Abstract
Biofilm formation in Staphylococcus aureus is subject to phase variation, and biofilm-negative derivatives emerge sporadically from a biofilm-positive bacterial population. To date, the only known mechanism for generating biofilm phenotypic variation in staphylococci is the reversible insertion/excision of IS256 in biofilm-essential genes. In this study, we present evidence suggesting that the absence of the σB transcription factor dramatically increases the rate of switching to the biofilm-negative phenotype in the clinical isolate S. aureus 15981, under both steady-state and flow conditions. The phenotypic switching correlates with a dramatic increase in the number of IS256 copies in the chromosomes of biofilm-negative variants, as well as with an augmented IS256 insertion frequency into the icaC and the sarA genes. IS256-mediated biofilm switching is reversible, and biofilm-positive variants could emerge from biofilm-negative σB mutants. Analysis of the chromosomal insertion frequency using a recombinant IS256 element tagged with an erythromycin marker showed an almost three-times-higher transposition frequency in a ΔσB strain. However, regulation of IS256 activity by σB appears to be indirect, since transposase transcription is not affected in the absence of σB and IS256 activity is inhibited to wild-type levels in a ΔσB strain under NaCl stress. Overall, our results identify a new role for σB as a negative regulator of insertion sequence transposition and support the idea that deregulation of IS256 activity abrogates biofilm formation capacity in S. aureus.
Staphylococcus aureus is a leading cause of human diseases ranging from minor skin infections and food poisoning to life-threatening endocarditis, osteomyelitis, and postsurgical infections. In recent years, S. aureus has become one of the most important bacteria responsible for infections associated with the use of medical devices (47). The capacity of S. aureus to adhere to and form multilayered communities, termed “biofilms,” on the surfaces of implanted devices appears to be one of the major causes of persistent infection following surgery (11, 43). Inside the biofilm, bacteria are embedded in a self-produced matrix that provides a stable protective environment and also acts as a nidus for the dissemination of cells that initiate recurrent infections (20).
Although much effort has been made to identify the essential genetic elements and regulatory mechanisms of S. aureus biofilm formation, little is known about the mechanisms underlying the dispersion of the bacteria from the mature biofilm. In a pioneering study, Ziebuhr et al. (48) showed that insertion/excision of the insertion sequence IS256 in the icaADBC operon plays a direct role in the “on-to-off” switching of the biofilm formation capacity of Staphylococcus epidermidis. The icaADBC operon encodes proteins responsible for the synthesis of the PIA/PNAG (polysaccharide intercellular adhesin/polymeric N-acetyl glucosamine) exopolysaccharide, which is an important component of the staphylococcal biofilm matrix (12, 21, 35, 37). More recently, it was reported that IS256 also contributes to the production of biofilm-negative variants of S. epidermidis through insertion/excision events in the sarA and rsbU genes (9). SarA, which controls the expression of over 100 genes, is required for icaADBC operon expression, PIA/PNAG synthesis, and biofilm development in S. aureus and S. epidermidis (4, 44, 45). The RsbU protein is a positive activator of the transcription factor σB, and its disruption is associated with the loss of biofilm-forming capacity in S. epidermidis (29). One question that remains is how an S. epidermidis cell is able to alter its IS256 activity to allow adaptation of its biofilm-forming capacity to environmental conditions. Different host factors, including histone-like proteins (IHF, HU, HNS), Dam methylation, DNA supercoiling, proteins involved in DNA damage repair, proteases, and chaperones, have been described as important in regulating transposition (38). However, there is apparently no general rule governing transposition regulation, and each mobile element seems to have its own set of factors.
