized following staining with ethidium bromide.
Analysis of the Aap promoter regions in NCTC 11047 subpopulations.
Amplification of the aap promoter regions of strain NCTC 11047 WT, Fib+, and Fib− subpopulations and RP62A was carried out using primer 11 (Table 1), targeting a conserved upstream hypothetical protein (SE2400 in RP62A genome sequence, GenBank accession number NC_002976), and primer 6 (r.c.) (Table 1), located just after the N-terminal signal sequence of aap (Table 1). PCR was carried out using the Expand High Fidelity PCR system (Roche) according to the manufacturer's instructions.
The products generated by PCR were sequenced on both strands using Big Dye Terminator mix, version 3.1 (Applied Biosystems), with primers 11 and 6 (r.c.). Reaction products were analyzed using an ABI Prism 3100 Genetic Analyzer. The resulting chromatograms were analyzed using Chromas Pro version 1.33, and a consensus sequence was derived from trace files using VectorNTI and aligned with corresponding regions from strains RP62A and 12228.
Statistical analysis.
Statistical analysis was carried out using SPSS (version 11.5), significant differences were tested with one-way analysis of variance with Tukey post hoc tests, and the mean difference was significant at the 0.05 level. For the percent hydrophobicity values the analysis of variance was performed on arcsine square root-transformed data.
RESULTS
Negative staining of S. epidermidis NCTC 11047 subpopulations.
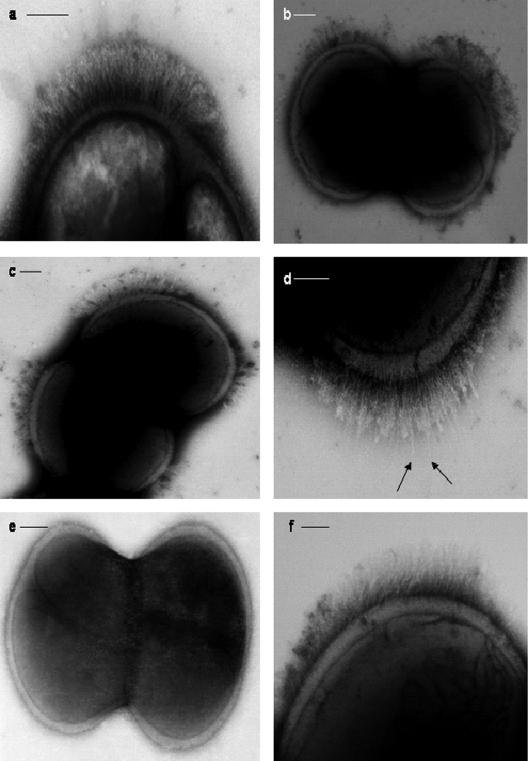
Stationary-phase cells of S. epidermidis NCTC 11047 carried short fibrillar appendages, localized on the cell surface in a tuft of fibrils projecting away from the cell wall (Fib+ cells) (Fig. 1a to d). Counts on different batches of 18-h stationary-phase cells always detected tufts on approximately 25% of cells, and tufts were also present on a subpopulation of cells during logarithmic phase. The mean length of the fibrils in the tufts was 122.2 ± 10.8 nm as measured from the fibril base to the fibril tip on three batches of cells (20 to 30 cells per batch). The position of tufts in relation to the septum varied from cell to cell (represented diagrammatically in Fig. 2a to f). Tufts never extended over more than a quarter of the cell circumference, and they were usually in a lateral position but could be located in a polar position. Typically a dividing Fib+ cell had one visible tuft in a lateral position on one daughter cell only (Fig. 1a and 2a), but on a very small number of cells two lateral tufts were present on one cell as it divided (Fig. 1b and 2b). Very rarely, four tufts were detected on a dividing cell (Fig. 1c and 2d). Counts of cells with multiple tufts were not possible due to the very low numbers of such cells detected. The tufts comprised of densely packed fibrils and separate fibrils could be seen only rarely (Fig. 1d). The majority of stationary-phase cells (75%) showed no detectable fibrils over the cell circumference (Fig. 1e). Treatment of WT cells of S. epidermidis NCTC 11047 with protease (1 mg ml−1 for 2 h at 37°C) and lysostaphin (100 μg ml−1) completely removed all tuft fibrils as detected by negative staining (data not shown). Also, cells were still intact after both enzyme treatments. Loss of the fibrils after protease treatment indicated that tuft fibrils either were comprised of protein or were attached to the cell surface by a protein component.
FIG. 1.
Micrographs of S. epidermidis NCTC 11047 cells (a to e) and S. epidermidis RP62A (f) negatively stained with 2% (wt/vol) methylamine tungstate. (a) One lateral tuft of fibrils on one side of the septum. No fibrils were detected on the remaining cell surface (not shown in the image). (b) Two lateral tufts of fibrils, one on either side of the septum. (c) Four lateral tufts symmetrically positioned on either side of the septum. (d) Tuft of fibrils showing a few longer individual fibrils (arrows) projecting through the mass of shorter fibrils. (e) Dividing cell without any tufts of fibrils. (f) Cell of strain RP62A carrying a tuft of fibrils. Bars, 100 nm.
FIG. 2.
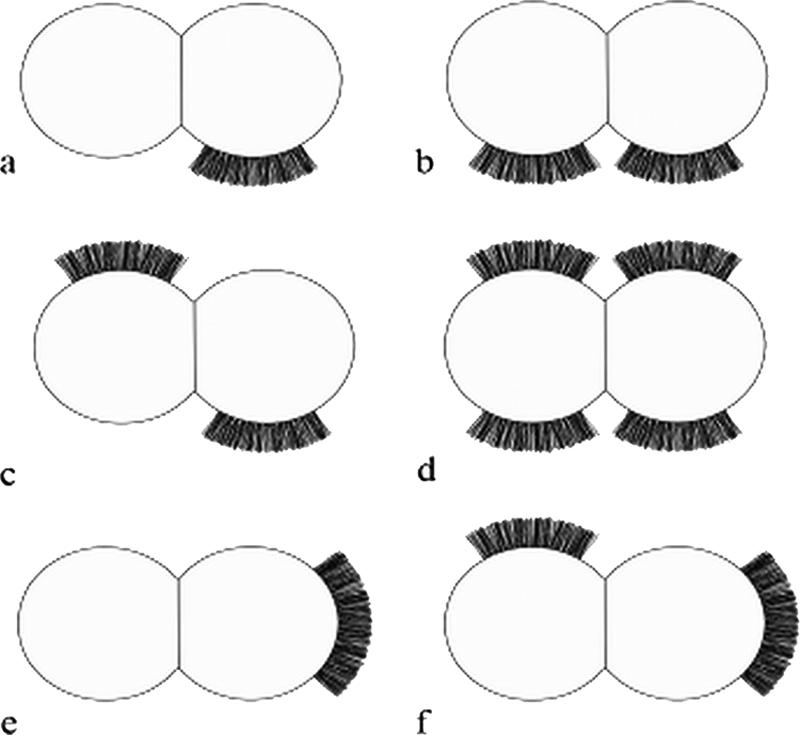
Schematic diagram of the different positions occupied by fibrillar tufts on 25% of S. epidermidis NCTC 11047 cells in relation to the septum of the dividing cell. (a to d) The majority of tufts were in the lateral position. (e and f) A minority of tufts were in a polar position.
