Abstract
Pseudomonas putida uses l-lysine as the sole carbon and nitrogen source which preferentially requires its metabolism through two parallel pathways. In one of the pathways δ-aminovalerate is the key metabolite, whereas in the other l-lysine is racemized to d-lysine, and l-pipecolate and α-aminoadipate are the key metabolites. All the genes and enzymes involved in the d-lysine pathway, except for those involved in the conversion of d-lysine into Δ1-piperideine-2-carboxylate, have been identified previously (30). In this study we report that the conversion of d-lysine into Δ1-piperideine-2-carboxylate can be mediated by a d-lysine aminotransferase (PP3590) and a d-lysine dehydrogenase (PP3596). From a physiological point of view PP3596 plays a major role in the catabolism of d-lysine since its inactivation leads to a marked reduction in the growth rate with l- or d-lysine as the sole carbon and nitrogen source, whereas inactivation of PP3590 leads only to slowed growth. The gene encoding PP3590, called here amaC, forms an operon with dpkA, the gene encoding the enzyme involved in conversion of Δ1-piperideine-2-carboxylate to l-pipecolate in the d-lysine catabolic pathway. The gene encoding PP3596, called here amaD, is the fifth gene in an operon made up of seven open reading frames (ORFs) encoding PP3592 through PP3597. The dpkA amaC operon was transcribed divergently from the operon ORF3592 to ORF3597. Both promoters were mapped by primer extension analysis, which showed that the divergent −35 hexamers of these operon promoters were adjacent to each other. Transcription of both operons was induced in response to l- or d-lysine in the culture medium.
Pseudomonas putida KT2440, a derivative of P. putida mt-2 from which the TOL plasmid was cured (15, 25), can grow on proline, lysine, glutamate, and other amino acids as the sole carbon and nitrogen source. The ability to assimilate these compounds confers a selective advantage by enabling the strain to grow in the rhizosphere of a number of plants, where these amino acids form part of the root exudates (4, 13, 21, 23).
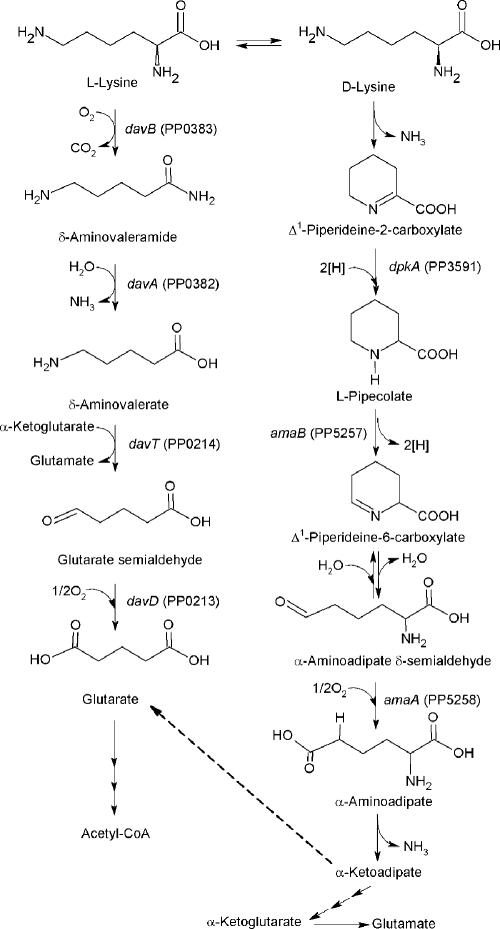
Pioneering work on bacterial lysine metabolism (17) helped establish two parallel catabolic pathways (Fig. 1), starting with the interconversion by a racemase of both enantiomers of lysine. l-Lysine catabolism proceeds through the so-called aminovalerate (AMV) pathway and involves the following steps: l-lysine → δ-aminovaleramide → δ-AMV → glutarate semialdehyde → glutarate, which is then channeled into the Krebs cycle via glutaryl- and acetyl-coenzyme A (CoA). On the other hand, d-lysine catabolism proceeds via a series of cyclized intermediates which are necessary to regenerate an α-amino acid and comprise the following metabolites (aminoadipate [AMA] pathway): d-lysine → α-keto-ɛ-amino caproate → Δ1-piperideine-2-carboxylate → pipecolate → Δ1-piperideine-6-carboxylate → α-amino-δ-formylcaproate → α-AMA → α-ketoadipate. The latter is transformed to α-ketoglutarate via a series of CoA derivatives. These steps were established at the biochemical level in the early 1970s (6, 7, 8, 9, 14, 20), whereas the genetic organization of the pathway remained unknown in P. putida and other microorganisms. We identified some of the catabolic genes and found that the expression of davT encoding δ-AMV aminotransferase was induced in the rhizosphere of maize (13). This led us to further study l-lysine catabolism in this strain. To identify the genes encoding the enzymes involved in the catabolism of l-lysine by P. putida KT2440, we isolated mini-Tn5 mutants unable to use this amino acid as the sole C and N source. The sites of transposon insertions in davB, davA, davD, and davT were determined by DNA sequencing, and the nature of the interrupted metabolic steps was identified by determining the accumulation of pathway intermediates with isotopically labeled l-lysine (13, 29, 30).