In a previous study in which we analyzed the biofilm formation capacity of the clinical strain S. aureus 15981, we observed that disruption of σB triggered the appearance of smooth colonies in the population (45). To corroborate this observation, in the present study we have investigated the biofilm phenotypic variation of the σB mutant under different growth conditions. Our results revealed that the absence of σB dramatically increased the rate of phenotypic switching to the biofilm-negative phenotype due to the increased transposition activity of IS256. Osmotic stress counteracted the absence of σB and restored the phenotypic switching of the σB mutant to levels similar to that of the wild-type strain. These results reveal strategies of the host cell for avoiding the adverse effects of the excessive transposition of mobile elements and provide novel insights into the regulation of the transition from the planktonic to the biofilm lifestyle in S. aureus.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The most relevant bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli XL1-Blue cells were grown in Luria-Bertani (LB) broth or on LB agar (Pronadisa, Spain) with appropriate antibiotics. Staphylococcal strains were cultured on Trypticase soy agar (TSA), in Trypticase soy broth (TSB) supplemented with 0.25% glucose (TSB-gluc), 0.5 M NaCl (TSB-NaCl), or 0.5 M KCl (TSB-KCl) where indicated. Media were supplemented with the following appropriate antibiotics at the indicated concentrations: erythromycin at 20 μg ml−1 or 1.5 μg ml−1, ampicillin at 100 μg ml−1, and chloramphenicol at 20 μg ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| 15981 | Biofilm-positive clinical strain | 45 |
| ΔσB strain | 15981 ΔσB | 45 |
| ΔσB+ strain | 15981 ΔσB with a restored σB gene | This study |
| icaC::Tn917 strain | 15981 icaC::Tn917 | 45 |
| E. coli XL1-Blue | Used for cloning assays | |
| Plasmids | ||
| pBT2 | E. coli-S. aureus shuttle vector with a thermosensitive origin of replication for gram-positive bacteria | 5 |
| pMAD | E. coli-S. aureus shuttle vector with a thermosensitive origin of replication for gram-positive bacteria and the bgaB gene from Bacillus stearothermophilus encoding a thermostable β-galactosidase | 1 |
| pLA1 | Plasmid that carries the IS256 element containing the erythromycin cassette (IS256r) | This study |
| pLA1-ΔTnp | Plasmid that carries the IS256r element with a site-specifically mutated transposase | This study |
Genetic manipulations.
General DNA manipulations were performed by standard procedures (3). For σB restoration, the σB gene was amplified using the sigB-A and sigB-D primers from S. aureus 15981 (Table 2). The 1,064-bp fragment was cloned in the pGEM-T Easy vector (Promega). The fragment was then cloned into the BamHI site of the shuttle plasmid pMAD (1), and the resulting plasmid was transformed into the ΔσB strain by electroporation, using a previously described protocol (13). Allelic exchange in the absence of a selection marker was performed as previously described (2). Briefly, the pMAD plasmid harboring a wild-type copy of the σB gene was integrated into the chromosome through homologous recombination at a nonpermissive temperature (43.5°C). From the 43.5°C plate, one to five colonies were picked, placed into 3 ml of TSB-gluc, and incubated for 24 h at 30°C. Tenfold serial dilutions of this culture were plated on TSA containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (1.5 μg/ml). White colonies, which no longer contained the pMAD plasmid, were tested to confirm the replacement by PCR using the sigB-A and sigB-D primers.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| icaA-1 | CTAACGAAAGGTAGGTA |
| icaA-2 | AAGTGCTGTTTCTCTTAC |
| icaC-1 | GGGACGGATTCCATGAAA |
| icaC-2 | CGAAGTCGTCAATTCAAAC |
| icaB-1 | CCGAACTCCAATGATTAT |
| IcaD-2 | CTTATACAAATGCGACGG |
| sigB-A | GCAGTGTTAATACTGCTTC |
| sigB-D | GTTAATGAAGGAACGGAGG |
| sar1 | GCTTTGAGTTGTTATCAAT |
| sar2 | CGTTGTTTGCTTCAGTGA |
| sarC | TTCACCAAATTGCGCTAAACCCTCCCTATTTGATGCATCTTGCT |
| sarD | ACCCGTTATCAATCGG |
| IS-1 | CCGACAAAGTCAACGAAA |
| IS-2 | GGCTGATGTTTGATTGGG |
| ISica-1 | CATTGTATTATCCTCAAATATATTTGATAAAGTCCGTATAATTG |
| ISeri-2 | GTCGACCTATACAATGTTTTTACCA |
| ISeri-3 | GGGTCGACCTATTGTGAGTTATTAGTG |
| ISica4 | TCATTGTTTGCATATACAAATATATAGTCAAGTCCAGACTCCTGTGTAAAATAGTATAACTATTAGG |
| Iscir-1 | CTCATAATAGCCATTTCGTTG |
| Iscir-2 | GCTTGCGCATCATTGGATG |
| Tnp-Fw | GGATTCGAAGACGCCTTTCAA |
| Tnp-Rv | GATTCAGTCGTTCAATTAGATTGGTACTC |
| Gyr-Fw | ACGTATGAAGGTGGTACGCATG |
| Gyr-Rv | ACGCGTTAATGCACGTTTGA |
| icaTaq-Fw | ATCGGTGGCGACTTTGATCT |
| icaTaq-Rv | CGGCATCAGTCATAATGACGATT |
| GyrTaq-Fw | TGTGATTTTTGGTACACCTTGCTT |
| GyrTaq-Rv | AGGTGACTTTTTGGTAATTCAAGGTT |
| Tnp-3 | CCATGGCTAAGTTAATATCTGTGAAC |
| Tnp-4 | CCATGGCGCATCATTGGATGATGG |
DNA hybridization for the detection of the icaADBC operon, IS256, and a recombinant IS256 element (IS256r) was performed according to the protocol supplied with a PCR-digoxigenin DNA-labeling and chemiluminescence detection kit (Roche). The oligonucleotide pairs IS-1/IS-2, ISeri-3/ISica-4, and IcaA1-IcaA2 were used to generate the specific IS256, erythromycin gene, and ica operon probes.