Only very small numbers of cells of S. epidermidis RP262A carried detectable fibrils, and extensive screening of many batches of cells stained by methylamine tungstate showed that <1% of the cells carried visible tufts of fibrils (Fig. 1f). The fibrils in the RP62A tufts were 159 ± 35 nm long and projected out laterally from one side of a cell. The RP62A tufts were indistinguishable from those on NCTC 11047 cells, and there was no significant difference in the lengths of the tuft fibrils on the two strains. Since there was a much higher number of tufted cells detected in the WT population of NCTC 11047 than with RP62A, S. epidermidis NCTC 11047 was selected for the subsequent work on fibril characterization.
Enrichment of the tufted and nontufted subpopulations of Staphylococcus epidermidis NCTC 11047.
Since tufts of fibrils on species of oral streptococci have hydrophobic properties (16, 18), it was predicted that the NCTC 11047 fibrils would also have hydrophobic properties. Therefore, partitioning between the inert hydrocarbon hexadecane and a buffer (49) was used to enrich for the S. epidermidis NCTC 11047 cells carrying the tufts of fibrils. We had previously used the hexadecane assay to successfully separate tufted and nontufted cells of the oral bacterium Streptococcus sanguis (4), and it was used in preference to any other hydrophobic partitioning method due to the speed and simplicity with which each enrichment step could be carried out. Also it is already known that contact of S. epidermidis NCTC 11047 cells with hexadecane does not significantly reduce their viability (40).
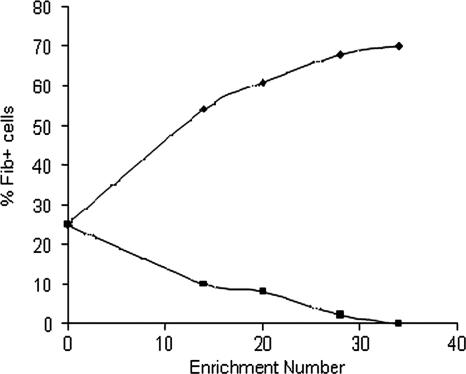
When the NCTC 11047 WT cells were partitioned, the more hydrophobic cells were recovered from the hexadecane phase and the more hydrophilic cells were recovered from the buffer phase. At intervals during the enrichment process both the putative “hydrophobic” and “hydrophilic” cultures were grown to stationary phase (18 h) and the relative numbers of “tufted” (Fib+) and “nontufted” (Fib−) cells in a population were counted using negative staining (Fig. 3). Fib+ cells were successfully enriched using this approach, and the percentage of tufted cells in the culture enriched from the hexadecane phase began to plateau at a maximum of 70% after 34 enrichment steps. Conversely the percentage of Fib+ cells in the subpopulation enriched from the buffer phase was reduced to zero after 34 enrichment steps (Fig. 3). The mean fibril length on the enriched tufted subpopulation (121.8 ± 10.5 nm) was not significantly different (P > 0.05) from the mean length of fibrils on the WT (122.2 ± 10.8 nm). Each measurement was taken from three batches of stationary-phase cells. Both Fib− and Fib+ strains have the same colony morphology, so they could be distinguished from each other only by negative staining.
FIG. 3.
Change in percentages of Fib+ cells (triangles) and Fib− cells (squares) of S. epidermidis NCTC 11047 subpopulations after repeated hydrophobic and hydrophilic enrichments in the hexadecane partition assay.
Quantitative assessment of cell surface hydrophobicity.
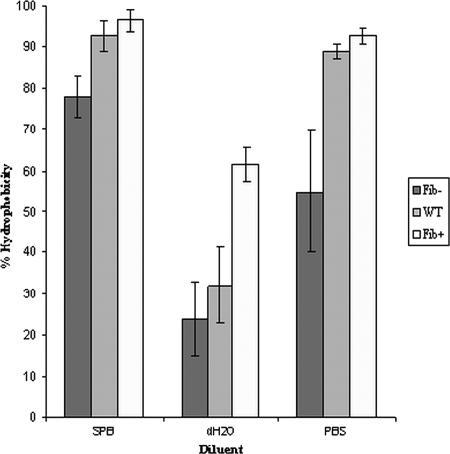
WT NCTC 11047 cells and the Fib+ and Fib− enriched subpopulations were tested in the hexadecane assay (49) to give an estimate of their relative cell surface hydrophobicity. Stationary-phase cells of the three strains were partitioned from SPB, PBS, and dH2O into hexadecane (Fig. 4). For all diluents the Fib+ cells always showed a higher affinity for hexadecane than the WT did, but the difference was significant only when cells were partitioned from water (P < 0.05). The Fib− cells showed a reduced affinity for the hexadecane compared to the WT cells, but the difference was significant only for SPB and PBS (P < 0.05). However, for all diluents the Fib+ cells always adhered in significantly greater numbers to the hexadecane droplets than did the Fib− cells (Fig. 4) (P < 0.05), indicating that the presence of fibrils correlated with increased cell surface hydrophobicity.
FIG. 4.
Comparison of cell surface hydrophobicities of S. epidermidis NCTC 11047 WT, Fib+, and Fib− subpopulations after partitioning between hexadecane and SPB, dH2O, or PBS. Error bars are standard errors of the means.