FIG. 1.
Catabolic pathways for the mineralization of l- and d-lysine in P. putida. The diagram is based on Fig. 1 in Revelles et al. (30) and adds the new enzymes identified in this study as involved in the catabolism of d-lysine into Δ1-piperideine-2-carboxylate.
This type of study led us to confirm that l-lysine catabolism indeed proceeds via two parallel pathways in P. putida and that the operation of the AMA pathway requires the initial lysine racemase, which converts l-lysine into d-lysine (30). The fate of l-[13C]lysine in mutants blocked in the AMV pathway has allowed us to show that AMA pathway intermediates can be channeled into the AMV pathway via α-ketoadipate and glutarate (30).
The davB promoter was silent in the absence of l-lysine but was induced to a high level in response to this amino acid (30). In contrast, the davD promoter exhibited a low basal level that increased about fourfold in response to the addition of exogenous l-lysine to the culture medium (13).
As mentioned above, the efficient utilization of l-lysine as the sole C and N source requires it to be partially metabolized via the d-lysine pathway upon racemization. In previous studies we along with another group identified the amaA, amaB, and dpkA genes involved in the steps from Δ1-piperideine-2-carboxylate down to AMA and showed that mutants in each step accumulated the expected metabolites (24, 30).
This study was undertaken to identify the gene(s) responsible for the first step in d-lysine catabolism in KT2440. Our results revealed that downstream from dpkA, and forming an operon with it, is the amaC gene, whose gene product is an aminotransferase involved in the first step in d-lysine catabolism. A mutant in amaC still grew with d-lysine, albeit at a lower rate, indicating that this 6-aminotransferase was not the only enzyme involved in d-lysine catabolism. After massive mini-Tn5 mutagenesis of KT2440, we searched for mutants unable to grow with d-lysine but able to grow with l-pipecolate. A transposon mutant in a locus different from that of dpkA had a mini-Tn5 inserted in the open reading frame (ORF) encoding PP3596, which is called d-amino acid dehydrogenase although it is, in fact, a deaminating oxidase that yields α-keto-ɛ-aminocaproate. This acid is cyclized to Δ1-piperideine-2-carboxylate, which in turn is channeled to l-pipecolate and AMA. The corresponding gene was called amaD, and its inactivation severely inhibited growth of P. putida KT2440 on d-lysine.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
P. putida KT2440 and its mutant derivatives are shown in Table 1. The P. putida KT2440 cosmid library used in this study was described before (31).
TABLE 1.
Growth rate of KT2440 and its isogenic mutants with l-lysine as the sole C and N source and l-pipecolate as a C source
| Strain | Mutant genea | Doubling time (min) in indicated culture mediumb
|
||
|---|---|---|---|---|
| M8 + l-lysine | M8 + d-lysine | M9+ l-pipecolate | ||
| KT2440 | None | 90 ± 10 | 85 ± 5 | 65 ± 5 |
| AmaC | amaC | 110 ± 5 | 120 ± 5 | 65 ± 5 |
| DpkA | dpkA | 420 ± 10 | No growth | 65 ± 5 |
| Lys2 | amaD | 325 ± 5 | No growth | 65 ± 5 |
| EEZ-PP3592 | ORF3592 | 90 ± 15 | 87 ± 35 | 66 ± 7 |
Inactivated gene in the isogenic mutant.
Doubling times were determined in exponentially growing cells. M8 medium was identical to M9 medium except that NH4Cl was omitted. Values are ± standard deviation.
Escherichia coli strain DH5α was used for cloning experiments and was grown at 37°C in Luria-Bertani medium (32). P. putida strains were grown at 30°C either in Luria-Bertani or in M9 minimal medium supplemented with Fe-citrate, MgSO4, and trace metals, as described previously (1), and with benzoate (15 mM), glucose (0.5% [wt/vol]), l-lysine (10 mM), d-lysine (10 mM), or l-tyrosine (10 mM) as a carbon source. When appropriate, antibiotics were added at the following concentrations (in μg/ml): chloramphenicol, 30; kanamycin, 50; and tetracycline, 15.
DNA techniques.
Standard methods were used for DNA manipulation (3). Hybridizations were done with a digoxigenin DNA labeling and detection kit (Roche, Manheim, Germany) according to the manufacturer's instructions. Plasmid sequencing was done with universal or reverse pUC19/M13 oligonucleotides as primers. Sequences were analyzed with Omiga 2.0 software (Oxford Molecular) and ORFFINDER (available at NCBI) and compared with data from the P. putida KT2440 genome (26) using BLAST programs (2).
Preparation of RNA, primer extension analysis, and RT-PCR.