For the measurement of IS256 insertions in the ica operon, single rough colonies of the wild-type strain and the ΔσB mutant were cultured overnight in 5 ml of TSB-gluc. Then, dilutions were plated on Congo red agar (CRA). Biofilm-positive variants display a rough colony morphology when grown on this medium, whereas biofilm-negative variants exhibit a smooth colony morphology. One hundred ten smooth colonies of the wild-type strain and the ΔσB mutant were subjected to PCR for their ica genes using primers icaA-1/icaA-2, icaC-1/icaC-2, and icaB-1/icaD-2. The nucleotide sequencing was determined by the dideoxy chain termination method, using an ABI PRISM 310 genetic analyzer (Applied Biosystems).
Screening for biofilm-negative variants.
Screening for biofilm-negative variants was performed on CRA (16). Strains were incubated overnight in 5 ml of TSB-gluc. Then, cultures were diluted 1:1,000 in 5 ml of TSB-gluc to continue the serial passage. After 5 days, a dilution was plated on CRA and incubated for 24 h at 37°C. To analyze the effect of environmental conditions on biofilm phenotypic variation, the strains were serially diluted in 5 ml of TSB, TSB-gluc, TSB-NaCl, and TSB-KCl at both 30 and 37°C for 5 days, and then the morphology on CRA plates was determined. The phenotypic variation on solid media was analyzed as follows. A single colony from each strain was resuspended in 20 μl of TSB. Five microliters of the resuspension was spotted on a TSA plate and incubated overnight. The next day, all of the colony was resuspended in 20 μl of TSB, and again 5 μl of the resuspension was spotted on a TSA plate to continue the serial passage. After 5 days, dilutions were plated on CRA and incubated for 24 h at 37°C. The ratio of smooth to rough colonies was determined. The frequency at which biofilm-negative variants were produced from a biofilm-forming colony (number of CFU per cell) was determined and generation was calculated as previously described (30).
Selection of biofilm-positive revertants.
A single colony of a biofilm-negative variant was isolated and incubated at 37°C in TSB-gluc in 96-well polystyrene microtiter plates. After 24 h, the medium was replaced. This procedure was repeated until a biofilm of adhering bacteria became visible (around day 4). The adhering bacterial cells were scratched from the bottom and streaked on CRA. Rough colonies were isolated, and their capacity to produce a biofilm was determined by the quantitative adherence assay.
PNAG detection.
PNAG production in S. aureus 15981, in the ΔσB strain, and in the biofilm-negative variants was detected as described previously (12). Cells were grown overnight in TSB-gluc, the optical density was determined, and the same number of cells of each culture was resuspended in 50 μl of 0.5 M EDTA (pH 8.0). Cells were incubated for 5 min at 100°C and then centrifuged to pellet them. Forty microliters of the supernatant was incubated with 10 μl of proteinase K (20 mg/ml; Sigma) for 30 min at 37°C. After the addition of 10 μl of Tris-buffered saline (20 mM Tris-HCl, 150 mM NaCl [pH 7.4]) containing 0.01% bromophenol blue, 5 μl was spotted on a nitrocellulose filter using a Bio-Dot microfiltration apparatus (Bio-Rad), blocked overnight with 5% skim milk in phosphate-buffered saline with 0.1% Tween 20, and incubated for 2 hours with an anti-S. aureus PNAG antibody diluted 1:10,000 (36). Bound antibodies were detected with peroxidase-conjugated goat anti-rabbit immunoglobulin G antibodies (Jackson ImmunoResearch Laboratories, Inc., PA) diluted 1:10,000 and the Amersham ECL Western blotting system.
Biofilm formation assays.