HIC with octyl-Sepharose confirmed that cells of the tufted, Fib+ subpopulation were more hydrophobic than the nonfibrillar Fib− subpopulation as there was an 89% ± 1% retention for the Fib+ cells and an 80% ± 2% retention for the Fib− cells on the octyl-Sepharose. This difference was statistically significant (P < 0.05). The overall retention of both strains by the nonhydrophobic Sepharose was not significantly different, and Fib+ and Fib− cells showed an overall mean retention of 20% ± 2%. This value is a measure of the entrapment by the agarose gel itself (50). Thus, two separate methods for quantifying cell surface hydrophobicity showed that the cells with tufts of fibrils are more hydrophobic than cells without any tufts. These results suggest that the fibrils contribute to cell surface hydrophobicity and therefore are likely to have hydrophobic properties.
Assessment of biofilm-forming ability.
Biofilm formation abilities of the S. epidermidis NCTC 11047 strains were compared and found to be low in all three strains. Biofilm formation by the Fib+ strain and that by the WT strain were not significantly different, with crystal violet OD490s of 0.205 ± 0.009 and 0.195 ± 0.003, respectively. However, biofilm formation by the Fib− strain gave an OD490 of 0.168 ± 0.003, which was only slightly less biofilm than that formed by the WT and Fib+ strains, although due to very low standard error there was a very small but significant difference (P < 0.05) in the OD490s. The biofilm-negative strain S. epidermidis ATCC 12228 was included in the study and gave an OD of 0.135 ± 0.005, which is comparable to the values for the NCTC 11047 strains. In comparison, the biofilm-positive strain S. epidermidis RP62A gave an OD490 reading of 0.942 ± 0.047, which is a factor of 4.6 greater than that formed by the Fib+ subpopulation of NCTC 11047. These data suggest that the S. epidermidis NCTC 11047 strains are all poor biofilm formers and can be described as biofilm-negative strains. Thus, the increase in the presence of fibril tufts on the Fib+ subpopulation to 70% compared to 25% on the WT cells does not significantly enhance biofilm formation.
Assessment of adhesion to polystyrene.
The abilities of S. epidermidis NCTC 11047 WT, Fib−, and Fib+ strains to adhere to polystyrene for 2 h were compared using the 96-well plate assay, and Fib+ cells attached themselves in much higher numbers than the Fib− cells. After 2 h of incubation on polystyrene, Fib+ cells gave a final OD490 of 0.126 ± 0.002, which was slightly, but significantly (P < 0.05), higher than that of the WT population (0.112 ± 0.002). However, adhesion of the nontufted Fib− cells to polystyrene (0.062 ± 0.001) was significantly lower (by 50%) than adhesion of the tufted Fib+ cells (P < 0.05). These results show that a higher number of fibrillar tufts is correlated with higher adhesion to polystyrene, indicating that the fibrils may play a role in adhesion to polystyrene for S. epidermidis NCTC 11047.
Analysis of cell surface proteins on S. epidermidis by SDS-PAGE.
To identify protein components of the fibrils, lysostaphin was used to solubilize the cell surface polypeptides (51). SDS-PAGE profiles of the protein extracts revealed two polypeptides with estimated molecular masses of 280 and 230 kDa, which were present on the WT and from the Fib+ populations (Fig. 5, lanes B and D) but absent on the Fib− population (Fig. 5, lane C). As far as it was possible to discern, all other polypeptide bands were similar for the three NCTC 11047 strains, although a very faint band (∼200 kDa) could sometimes be detected in the Fib+ protein extract (Fig. 5, lane D).
FIG. 5.
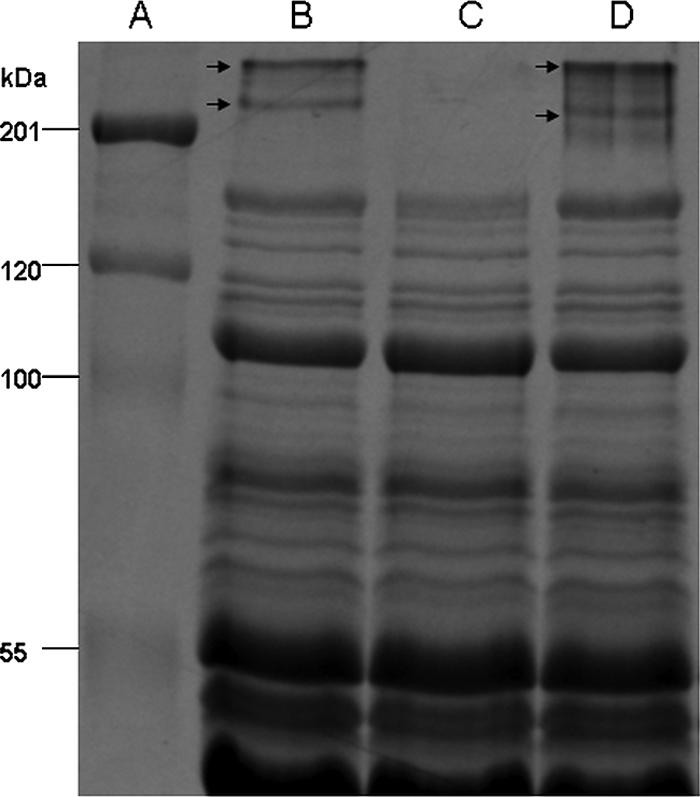
SDS-PAGE analysis of cell surface polypeptides of S. epidermidis strains, extracted using lysostaphin (100 μg ml−1, 4 h, 37°C) and run on a 10% SDS-polyacrylamide gel stained with Coomassie brilliant blue. (A) Standard molecular mass markers (kDa). (B) S. epidermidis NCTC 11047 WT cells. (C) S. epidermidis NCTC 11047 Fib− subpopulation. (D) S. epidermidis NCTC 11047 Fib+ subpopulation. Both NCTC 11047 WT and Fib+ subpopulations express two high-molecular-mass (280- and 230-kDa) polypeptides (indicated by arrows), which are absent from the Fib− subpopulation.
Fibrils on Streptococcus gordonii DL1 are comprised of the high-molecular-mass protein CshA (259 kDa) (37), so in order to identify putative fibril polypeptides on S. epidermidis, the two high-molecular-mass bands from S. epidermidis NCTC 11047 WT and Fib+ strains were excised and identified by electrospray-tandem mass spectrometry. The faint ∼200-kDa protein on some batches of Fib+ cells could not be sequenced. Five fragments from each of the 280- and 230-kDa proteins from NCTC 11047 shared 100% identity with Aap of RP62A (accession number NC_002976). The fully sequenced strain RP62A also showed two high-molecular-mass bands (280 and 230 kDa) on SDS-polyacrylamide gels (data not presented), which were sequenced and found to be Aap. Therefore, both NCTC 11047 and RP62A expressed two high-molecular-mass bands comprising Aap that correlated with the presence of tufts of fibrils.