P. putida KT2440 and its isogenic mutants were grown in duplicate in minimal medium M9 in the absence and in the presence of l-lysine with benzoate as the carbon source until the culture reached a turbidity of about 1.0 at 660 nm. Cells (30 ml for primer extension and 1.5 ml for reverse transcriptase PCR [RT-PCR]) were harvested by centrifugation (5,000 × g for 10 min) and processed for RNA isolation according to the method of Marqués et al. (19). For primer extension analysis we used as a specific primer an oligonucleotide complementary to the dpkA mRNA (5′-GGCGCTGGCGCAGTTGTGG-3′) and a primer complementary to the mRNA of the ORF encoding PP3592 (5′-CCAGGCGTTGCTTGATCAGT-3′). Primers were labeled at their 5′ ends with [γ-32P]ATP and T4 polynucleotide kinase as described previously (3). An amount corresponding to about 105 cpm of the labeled primer was hybridized to 50 to 100 μg of total RNA, and extension was carried out using avian myeloblastosis virus reverse transcriptase as described previously (19). Electrophoresis of cDNA products was done in a urea-polyacrylamide sequencing gel to separate the reaction products. RT-PCR was done with 1 μg of RNA in a final volume of 20 μl using the Titan OneTube RT-PCR system according to the manufacturer's instructions (Roche Laboratories). The annealing temperature used for RT-PCR was 60°C, and the cycling conditions were as follows: 94°C for 30 s, 60°C for 30 s, and 68°C for 1 min. Positive and negative controls were included in all assays. The primers used to test contiguity in the mRNA are available on request.
Construction of a P. putida amaC mutant strain.
A mutant strain bearing an inactivated chromosomal amaC was constructed as follows. Plasmid pCHESIΩKm is a pUC18 derivative containing the origin of transfer oriT of RK2 and the Ω-Km interposon of plasmid pHP45ΩKm (where Km indicates kanamycin resistance) cloned as a HindIII fragment (18). To generate the amaC mutation, an internal 593-bp fragment of P. putida amaC (ORF encoding PP3590) was amplified by PCR using primers provided with EcoRI and BamHI sites and subsequently cloned between the EcoRI and BamHI sites of pCHESIΩKm in the same transcriptional direction as the Plac promoter. The resulting plasmid, pCHESI-ΔamaC, was mobilized from E. coli DH5α into P. putida KT2440 by triparental mating using the E. coli HB101 (pRK600) helper strain (16). P. putida transconjugants bearing a cointegrate of the plasmid in the host chromosome were selected on M9 minimal medium with benzoic acid (10 mM) as the sole carbon source and kanamycin. A clone carrying an inactivated amaC gene was randomly chosen and called P. putida ΔamaC Km.
Identification of a mini-Tn5 inserted in the ORF encoding PP3592.
A clone carrying a mini-Tn5 insertion in the 34th codon of the ORF encoding PP3592 was obtained from material produced in the Pseudomonas Reference Culture Collection (12).
Enzymatic assays.
P. putida strain KT2440 was grown on 500 ml of M9 with 5 mM benzoate as the carbon source at 30°C on a rotating shaker at 200 rpm until an optical density at 600 nm of 0.8 to 1 was reached. Cells were harvested by centrifugation (5,000 × g for 10 min) at 4°C, washed twice with M9 medium (pH 7.0), resuspended in 3 ml of 100 mM phosphate buffer (pH 8.0), and disrupted in a French press at 120 MPa. Cell debris was removed by centrifugation (9,000 × g for 45 min), and the clear supernatant was used for enzymatic assays. Protein concentration was determined with the Bradford method (5).
d-Lysine oxidase activity was assayed with a Clark-type oxygen electrode (model DW-1; Hansatech-Instruments, King's Lynn, Norfolk, United Kingdom) in 100 mM phosphate buffer (pH 8.0) in the presence of the protease inhibitor cocktail (at the concentration recommended by the supplier) and 1 mM dithiothreitol at 25°C, and oxygen consumption was measured with 5 mM d-lysine as the substrate. Specific enzyme activity is given as micromoles of oxygen consumed per minute per milligram of protein. A mutant defective in d-lysine oxidase was used as a control to discriminate nonspecific oxidase reactions.