A biofilm formation assay in microtiter wells was performed as described previously (21). To analyze the biofilm formation under flow conditions, we used 60-ml microfermentors with a continuous flow of 40 ml h−1 of TSB-gluc. Submerged Pyrex slides served as the growth substratum. Bacteria (108) from overnight precultures of wild-type 15981 and the ΔσB strain grown in TSB-gluc were used to inoculate microfermentors and subsequently cultivated for 5 days at 37°C. Biofilm development was recorded with a Nikon Coolpix 950 digital camera. To quantify the biofilm, bacteria that adhered to the Pyrex slide were resuspended in 10 ml of TSB. The optical density of the suspension was determined at 650 nm.
Detection of phenotypic variants in coagulases, lipases, proteases, and hemolysins.
Strain 15981 and the ΔσB strain were incubated overnight in 5 ml of TSB-gluc. Then, cultures were diluted 1:1,000 in 5 ml of TSB-gluc to continue the serial passages. After 5 days of subcultivation, a dilution was plated on Baird Parker agar (Difco), Spirit Blue agar (Difco), TSA supplemented with skim milk (5%, wt/vol), and TSA with 5% sheep blood (Difco) to determine the number of colonies with phenotypic variations in coagulase, lipase, protease, and hemolysin production, respectively. For the detection of IS256 insertions in the sarA gene, a dilution of an overnight culture of the wild type and the ΔσB mutants was plated on TSA-milk. After 24 h of incubation, the protease-overproducing variants were selected and PCR for the sarA gene performed using the sar1/sar2 and sarC/sarD primers.
Real-time quantitative PCR.
Total S. aureus RNA was isolated using the Fast RNA-Blue kit (Bio 101) according to the manufacturer's instructions. Two micrograms of each RNA was subjected in duplicate to DNase I (Gibco-BRL) treatment for 30 min at 37°C. The enzyme was then inactivated at 65°C in the presence of EDTA. To verify the absence of genomic DNA in every sample, the RNA duplicates were reverse transcribed in the presence and absence of Moloney murine leukemia virus reverse transcriptase (Gibco-BRL) (RT-PCR). All preparations were purified using CentriSep spin columns (Princeton Separations, Adelphia, NJ). To measure IS256 transposase (tnp) expression, 1/20 of each reaction mixture was used for real-time quantitative PCR (RT-PCR) using an ABI PRISM model 9700HT apparatus (Applied Biosystems). The tnp transcript was amplified using primers Tnp-Fw and Tnp-Rv (Table 2). The gyrB transcript was amplified as the endogenous control using primers Gyr-Fw and Gyr-Rv. SYBER green was used to label the amplified DNA. To monitor the specificity, final PCR products were analyzed by determining melting curves and by electrophoresis. Only samples with no amplification of the minus reverse transcriptase aliquot were considered in the study. The amount of tnp transcript was expressed as the difference (n-fold) from the amount of the control gene (2−ΔCT, where ΔCT represents the difference in threshold cycle between the target and control genes). For ica expression, TaqMan probes were used (Applied Biosystems). The icaC transcripts were amplified using the primers icaTaqFw and icaTaqRv and the icaC-specific TaqMan probe.
Transposition activity of the IS256r element.
To generate the recombinant element IS256r, the chromosomal icaC::IS256 insertion from the ΔσB biofilm variant L-24 was amplified by PCR using primers ISica-1 and ISeri-2 (Table 2). The erythromycin resistance gene was amplified using primers ISeri-3 and ISica-4 (Table 2). Both fragments were fused, generating a recombinant element (IS256r). IS256r contains an erythromycin resistance gene as a selected marker and maintains intact both inverted repeats and the transposase gene. IS256r was cloned into the thermosensitive E. coli-Staphylococcus shuttle vector pBT2 (5), generating plasmid pLA1. Plasmid pLA1 was introduced into the S. aureus 15981 wild type and ΔσB mutant by electroporation. To construct the plasmid pLA1-ΔTnp, the tnp gene of the IS256r element was mutated using primers tnp-3 and tnp-4, which generate a deletion of 64 bp (Table 2). Mutated IS256 was fused with the erythromycin resistance gene, generating the IS256rΔTnp recombinant element. IS256rΔTnp was cloned into pBT2, generating plasmid pLA1-ΔTnp.
Extrachromosomal template DNA from E. coli strains carrying pLA1 and pLA1-ΔTnp was prepared as previously described (34). IS256 circle junctions were detected using primers Iscir1 and Iscir2 (Table 2), which are directed in opposite orientations.
To analyze the transposition activity of IS256r in each strain, transformants were grown at 28°C for 5 to 7 h to allow recombinant element transposition and then spread at 43°C (temperature nonpermissive for plasmid replication). The frequency of transposition was determined as the number of erythromycin-resistant and chloramphenicol-sensitive colonies.