The two Aap bands on NCTC 11047 (WT and Fib+ strains) and RP62A did not stain with the periodic-Schiff reagent (data not presented), indicating the lack of glycosylation of Aap. No fibrils could be detected by negative staining on Fib+ cells of NCTC 11047 treated with lysostaphin. Protease treatment of collected cell surface proteins after lysostaphin extraction resulted in the loss of the 230-kDa and 280-kDa proteins from the gel (data not shown). Protease treatment also removed fibrils from the cell surface of Fib+ cells. These data indicate that the presence of fibrils correlates with the expression of Aap, detected as two high-molecular-mass polypeptides.
Stability of Fib+ and Fib− populations.
In order to test whether the presence or absence of the tufts was a stable trait, the Fib+ and Fib− populations were serially subcultured to test for reversion by sequentially plating cells on tryptone soy agar 34 times. The percentage of cells with tufts was recorded, after negative staining, in the TEM. The percentages of Fib+ and Fib− cells in both populations showed very little change after 34 subcultures. No tufts were detected on the Fib− population, and the “tuft count” decreased only from 75% to 60% on the Fib+ population.
Counting the numbers of Fib+ cells is extremely difficult, and it was not possible to count enough cells to do a statistical comparison of the presence of tufts before and that after subculture of the Fib+ strain. However, we can conclude that no major reduction in fibril expression has occurred after extensive subculture of the Fib+ subpopulation. In addition SDS-polyacrylamide gels of surface proteins from the serially subcultured Fib− cells showed no reappearance of the Aap bands (280 and 230 kDa) (data not presented). These data indicate that the presence or absence of the tuft on the two subpopulations is an apparently stable property and that reversion to the WT ratio of 75% Fib− to 25% Fib+ does not easily occur on subculture. In addition when the Fib+ and Fib− strains were grown as biofilms on cellulose filters overnight they did not respectively lose or regain the two high-molecular-weight bands or the fibrils (data not presented).
Immunogold negative staining with anti-Aap antibodies.
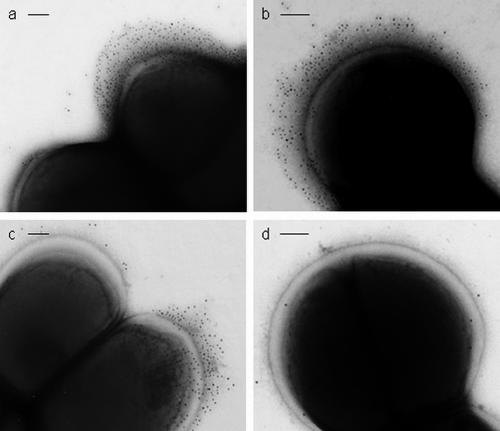
In order to determine whether the localized fibrillar tufts of S. epidermidis NCTC 11047 were comprised of Aap, immunogold negative staining was carried out. Cells were incubated with antisera specific to either the A region of Aap or the B repeat of Aap. With both antibodies, gold particles were found to label the fibrillar tufts of Fib+ cells (Fig. 6a and b) and the WT cells (Fig. 6c). There was no apparent difference in the distribution of the gold on the tufts for either antibody. In order to show the gold particles, cells have been selected with a low level of negative staining (Fig. 6a to c) and fibrils are not heavily stained. However, on more darkly stained cells the fibrils could always be seen underneath the gold (data not presented). The outline of gold particles always followed the outline of the shape of the fibrillar tuft. No label (above background) was detected with either antiserum on Fib− cells (Fig. 6d), proving a complete lack of Aap on these cells. It was possible to find cells that had gold label on one daughter cell (on the tuft), but there was no tuft and no label on the other daughter cell. Thus, it is possible to detect the cells switching expression of the fibrillar tuft as the cells divide.
FIG. 6.
Electron micrographs of S. epidermidis NCTC 11047 cells labeled with anti-Aap antibodies and 10-nm colloidal gold-conjugated secondary antibody negatively stained with 2% (wt/vol) methylamine tungstate. (a) Fib+ cell labeled on the tuft fibrils with anti-Aap A-region antibody (top cell) attached to a Fib− cell with no tuft and no gold labeling. (b) Fib+ cell labeled with anti-Aap B-region antibody revealing a very extensive fibrillar tuft. (c) WT cells labeled with anti-A antibody, showing a labeled cell with a tuft (Fib+) attached to a cell with no labeling and no tuft (Fib−). (d) Fib− cell showing no specific gold attached after labeling with anti-A antibody. Bars, 100 nm.
PCR amplification of aap and genes of the ica operon.
Genomic DNA was isolated from S. epidermidis NCTC 11047 WT, Fib−, and Fib+ cells and S. epidermidis RP62A. PCR was carried out in order to amplify the whole aap gene of these populations. All three NCTC 11047 populations gave an amplicon of approximately 7 kb, which corresponded to the size of the aap amplicon of RP62A, the positive control. Fib− cells gave the same-sized amplicon as the WT and Fib+ populations, implying that there is no insertion or deletion in this gene that prevents Aap from being expressed on the cell surface of Fib− cells.
S. epidermidis NCTC 11047 was screened for the ica operon, which encodes polypeptides involved in the synthesis of PIA (21, 34). All three 11047 strains were found to be negative for the ica operon with all six primer pairs used, whereas the control strain RP62A was positive for all. Consistently, synthesis of PIA could not be detected on Congo red agar. The NCTC 11047 strains all produced large, smooth, red colonies, indicating a lack of PIA, whereas RP62A produced dry, black, highly crenated colonies due to the presence of PIA (58).
Detection of aap transcription products by RT-PCR.
RT-PCR was carried out in order to determine whether the aap gene was being transcribed by the Fib− cells as well as the Fib+ cells. All NCTC 11047 strains produced mRNA corresponding to nucleotide positions 1357 to 1756 and 6657 to 7121 of aap. As a 400-bp fragment of the A region of aap and a 500-bp region at the C terminus of aap were both amplified, it can be deduced that full-length aap is transcribed in all four strains tested. Equal amounts of RNA were used in each reaction, and amplified PCR products from cDNA of all four strains (S. epidermidis NCTC 11047 WT, Fib−, and Fib+ strains and S. epidermidis RP62A) showed the same band intensity on agarose gels, indicating that the level of aap transcription was equal in each strain.
DNA sequence analysis of the Aap promoter region.