d-Lysine aminotransferase activity was assayed in a mixture of 25 μmol of d-lysine, 25 μmol of α-ketoglutarate or pyruvate, and crude extract in 100 mM potassium phosphate buffer (pH 8.0) in a final volume of 1.0 ml. The reaction was initiated by adding d-lysine, and the mixture was incubated at 25°C for 30 min. Enzyme activity was assayed by sampling 100-μl aliquots every 5 or 10 min, which were pipetted into 10% trichloroacetic acid to stop the reaction, and by subsequent determination of the substrates and glutamate or alanine formed from the cosubstrates upon reaction with dabsyl chloride (11). A 50-μl sample was injected into a model Hewlett Packard 1050 system (Böblingen, Germany) and separated on a Novapak C8 column of 150 by 3.9 mm (Waters S.A., Barcelona, Spain) at a flow rate of 0.9 ml min−1. Compounds were eluted in the following gradient system: solvent A consisted of 25 mM sodium acetate buffer (pH 6.6) and acetonitrile (80:20, vol/vol); solvent B was acetonitrile (gradient grade). The gradient started at 0% solvent B, rose to 20% solvent B at 5 min and to 80% B at 15 min and dropped back to 0% at 16 min; reequilibration was for 7 min. Detection was at 435 nm. The retention time of dabsyl-alanine was 5.6 min and that of dabsyl-lysine was 8.3 min; dabsyl-glutamate eluted at 9.5 min. Enzyme activities were assayed in the wild-type P. putida and with the amaD mutant blocked in the oxidase reaction.
RESULTS
Genetic analysis of the P. putida chromosomal region encoding PP3589 through PP3598.
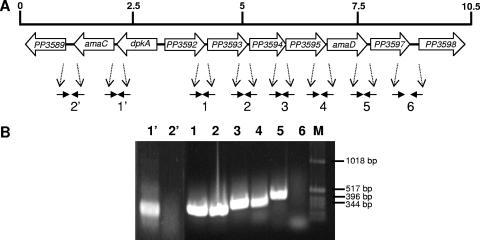
Previous work indicated that the dpkA gene encodes Δ1-piperideine-2-carboxylate reductase (PP3591), the second enzyme in the d-lysine catabolic route (24, 30). Adjacent to dpkA is a gene encoding PP3590, which is oriented in the same transcriptional direction as dpkA in the chromosome of P. putida KT2440 (Fig. 2A). The intergenic space between dpkA and the ORF encoding PP3590 is 166 bp long. The translated gene product of the latter ORF showed high similarity with amino acid aminotransferases and had been annotated as tyrB2; its corresponding gene product was proposed to be potentially involved in l-tyrosine catabolism (26). Downstream from the ORF encoding PP3590 and in the same transcriptional direction is located the gene encoding PP3589, which is 1,278 bp long, and its translated gene product exhibits a high degree of similarity (60 to 80%) to serine transporter proteins. The distance between the start codon of this gene (ORF3589) and the stop codon of the ORF encoding PP3590 is 387 bp.
FIG. 2.
Physical organization of the dpkA/amaC genes and the ORFs encoding the PP3592 to PP3597 clusters involved in d-lysine catabolism. (A) Map of the two operons involved in the early steps of d-lysine catabolism. (B) RT-PCR to probe the operon structure of dpkA-amaC and ORF PP3592 through ORF PP3597. Primers used were designed based on the 3′ end of the upstream gene and the 5′ end of the downstream gene. Primer sequences are available on request. Lane 1′, RT-PCR based on dpkA-amaC primers; lane 2′, RT-PCR based on amaC and ORF PP3589 primers; lane 1, RT-PCR based on ORF PP3592 and ORF PP3593 primers; lane 2, RT-PCR based on ORF PP3593 and ORF PP3594 primers; lane 3, RT-PCR based on ORF PP3594 and ORF PP3595 primers; lane 4, RT-PCR based on ORF PP3595 and ORF PP3596 primers; lane 5, RT-PCR based on ORF 3596 and ORF PP3597 primers; lane 6, RT-PCR based on ORF PP3597 and ORF PP3598 primers; lane M, size markers.
Upstream from dpkA is an ORF that is transcribed divergently with respect to dpkA and whose translated product exhibits similarity (50 to 60%) to DNA-binding proteins belonging to the RpiR family of regulators. The distance between the start codon of dpkA and the start codon of ORF3592 is 119 bp. Downstream from the ORF encoding PP3592 are five additional ORFs that seem to form an operon together with the regulatory gene, because the different ORFs overlap (−3) or are separated by very short intergenic distance (from +4 to +7 nucleotides). BLAST analyses revealed that PP3593, PP3594, PP3595, and PP3597 are members of ABC transporter systems and that PP3596 exhibits high similarity to d-amino acid dehydrogenases.
The dpkA-amaC genes form an operon, and the ORF encoding PP3592 through PP3597 forms another operon.
To elucidate whether the two sets of clusters of genes with the same orientation were transcribed as a single mRNA, P. putida KT2440 cells were grown in M9 minimal medium with glucose as the carbon source in the absence and in the presence of 5 mM l-lysine (this was added because the dpkA gene product was shown before to be necessary for l-lysine catabolism), mRNA from cells in the late-logarithmic growth phase was isolated, and RT-PCRs were carried out as described in Materials and Methods (Fig. 2A). In the absence of l-lysine we found no amplification product, indicating that neither of the two sets of genes was expressed. When cells had been grown with l-lysine, we found cDNA products of the expected sizes with the primers corresponding to dpkA and the ORF encoding PP3590 (Fig. 2B, lane 1′) but not with primers designed for the ORFs encoding PP3590 and PP3589 (Fig. 2B, lane 2′). This suggested that dpkA and the ORF encoding PP3590 are organized in the same transcriptional unit, whereas the ORF encoding PP3589 seems to be transcribed independently. Controls with internal primers for all three genes of this cluster showed that all of them were transcribed during growth on l-lysine (data not shown).