To investigate any possible variation in the aap promoter region for the different NCTC 11047 subpopulations, the region was sequenced and compared with the aap promoter region of strain RP62A. The aligned regions of DNA sequence upstream of aap in NCTC 11047 WT and the Fib+ and Fib− subpopulations shared 100% identity (data not shown). In addition, a putative ribosome binding site (GGGAG) and −10 and −35 regions were identified. This proof of integrity of the promoter regions for all three NCTC 11047 strains is consistent with the results of RT-PCR analyses which together suggest that aap is transcribed in Fib− cells as well as in the Fib+ cells.
DISCUSSION
Fibril tufts correlate with high cell surface hydrophobicity.
This study has shown that a subpopulation of cells of S. epidermidis NCTC 11047 carries fibrillar appendages that have not been previously reported on any strain of S. epidermidis. It is proposed that the fibrils have hydrophobic properties, as serial enrichment in a hydrophobic partition assay using hexadecane enabled separation of the more hydrophobic Fib+ cells from the less hydrophobic Fib− cells. This enrichment using hexadecane was previously successful as a method for enriching for tufted cells of Streptococcus cristatus NCTC 12479 (formerly Streptococcus sanguis CR311) (4, 18), and the more hydrophobic streptococci with long fibrils in the tufts also preferentially partitioned into the hexadecane layer. However, attachment of bacteria to hexadecane droplets results from a combination of both hydrophobic and electrostatic interactions (12). Therefore, HIC using octyl-Sepharose was used as an independent method (40, 50) to more accurately compare the relative hydrophobicities of the two subpopulations. Since the Fib+ cells attached in higher numbers than the Fib− cells to octyl-Sepharose, the results are consistent with the hypothesis that the fibrils have hydrophobic properties. Because the fibrils are localized, they are predicted to contribute to microscale heterogeneity of cell surface hydrophobicity. Thus, the cell surface is likely to be comprised of a smaller, more hydrophobic, fibril tuft zone surrounded by a more extensive, less hydrophobic, nonfibrillar cell surface area. However, since adhesion of the Fib− cells to octyl-Sepharose is still fairly high (80% for Fib− cells compared to 89% for Fib+ cells), there are clearly many other cell surface molecules on the nonfibrillar part of the cell surface that have hydrophobic properties.
It was not possible to increase the relative numbers of tufted S. epidermidis NCTC 11047 cells to above 70%, even after 34 enrichment steps through hexadecane, although there is no apparent reason why 100% Fib+ cells could not be achieved by enrichment. Counting the numbers of “tufted” cells in the TEM is likely to result in an underestimate. Tuft fibrils are difficult to detect, and if the tuft was underneath or above the visible circumference, it would not have been counted, as the cell would have appeared to be nontufted. In addition the fibrils often do not show a pronounced contrast in the negative stain. For both these reasons it is possible that 100% of the Fib+ cells carry fibrils but 30% of Fib+ cells were not detectable. It is therefore possible that the parent strain of S. epidermidis NCTC 11047 contains approximately 50% Fib+ cells rather than the 25% detected. The Fib+ and Fib− subpopulations are clonal descendants of the NCTC 11047 WT, as it is highly unlikely for a number of reasons that the enrichment procedure led to the selection of a subpopulation of contaminants. Firstly, multiplication of cells occurred only in the nonselective medium TSB; secondly the mean lengths of tuft fibrils from WT NCTC 11047 and the Fib+ cells were not significantly different; and thirdly, the SDS-PAGE profiles were identical apart from differences in the presence of the 280- and 230-kDa proteins (see below).
Organization of fibrils into localized tufts.
This is the first description of localized fibrillar appendages on a staphylococcus, but tufts of fibrils are common on oral streptococci and have been found on Streptococcus oralis (28), Streptococcus mitis (formerly Streptococcus sanguis biotype II) (17), and S. cristatus (15, 18). The functions of these prominent structures are not well understood, although S. cristatus cells adhere specifically via the tuft of fibrils to other plaque bacteria (30) in coaggregation interactions between different genera in mature dental plaque.
Negative staining of the enriched Fib+ subpopulation revealed that tufts could sometimes be present at a number of locations simultaneously on the cell surface (Fig. 1b to d and Fig. 2b to d and f). The majority of tufts were in the lateral position, but some were polar. This could mean that the fibrils can migrate around the cell surface (from lateral to polar), or it may be a result of the septum always occurring at right angles to the previous septum, resulting in the lateral surface becoming the apparent pole after two cell division rounds. Localized protein export is necessary to produce these patches of localized fibrils, and recently a laterally positioned microdomain for protein secretion via the general secretory (Sec) pathway has been discovered in Streptococcus pyogenes (48). A similar localized export process may operate for fibril proteins in S. epidermidis NCTC 11047.
The fibrils detected on NCTC 11047 are clearly distinct from the much longer, very thin “fimbria-like” appendages found on S. epidermidis 354 that mediate in adhesion to polystyrene (55). These appendages are comprised of the proteins SSP-1 and SSP-2, and both have a molecular mass in excess of 200 kDa. Since the proteins have not been sequenced and the genes encoding these proteins have not been identified, it is not possible to relate the two morphologically distinct structures on the two strains.
Since only a subpopulation of cells of the WT S. epidermidis strain NCTC 11047 carries tufts of fibrils, it is possible that fibril expression is subject to phase variation. Phase variation of fimbriae (pili) on gram-negative bacteria is a very common phenomenon (2, 11, 42, 54), and phase variation mechanisms exist that alternate between expression (phase on) and nonexpression (phase off) of fimbriae (23). Phase variation in gram-positive bacteria is less well studied, although several surface proteins linked with Streptococcus pyogenes virulence are known to phase vary (7).
Currently we have no experimental evidence to support the hypothesis that fibrils are phase variable, as in this study no reversion of either the Fib+ or the Fib− phenotype was detected. Even after 34 subcultures the Fib− cells did not regain fibrillar tufts or the high-molecular-weight protein bands of Aap (see below), and the Fib+ cells did not apparently lose them. This lack of ability to revert to the WT ratio of 3:1 Fib− to Fib+ cannot currently be explained. However, the possibility of a point mutation having occurred in the Fib− subpopulation, leading to some as-yet-unknown effect on translation, cannot be ruled out. However, the Fib− subpopulation within the WT population clearly has a mechanism which prevents fibril expression on the cell surface, whereas the Fib+ cells can express the fibrils. In addition, S. epidermidis NCTC 11047 WT cells often have Fib+ and Fib− daughter cells attached to each other as they divide (Fig. 2a), implying that the mechanism controlling fibril expression is switched off in one cell while it remains switched on in the other cell as the DNA replicates.
All three NCTC 11047 strains carry the aap gene, and PCR products of the whole aap gene of each population are the same length, indicating that there is no major insertion or deletion in the aap gene of Fib− cells that is inhibiting expression of Aap on the cell surface. In addition, using RT-PCR, we have shown that WT, Fib−, and Fib+ cells all transcribe aap, indicating that expression of Aap is regulated at the posttranscriptional level, although as yet it is not clear what this mechanism of regulation is. If translation also occurs, it is possible that the fibrils could be retained in the cell or that cell wall anchoring is prevented by some unknown mechanism. These possibilities are currently under investigation.
Correlation between fibrils, high-molecular-mass proteins, and Aap.
The 280- and 230-kDa polypeptides present in CSP extracts of Fib+ cells and WT cells were the only high-molecular-mass proteins absent from Fib− cells. Both bands were identified as Aap by mass spectrometric analysis, as fragments from them share 100% identity with Aap from the already sequenced S. epidermidis strains RP62A and ATCC 12228 (14, 57). Antibodies raised against the N-terminal A region of Aap and the single B-repeat unit (128-aa repeat) both labeled the fibrillar tufts, conclusively showing that Aap is expressed as a full-length molecule on the cell surface of S. epidermidis NCTC 11047. Therefore, it is proposed that the 280-kDa molecule of Aap is the full-length protein represented on the cell surface as 122-nm fibrillar appendages localized in a tuft structure.
Thus, each fibril in the tuft of NCTC 11047 is proposed to be a molecule of Aap. This is similar in principle to the 60-nm peritrichous fibrils carried by Streptococcus gordonii DL1, the dental plaque primary colonizer. Each fibril is one molecule of the 259-kDa cell wall-anchored protein CshA (37). CshA also has a domain structure and is anchored via an LPXTG motif to peptidoglycan. It is a multifunctional fibrillar adhesive protein involved in coaggregation to other oral bacteria and in adhesion to fibronectin (36). Aap has not been reported to be glycosylated, and the 280- and 230-kDa Aap bands of NCTC 11047 do not stain with periodic-Schiff reagent, confirming the lack of glycosylation.
Biofilm formation by S. epidermidis 5179 depends on proteolytic cleavage of the full-length (220-kDa) Aap to a 140-kDa truncated form, which consists of the B domain of Aap without the A region (47). Strain 5179 also expresses a 180-kDa Aap thought to be a partially truncated form of the 220-kDa Aap, although its function has not yet been determined. Like S. epidermidis 5179, strain NCTC 11047 expresses two high-molecular-mass Aaps (280 and 230 kDa), although it is not yet clear why these two forms are expressed and what the significance is in the difference between these two Aap bands. As both anti-A and anti-B antibodies labeled the tufts of fibrils, it is clear that Aap is expressed as a full-length molecule on the cell surface of S. epidermidis NCTC 11047, and so according to the mechanism of biofilm formation proposed by Rohde et al. (47), this strain would be biofilm negative. In support of this there is only a very small difference in biofilm-forming ability between the Fib+ and Fib− subpopulations, as biofilms formed by NCTC 11047 and its Fib+ and Fib− subpopulations are very similar in thickness to the nonbiofilm-forming strain S. epidermidis ATCC 12228. Additional support for the proposal that NCTC 11047 has a biofilm-negative phenotype is provided by the fact that it lacks the ica operon, which is responsible for biosynthesis of PIA, the polymer responsible for the biofilm-positive phenotype of many strains, including RP62A (31).
The crystal violet assay for biofilm formation was used in this study because, since it was developed by Christensen et al. (6), it has been used consistently in the majority of biofilm quantification studies for a very wide range of gram-positive and gram-negative bacteria. Christensen et al. (6) proved that the numbers of cells (by weight) in a biofilm correlated with the OD of the crystal violet taken up by the cells in the biofilm.
The presence of tufts of fibrils on a very small number of cells on RP62A also correlated with the presence of the aap gene in this strain, but its ability to form a copious biofilm is due to the production of the PIA polymer as already discussed (31). Aap of strain RP62A is not truncated, as SDS-polyacrylamide gels show the same two high-molecular-weight Aap bands as shown by NCTC 11047. S. epidermidis 12228 carries the aap gene (57), but we do not yet know whether it carries tufts of fibrils. On the basis of the biofilm formation mechanism proposed by Rohde et al. (47), it is predicted that proteolytic cleavage of Aap on NCTC 11047 would result in an increase in the ability of this strain to form biofilms, although this hypothesis is yet to be tested. A bigger study to correlate the presence of the aap gene with the expression of fibrils on a large number of S. epidermidis strains is under way.
Although there is very little difference in the amounts of biofilm formed by Fib+ and Fib− cells of strain NCTC 11047 after 24 h, the presence of the hydrophobic fibrillar tufts does enhance initial adhesion of the Fib+ cells to polystyrene after 2 h. At this early stage in adhesion, the level of Fib+ adhesion is twice as high as that of the Fib− cells. It is possible that the fibrillar tufts mediate adhesion to polystyrene by a specific mechanism in a process similar to that mediated by the high-molecular-mass SSP-1 and SSP-2 proteins present on S. epidermidis 354 (51, 55). By the time the 24-h biofilm of strain NCTC 11047 is formed, the differences in initial adhesion of the Fib+ and Fib− cells have been almost lost as the numbers of cells of these two subpopulations build up in the biofilms to similar levels in the accumulation phase. The reasons for this are not clear, as the two strains have very similar doubling times and reach the same final OD in the wells (data not presented).
This study has demonstrated an unequivocal correlation between the presence of localized tufts of fibrils and the presence of Aap on the cell surface. It also predicts that the localized fibrils of Aap contribute to the overall cell surface hydrophobicity for S. epidermidis strain NCTC 11047. Revealing the localization and molecular architecture of Aap on the cell surface will allow a more detailed understanding of its function in processes relating to colonization and infection.
Acknowledgments
This study was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) CASE award supported by NeuTec Pharma PLC. T.J.F. was supported financially by a Programme Investigator Grant from Science Foundation Ireland.