The set of primer extension assays with primers covering genes encoding PP3592 through PP3597 were always positive (Fig. 2B, lanes 1 to 5), indicating that this cluster of genes is expressed in response to l-lysine. However, no amplification was obtained with primers based on the ORF3597 and ORF3598 sequences, implying that they are not part of the same transcriptional unit.
Analysis of the divergent promoters in front of dpkA and the ORF encoding PP3592.
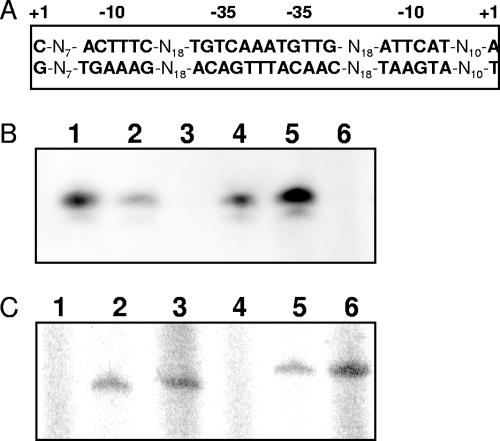
To identify the transcription start point of the dpkA-ORF3590 operon and the divergently oriented ORF PP3592 through ORF PP3597, we isolated mRNA from KT2440 growing on M9 with glucose as a carbon source without lysine or with 5 mM l-lysine or 5 mM d-lysine and used a 20-mer primer complementary to a position located 100 bp within the dpkA gene for primer extension or a 20-mer primer complementary to a position located 85 nucleotides within the ORF3592. Our results revealed no significant amount of transcripts in the absence of lysine but a significant transcription band when cells had been grown with l- or d-lysine. The transcription start point of dpkA was located 69 nucleotides upstream with respect to the G of the first GTG in dpkA (Fig. 3B). Analysis of the sequence upstream revealed that the hexamers for the −10 region (5′-TGAAAG-3′) and −35 region (5′-ACAGTT-3′) (Fig. 3A, left bottom strand) had a certain degree of similarity with P. putida sigma-70 dependent promoters (10). The transcription start point of ORF3592 was represented by an A located 52 nucleotides upstream of the first A of the first ATG start codon (Fig. 3C). The −10 region (5′-ATTCAT-3′; top strand) and −35 region (5′-ATGTTG-3′) of this promoter exhibited a good degree of similarity to promoters recognized by RNA polymerase with sigma-70 as well. Because of the short intergenic distance (119 nucleotides) and the position of the corresponding transcriptional start points, the −35 hexamers of both promoters appeared adjacent to each other.
FIG. 3.
Analysis of mRNA expression from the dpkA and ORF3592 promoters in the wild-type and a mutant background with a nonpolar insertion in ORF3592. (A) Promoter structure of the dpkA and ORF3592 genes. The transcription start points were determined by primer extension as detailed in Materials and Methods. The +1 and the potential −10 and −35 hexamers of each promoter are shown. (B and C) Expression of dpkA and ORF3592 promoters in the wild type (lanes 1 to 3) and the mutant deficient in the synthesis of PP3592 (lanes 4 to 6). Lanes 1 and 4, cells grown on M9 minimal medium; lanes 2 and 5, cells grown on M9 minimal medium with 5 mM l-lysine; lanes 3 and 6, cells grown on M9 minimal medium with 5 mM d-lysine.
Limited role of PP3590 in d-lysine catabolism.
Since the dpkA gene product is induced in l- and d-lysine catabolism and the mRNA of the ORF encoding PP3590 forms part of the same transcript, we decided to explore whether PP3590, that exhibits similarity to amino acid aminotransferases (7, 27), played a major role in d-lysine catabolism. We knocked out ORF3590 using the pCHESI derivative as described in Materials and Methods and tested growth of the mutant on l- and d-lysine as the sole carbon or nitrogen source. The results showed that the PP3590 mutant used l-lysine or d-lysine less efficiently than the parental strain (doubling times in the range of 120 min) whereas the parental P. putida KT2440 grew with doubling times of around 90 min (Table 1). Growth of the mutant defective in ORF3590 contrasted with that of a mutant deficient in dpkA, which did not grow at all when this amino acid was the sole carbon and nitrogen source (Table 1). We concluded that the role of this amino acid transferase was probably of minor importance in d-lysine catabolism and that another enzyme leading to the same intermediate must by functionally active.