We thank Lee Cosgrove for Fig. 2 and Wiesia Woodyatt for measuring the fibril lengths of S. epidermidis NCTC 11047. We thank the staff in the EM Facility in the Faculty of Life Sciences (University of Manchester) for their assistance and the Wellcome Trust for equipment grant support to the EM Facility.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Arciola, C. R., S. Gamberoni, D. Capoccia, L. Visai, P. Speziale, L. Baldassarri, and L. Montanaro. 2005. A multiplex PCR method for the detection of all five individual genes of ica locus in Staphylococcus epidermidis. A survey on 400 clinical isolates from prosthesis-associated infections. J. Biomed. Mater. Res. A 75:408-413. [DOI] [PubMed] [Google Scholar]
- 2.Bayliss, C. D., W. A. Sweetman, and E. R. Moxon. 2004. Mutations in Haemophilus influenzae mismatch repair genes increase mutation rates of dinucleotide repeat tracts but not dinucleotide repeat-driven pilin phase variation rates. J. Bacteriol. 186:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowden, M. G., W. Chen, J. Singvall, Y. Xu, S. J. Peacock, V. Valtulina, P. Speziale, and M. Hook. 2005. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 151:1453-1464. [DOI] [PubMed] [Google Scholar]
- 4.Busscher, H. J., P. S. Handley, P. G. Rouxhet, L. M. Hesketh, and H. C. van der Mei. 1991. The relationship between structural and physicochemical surface properties of tufted Streptococcus sanguis strains, p. 317-338. In P. G. Rouxhet (ed.), Microbial cell surface analysis, structural and physicochemical methods. VCH Publishers Inc., New York, NY.
- 5.Christensen, G. D., A. L. Bisno, J. T. Parisi, B. McLaughlin, M. G. Hester, and R. W. Luther. 1982. Nosocomial septicaemia due to multiply antibiotic-resistant Staphylococcus epidermidis. Ann. Intern. Med. 96:1-10. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleary, P. P., l. McLandsborough, L. Ikeda, D. Cue, J. Krawczak, and H. Lam. 1998. High frequency intracellular infection and erythrogenic toxin A expression undergo phase variation in M1 group A streptococci. Mol. Microbiol. 28:157-167. [DOI] [PubMed] [Google Scholar]
- 8.Deighton, M. A., J. Capstick, E. Domalewski, and T. van Nguyen. 2001. Methods for studying biofilms produced by Staphylococcus epidermidis. Methods Enzymol. 336:177-195. [DOI] [PubMed] [Google Scholar]
- 9.Finch, R. G., P. Hill, and P. Williams. 1995. Staphylococci—the emerging threat. Chem. Ind. 6:225-228. [Google Scholar]
- 10.Frebourg, N. B., S. Lefebvre, S. Baert, and J.-F. Lemeland. 2000. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 38:877-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gally, D. L., J. Leathart, and I. C. Blomfield. 1996. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol. Microbiol. 21:725-738. [DOI] [PubMed] [Google Scholar]
- 12.Geertsema-Doornbusch, G. I., H. C. van der Mei, and H. J. Busscher. 1993. Microbial cell surface hydrophobicity: the involvement of electrostatic interactions in microbial adhesion to hydrocarbons (MATH). J. Microbiol. Methods 18:61-68. [Google Scholar]
- 13.Gilbert, P., T. Maira-Litran, A. J. McBain, A. H. Rickard, and F. W. Whyte. 2002. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microb. Physiol. 46:202-256. [PubMed] [Google Scholar]
- 14.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madpu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handley, P., A. Coykendall, D. Beighton, J. M. Hardie, and R. A. Whiley. 1991. Streptococcus crista sp. nov., a viridans streptococcus with tufted fibrils, isolated from the human oral cavity and throat. Int. J. Syst. Bacteriol. 41:543-547. [DOI] [PubMed] [Google Scholar]
- 16.Handley, P. S., L. M. Hesketh, and R. A. Moumena. 1991. Charged and hydrophobic groups are localized in the short and long tuft fibrils of Streptococcus sanguis strains. Biofouling 4:105-111. [Google Scholar]
- 17.Handley, P. S., P. L. Carter, J. E. Wyatt, and L. M. Hesketh. 1985. Surface structures (peritrichous fibrils and tufts of fibrils) found on Streptococcus sanguis strains may be related to their ability to coaggregate with other oral genera. Infect. Immun. 47:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handley, P. S., F. F. Correia, K. Russell, B. Rosan, and J. M. DiRienzo. 2005. Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus. Oral Microbiol. Immunol. 20:131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartford, O., L. O'Brien, K. Schofield, J. Wells, and T. J. Foster. 2001. The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology 147:2545-2552. [DOI] [PubMed] [Google Scholar]
- 20.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 21.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 22.Heilmann, C., G. Thumm, G. S. Chhatwal, J. Hartleib, A. Uekotter, and G. Peters. 2003. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology 149:2769-2778. [DOI] [PubMed] [Google Scholar]
- 23.Hernday, A., M. Krabbe, B. Braaten, and D. Low. 2002. Self-perpetuating epigenetic pili switches in bacteria. Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16470-16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogt, A. H., J. Dankert, and J. Feijen. 1983. Encapsulation, slime production and surface hydrophobicity of coagulase-negative staphylococci. FEMS Microbiol. Lett. 18:211-215. [Google Scholar]
- 25.Hoyle, B. D., and J. W. Costerton. 1991. Bacterial resistance to antibiotics: the role of biofilms. Prog. Drug Res. 37:91-105. [DOI] [PubMed] [Google Scholar]
- 26.Hussain, M., C. Heilmann, G. Peters, and M. Herrmann. 2001. Teichoic acid enhances adhesion of Staphylococcus epidermidis to immobilized fibronectin. Microb. Pathog. 31:261-270. [DOI] [PubMed] [Google Scholar]
- 27.Hussain, M., M. Herrmann, C. von Eiff, F. Perdreau-Remington, and G. Peters. 1997. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65:519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jameson, M. W., H. F. Jenkinson, K. Parnell, and P. S. Handley. 1995. Polypeptides associated with tufts of cell-surface fibrils in an oral Streptococcus. Microbiology 141:2729-2738. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K., and M. Favre. 1973. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 80:575-599. [DOI] [PubMed] [Google Scholar]
- 30.Lancy, P., J. M. Dirienzo, Jr., B. Appelbaum, B. Rosan, and S. C. Holt. 1983. Corncob formation between Fusobacterium nucleatum and Streptococcus sanguis. Infect. Immun. 40:303-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mack, D., M. Haeder, N. Siemssen, and R. Laufs. 1996. Association of biofilm production of coagulase-negative staphylococci with expression of a specific polysaccharide intercellular adhesin. J. Infect. Dis. 174:881-884. [DOI] [PubMed] [Google Scholar]
- 32.Mack, D., M. A. Horstkotte, H. Rohde, and J. K.-M. Knobloch. 2006. Coagulase-negative staphylococci, p. 109-153. In R. G. Finch (ed.), Biofilms, infection, and antibiotic therapy. CRC Press, Taylor and Francis Group, Boca Raton, FL.
- 33.Mack, D., H. Rohde, L. G. Harris, A. P. Davies, M. A. Horstkotte, and J. K.-M. Knobloch. 2006. Biofilm formation in medical device-related infection. Int. J. Artif. Organs 29:343-359. [DOI] [PubMed] [Google Scholar]
- 34.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996b. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenney, D., J. Hubner, E. Muller, Y. Wang, D. A. Goldmann, and G. B. Pier. 1998. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66:4711-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNab, R., A. Holmes, J. Clarke, G. Tannock, and H. Jenkinson. 1996. Cell surface polypeptide CshA mediates binding of Streptococcus gordonii to other oral bacteria and to immobilized fibronectin. Infect. Immun. 64:4204-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNab, R., H. Forbes, P. S. Handley, D. M. Loach, G. W. Tannock, and H. F. Jenkinson. 1999. Cell wall-anchored CshA polypeptide (259 kilodaltons) in Streptococcus gordonii forms surface fibrils that confer hydrophobic and adhesive properties. J. Bacteriol. 181:3087-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller, E., S. Takeda, H. Shiro, D. Goldmann, and G. B. Pier. 1993. Occurrence of capsular polysaccharide/adhesin among clinical isolates of coagulase-negative staphylococci. J. Infect. Dis. 168:1211-1218. [DOI] [PubMed] [Google Scholar]
- 39.Nilsson, M., L. Frykberg, J. I. Flock, L. Pei, M. Lindberg, and B. Guss. 1998. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect. Immun. 66:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pembrey, R. S., K. C. Marshall, and R. P. Schneider. 1999. Cell surface analysis techniques: what do cell preparation protocols do to cell surface properties? Appl. Environ. Microbiol. 65:2877-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters, G., R. Locci, and G. Pulverer. 1982. Adherence and growth of coagulase-negative staphylococci on surfaces of intravenous catheters. J. Infect. Dis. 146:479-482. [DOI] [PubMed] [Google Scholar]
- 42.Power, P. M., L. F. Roddam, K. Rutter, S. Z. Fitzpatrick, Y. N. Srikhanta, and M. P. Jennings. 2003. Genetic characterization of pilin glycosylation and phase variation in Neisseria meningitidis. Mol. Microbiol. 49:833-847. [DOI] [PubMed] [Google Scholar]
- 43.Raad, I., A. Alrahwan, and K. Rolston. 1998. Staphylococcus epidermidis: emerging resistance and need for alternative agents. Clin. Infect. Dis. 26:1182-1187. [DOI] [PubMed] [Google Scholar]
- 44.Roche, F. M., M. Meehan, and T. J. Foster. 2003. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology 149:2759-2767. [DOI] [PubMed] [Google Scholar]
- 45.Roche, F. M., R. Massey, S. J. Peacock, N. P. Day, L. Visai, P. Speziale, A. Lam, M. Pallen, and T. J. Foster. 2003. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149:643-654. [DOI] [PubMed] [Google Scholar]
- 46.Rohde, H., M. Kalitzky, N. Kroger, S. Scherpe, M. A. Horstkotte, J. K.-M. Knobloch, A. R. Zander, and D. Mack. 2004. Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. J. Clin. Microbiol. 42:5614-5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohde, H., C. Burdelski, K. Bartscht, M. Hussain, F. Buck, M. A. Horstkotte, J. K. M. Knobloch, C. Heilmann, M. Herrmann, and D. Mack. 2005. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55:1883-1895. [DOI] [PubMed] [Google Scholar]
- 48.Rosch, J., and M. Caparon. 2004. A microdomain for protein secretion in Gram-positive bacteria. Science 304:1513-1515. [DOI] [PubMed] [Google Scholar]
- 49.Rosenburg, M., D. Gutnick, and E. Rosenburg. 1980. Adherence of bacteria to hydrocarbons: a simple method for measuring cell surface hydrophobicity. FEMS Microbiol. Lett. 9:29-33. [Google Scholar]
- 50.Smyth, C. J., P. Jonsson, E. Olsson, O. Soderlind, J. Rosengren, S. Hjerten, and T. Wadstrom. 1978. Differences in hydrophobic surface characteristics of porcine enteropathogenic Escherichia coli with or without K88 antigen as revealed by hydrophobic interaction chromatography. Infect. Immun. 22:462-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Timmerman, C. P., A. Fleer, J. M. Besnier, L. De Graaf, F. Cremers, and J. Verhoef. 1991. Characterization of a proteinaceous adhesin of Staphylococcus epidermidis which mediates attachment to polystyrene. Infect. Immun. 59:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Timmerman, C. P., J. M. Besnier, L. De Graaf, R. Torensma, A. J. Verkley, A. Fleer, and J. Verhoef. 1991. Characterisation and functional aspects of monoclonal antibodies specific for surface proteins of coagulase-negative staphylococci. J. Med. Microbiol. 35:65-71. [DOI] [PubMed] [Google Scholar]
- 53.Vandecasteele, S. J., W. E. Peetermans, R. Merckx, and J. V. Eldere. 2003. Expression of biofilm-associated genes in Staphylococcus epidermidis during in vitro and in vivo foreign body infections. J. Infect. Dis. 188:730-737. [DOI] [PubMed] [Google Scholar]
- 54.Van der Woude, M. W., and A. J. Baumler. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veenstra, G. J. C., F. F. M. Cremers, H. van Dijk, and A. Fleer. 1996. Ultrastructural organization and regulation of a biomaterial adhesin of Staphylococcus epidermidis. J. Bacteriol. 178:537-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams, R. J., B. Henderson, L. J. Sharp, and S. P. Nair. 2002. Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect. Immun. 70:6805-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, Y.-Q., S.-X. Ren, G. Fu, J. Yang, Z.-Q. Qin, Y.-G. Miao, W.-Y. Wang, R.-S. Chen, Y. Shen, Z. Chen, Z.-H. Yuan, G.-P. Zhao, D. Qu, A. Danchin, and Y.-M. Wen. 2003. Genome based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577-1593. [DOI] [PubMed] [Google Scholar]
- 58.Ziebuhr, W., C. Heilmann, F. Gotz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]