We determined d-lysine aminotransferase activity in vitro with cell extracts of the wild type and the mutant strain with a knockout in ORF3590 grown in the absence and in the presence of 5 mM d-lysine. In cell extracts of the mutant strain no activity was found regardless of the growth conditions. In the parental strain a low level of activity was measured (<50 nmol/mg of protein per min) when cells were grown with d-lysine but not in its absence. This low level of activity is in agreement with the relatively minor effect of the mutation on the growth rate on l- and d-lysine. Nonetheless, since the gene product of ORF3590 seemed to be involved at least to a certain extent in the AMA catabolic pathway for d-lysine metabolism and because it was shown biochemically to exhibit d-lysine aminotransferase activity, we termed this gene amaC.
PP3596 is a d-lysine oxidase necessary for d-lysine catabolism.
Since the dpkA mutant strain did not grow at all on d-lysine whereas the amaC mutant did, albeit at a reduced rate, we concluded that a second enzyme should exist to convert d-lysine via α-keto-∈-aminocaproate into Δ1-piperideine-2-carboxylate. P. putida KT2440 was mutagenized using a mini-Tn5-Km transposon as described in Materials and Methods, and among about 15,000 transconjugants we searched for mutants affected in growth on d-lysine but not on l-pipecolate. Several mutants were selected, and those exhibiting an insertion in dpkA were discarded. However, we found a mutant, called Lys2, that had an insertion outside dpkA and selected it for further study. Analyses showed that the mini-Tn5 was inserted in the PP3596 gene, which encoded a protein with high degree of similarity (51%) to d-amino acid dehydrogenases. The KT2440 mutant Lys2 did not grow with d-lysine but grew in medium containing l-pipecolate (Table 1). Because of its essential role in the catabolism of d-lysine, ORF3596 was called amaD. As shown by RT-PCR experiments (Fig. 2), ORF3596 is located within a cluster of genes with a putative regulatory protein and a series of genes that encode an ABC transport system. The specific enzyme activities of AmaD were compared in the wild type and the corresponding Lys2 mutant with d-lysine and l-lysine as substrates. The results revealed that in the wild type, AmaD activity (only active with d-lysine at 33 ± 1 μmol × mg protein−1 min−1) was inducible, whereas in the Lys2 mutant AmaD activity was negligible. Further analysis of the genes in the cluster showed that inactivation of ORF3597 did not significantly inhibit growth on l- or d-lysine (not shown). This suggests that the ABC transport system made up of PP3593, PP3594, PP3595, and PP3597, although inducible by l- or d-lysine (Fig. 2), is dispensable for growth with l- or d-lysine. This, however, was not too surprising because we had already identified two other transport systems for l-lysine uptake (29).
The RpiR family protein is not necessary for growth on l- or d-lysine.
Regulatory genes for catabolic pathways have often been found adjacent to the gene regulated by the cognate product (22, 28). Primer extension results revealed that the dpkA amaC operon, as well as the operon ORF3592 to ORF3597, was expressed in cells growing on l- and d-lysine (Fig. 3). To test whether the product of the ORF encoding PP3592 was involved in the expression of one or both of the two operons, we took advantage of the existence of a nonpolar mini-Tn5 mutant in the Pseudomonas Reference Culture Collection (mutant EEZ-PP3592) and assayed growth of the mutant strain in l- and d-lysine. Our results showed that this mutant grew with l- or d-lysine at a rate similar to that of the wild-type strain (doubling time, 90 ± 2 min). To further confirm that PP3592 was not involved in the control of these operons, we measured expression of the dpkA amaC operon and that of ORF3592 through ORF3597 in the mutant ORF3592::mini-Tn5. The level of expression was similar to that of the wild type, which ruled out the involvement of PP3592 in d- or l-lysine catabolism.
DISCUSSION
Earlier work by Revelles et al. (30) had established that the efficient use of l-lysine as the sole carbon and nitrogen source by KT2440 requires the simultaneous operation of two catabolic pathways: one that converts l-lysine into glutarate—and for which the genes involved in the four consecutive catabolic steps were identified—and another one that requires racemization of l-lysine to d-lysine and its subsequent metabolism to α-AMA and for which genes for three of the four steps of the pathway were identified (30). Several additional genes involved in l-lysine uptake had already been identified in a previous study (29).
The picture emerging from the genetic and physical organization of the genes involved in l- and d-lysine catabolism is that the genes are scattered along the chromosome and are organized in short transcriptional units. For δ-AMV metabolism down to glutarate, the davDT genes form one operon, whereas for l-lysine metabolism to δ-AMV the davBA genes form a separate operon (13, 29, 30). The same was found for the catabolism of l-pipecolate to 2-AMA, since the amaB and amaA genes form another operon (30).
The role of DpkA in d-lysine catabolism was not initially obvious because the gene was originally annotated as a potential malate dehydrogenase. However, the biochemical characterization of the enzyme by Muramatsu et al. (24) clearly showed that it encodes a Δ1-piperideine-2-carboxylate reductase involved in the conversion of Δ1-piperideine-2-carboxylate into l-pipecolate. In our original search for mutants of KT2440 that did not grow on d-lysine as the sole C and N source but which grew on l-pipecolate, we knocked out dpkA and showed that the resulting mutant indeed failed to grow on d-lysine as the sole C and N source (30), confirming the biochemical results of Muramatsu et al. (24). Because of the clustering of genes in the l- and d-lysine pathways as described above, we analyzed in detail the physical region in which dpkA was present and generated a series of mutants whose genetic analyses led to an unexpected finding: the conversion of d-lysine to Δ1-piperideine-2-carboxylate can be mediated by two enzymes in parallel. One of them (PP3590), encoded by ORF3590, represents the d-lysine amino transferase termed here AmaC; the other, a d-amino acid dehydrogenase, termed AmaD, is encoded by ORF3596. The latter oxidized d-lysine into 6-amino-2-oxo-hexanoate (α-keto-∈-aminocaproate), which is cyclized spontaneously to Δ1-piperideine-2-carboxylate. The significantly different growth rates obtained for the mutants in ORF3592 and ORF3596 characterized in the present study suggested that from a quantitative point of view the d-lysine dehydrogenase plays a much more important role in the metabolism of d-lysine than the reaction catalyzed by the amino transferase. Our enzymatic analyses confirmed the proposed reactions, and our results showed that in the corresponding mutants the expected enzymatic activity was fully lost. As far as we know this study represents the first report of a d-lysine dehydrogenase (oxidating) involved in the catabolism of l- and d-amino acids.
Analyses of the physical organization of the two clusters of genes induced in response to l-lysine revealed that an ORF that encodes a putative regulator belonging to the RpiR family was located upstream from dpkA, and expression of the ORF was also stimulated by l-lysine. However, inactivation of ORF3592 did not limit the growth rate of the strain with l- or d-lysine. We also found that in this mutant strain the pattern of expression of these operons was identical to that of the parental strain, a result that ruled out its role in the catabolism of d- or l-lysine. At present we have no clues to the potential transcriptional regulator(s) that is involved in stimulation of transcription of the promoters in front of ORF3592 and dpkA.
In agreement with earlier findings in our group (29, 30), the Lys2 mutant used l-lysine as the sole C and N source less efficiently than the wild-type strain. This is in agreement with the metabolism of l-lysine through the two branches shown in Fig. 1. At present we do not know the molecular basis for this observation, but one possibility is that if the conversion of l-lysine into acetyl-CoA via the AMV pathway depresses oxaloacetate levels in vivo, it might not only limit the operation of the Krebs cycle but also restrict the synthesis of amino acids such as methionine, aspartate, threonine, and isoleucine, which are directly derived from oxaloacetate. Therefore, α-ketoglutarate produced via the AMA pathway may replenish Krebs cycle intermediates. Since the AMA pathway is interconnected with the AMV pathway from α-ketoadipate to glutarate (Fig. 1), we suggest that this step functions as a valve that directs carbon flux to either branch of the catabolic pathway for optimal operation.
Acknowledgments
This study was supported by grant BIO2003-0515 and BIO2006-05668 from the CICYT.
We thank Manuel Espinosa-Urgel for critical reading of the manuscript. We thank Carmen Lorente, María del Mar Fandila and Inés Abril for secretarial assistance and K. Shashok for improving the use of English in the manuscript.
Footnotes
Published ahead of print on 26 January 2007.
REFERENCES
- 1.Abril, M. A., C. Michán, K. N. Timmis, and J. L. Ramos. 1989. Regulator and enzyme specificity of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 171:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, G. G. Seidmen, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. Green Publishing Associates and Wiley Interscience, New York, NY.
- 4.Basic, A., S. F. Moody, and A. E. Clarke. 1986. Structural analysis of secreted root slime from maize (Zea mays L.). Plant Physiol. 80:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y. F., and E. Adams. 1971. Induction of separate catabolic pathways for l- and d-lysine in Pseudomonas putida. Biochem. Biophys. Res. Commun. 45:570-577. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Y. F., and E. Adams. 1974. d-Lysine catabolic pathway in Pseudomonas putida: interrelations with l-lysine catabolism. J. Bacteriol. 117:753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. F., and E. Adams. 1977. Glutarate semialdehyde dehydrogenase of Pseudomonas. J. Biol. Chem. 252:7979-7986. [PubMed] [Google Scholar]
- 9.Chang, Y. F., and E. Adams. 1977. Factors influencing growth on l-lysine by Pseudomonas. J. Biol. Chem. 252:7987-7991. [PubMed] [Google Scholar]
- 10.Domínguez-Cuevas, P., and S. Marqués. 2004. Compiling sigma-70 dependent promoters, p. 319-344. In J. L. Ramos (ed.), Pseudomonas, vol. 2. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 11.Drnevich, D., and T. C. Vary. 1993. Analysis of physiological amino acids using dabsyl derivatization and reverse-phase liquid chromatrography. J. Chromatogr. 613:137-144. [DOI] [PubMed] [Google Scholar]
- 12.Duque, E., A. J. Molina-Henares, J. de la Torre, M. A. Molina-Henares, T. del Castillo, J. Lam, and J. L. Ramos. Towards a genome-wide mutant library of Pseudomonas putida strain KT2440. In J. L. Ramos and A. Filloux (ed.), Pseudomonas, vol. 5, in press. Springer, New York, NY.
- 13.Espinosa-Urgel, M., and J.-L. Ramos. 2001. Expression of a Pseudomonas putida aminotransferase involved in lysine catabolism is induced in the rhizosphere. Appl. Environ. Microbiol. 67:5219-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forthegill, J. L., and J. R. Guest. 1977. Catabolism of l-lysine by Pseudomonas aeruginosa. J. Gen. Microbiol. 99:139-155. [DOI] [PubMed] [Google Scholar]
- 15.Franklin, F. C. H., M. Bagdasarian, M. M. Bagdasarian, and K. N. Timmis. 1981. Molecular and functional analysis of the TOL plasmid pWW0 from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta-cleavage pathway. Proc. Natl. Acad. Sci. USA 78:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ichihara, A., M. Ogata, and M. Suda. 1960. Metabolism of l-lysine by bacterial enzymes: IV δ-aminovaleric acid-glutamic acid transaminase. J. Biochem. (Tokyo) 48:412-420. [Google Scholar]
- 18.Llamas, M. A., J. J. Rodríguez-Herva, R. E. W. Hancock, W. Bitter, J. Tommasen, and J. L. Ramos. 2003. Role of the Pseudomonas putida tol-oprL gene products in uptake of solutes through the cytoplasmic membrane. J. Bacteriol. 185:4707-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marqués, S., J. L. Ramos, and K. N. Timmis. 1993. Analysis of the mRNA structure of the Pseudomonas putida TOL meta fission pathway operon around the transcription initiation point, the xylTE and the xylFJ region. Biochim. Biophys. Acta 1216:227-237. [DOI] [PubMed] [Google Scholar]
- 20.Miller, D. L., and V. W. Rodwell. 1971. Metabolism of basic amino acids in Pseudomonas putida. Intermediates in 1-arginine catabolism. J. Biol. Chem. 246:5053-5058. [PubMed] [Google Scholar]
- 21.Molina, L., C. Ramos, E. Duque, M. C. Ronchel, J. M. García, L. Wyke, and J. L. Ramos. 2000. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol. Biochem. 32:315-321. [Google Scholar]
- 22.Molina-Henares, A. J., T. Krell, M. E. Guazzaroni, A. Segura, and J. L. Ramos. 2006. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol. Rev. 30:157-186. [DOI] [PubMed] [Google Scholar]
- 23.Mozafar, A. 1992. Effect of Pseudomonas fluorescens on the root exudates of tomato mutants differently sensitive to Fe chlorosis. Plant Soil 144:167-176. [Google Scholar]
- 24.Muramatsu, H., H. Mihara, R. Kakutani, M. Yasuda, M. Ueda, T. Kurihara, and N. Esaci. 2005. The putative malate/lactate dehydrogenase from Pseudomonas putida is an NADPH-dependent Δ1-piperideine-2-carboxylate/Δ1-pyrroline-2-carboxylate reductase involved in the catabolism of d-lysine and d-proline. J. Biol. Chem. 280:5329-5335. [DOI] [PubMed] [Google Scholar]
- 25.Nakazawa, T. 2002. Travels of a Pseudomonas, from Japan around the world. Environ. Microbiol. 4:782-786. [DOI] [PubMed] [Google Scholar]
- 26.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, and H. Hilbert et al. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 27.Phillips, A. T. 1986. Biosynthetic and catabolic features of amino acid metabolism in Pseudomonas, p. 385-438. In J. R. Sokatch (ed.), The bacteria: a treatise on structure and function, vol. X. The biology of Pseudomonas. Academic Press, Orlando, FL. [Google Scholar]
- 28.Ramos, J. L. M. Martínez-Bueno, A. J. Molina-Henares, W. Terán, K. Watanabe, X. Zhang, M. T. Gallegos, R. Brennan, and R. Tobes. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Revelles, O., M. Espinosa-Urgel, S. Molin, and J. L. Ramos. 2004. The davDT operon of Pseudomonas putida, involved in lysine catabolism, is induced in response to the pathway intermediate δ-aminovaleric acid. J. Bacteriol. 186:3439-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Revelles, O., M. Espinosa-Urgel, T. Führer, U. Sauer, and J. L. Ramos. 2005. Multiple and interconnected pathways for l-lysine catabolism in Pseudomonas putida KT2440. J. Bacteriol. 187:7500-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez-Herva, J. J., and J. L. Ramos. 1996. Characterization of an OprL null mutant of Pseudomonas putida. J. Bacteriol. 178:5836-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.