Statistical analysis.
Data corresponding to gene expression were compared using the Mann-Whitney test. Data corresponding to the frequency of transposition of the recombinant element IS256r were compared using the Pearson χ2 test. All the tests were two sided, and the significance level was 5%. The statistical analysis was performed with the SPSS program.
RESULTS
Effect of σB on biofilm phenotypic variation.
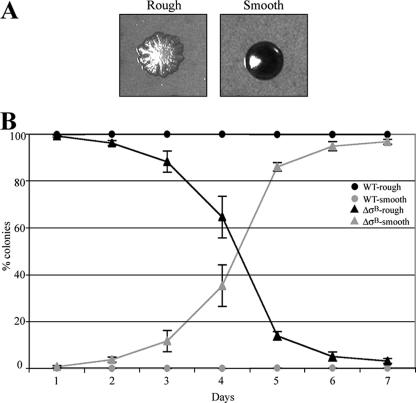
In a previous study, we demonstrated that the σB-null mutation in S. aureus did not impair the biofilm formation capacity of the 15981 clinical isolate (45). However, we realized that after several passages, the ΔσB mutant lost the capacity to produce a biofilm. To investigate whether σB regulates biofilm phenotype switching, a single rough colony each of the parent and ΔσB mutant strains was subcultured in TSB-gluc broth for 7 days as described in Materials and Methods. A dilution of each culture was plated daily on CRA plates, and the numbers of rough and smooth colonies were determined. S. aureus biofilm-negative derivatives are easily recognized on CRA plates because they produce smooth colonies, whereas biofilm-positive colonies are rough (Fig. 1A). In the ΔσB strain, the percentage of rough colonies changed dramatically from 97% at day 1 to 2 to 5% at day 7, while the percentage of rough colonies changed from 99.5% at day 1 to 95% at day 7 in the wild-type strain (Fig. 1B). To confirm that the biofilm phenotypic variation was a consequence of σB deletion, the σB gene was restored in the ΔσB mutant by allelic exchange. The restored strain (ΔσB+) exhibited switching frequencies identical to those of the wild-type 15981 strain after serial passage, confirming that the increased biofilm instability was indeed due to σB deficiency (data not shown).
FIG. 1.
Mutation of σB in a biofilm-positive strain causes a phenotype switch to a smooth morphology. (A) Colony morphology of biofilm-positive variants (rough morphology) and biofilm-negative variants (smooth morphology) of S. aureus 15981 after 24 h on CRA. (B) Percentages of rough colonies (black) and smooth colonies (gray) of the S. aureus 15981 strain and its isogenic ΔσB mutant on CRA after subcultivation in TSB-gluc for 7 days. Error bars represent the standard deviations from three independent experiments.
Analyses of the growth rate and the number of viable cells in smooth and rough variants indicated no difference in growth capacity that could explain the enrichment in smooth variants after 7 days (data not shown). However, to exclude the possibility that our experimental procedure in liquid media could artificially enrich smooth colonies in the ΔσB strain, we repeated the experiment on solid TSA plates as described in Materials and Methods. The results confirmed that the biofilm phenotype instability of the ΔσB strain also occurs after serial passage in solid medium (data not shown).
Biofilm phenotypic variation of the ΔσB mutant in microfermentors.
Biofilm formation in microfermentors occurs under flow conditions with a continuous replenishment of medium to avoid stationary phase. We used microfermentors to examine whether the rapid biofilm phenotypic switching of the ΔσB strain also occurs under flow conditions. The wild-type and ΔσB mutant strains were grown in microfermentors, and biofilm development was monitored over a period of 5 days. After 24 h, visual inspection of the biofilm revealed no difference in biofilm development between the parent and the ΔσB strain (Fig. 2A). However, after 5 days, the biofilm biomass produced by the ΔσB mutant was reduced twofold, and only a sparse layer of cells covered the surface of the slide (Fig. 2B). These data suggest that nutrient depletion and/or accumulation of waste products during steady-state incubation was not responsible for the induction of the rapid phenotypic switching observed in the ΔσB strain. These results also showed that the rapid enrichment of smooth bacteria in the ΔσB strain affects the three-dimensional structure of the biofilm.
FIG. 2.
Influence of σB deletion on biofilm stability after 5 days in continuous-flow microfermentors. (A) Biofilm development of the wild-type strain (WT) and the ΔσB mutant in microfermentors after 24 h (upper panel) and 5 days (lower panel). (B) Quantification of the biomass of a 5-day biofilm adhering to a glass slide. Cells were removed from the glass slide, placed into 10 ml of TSB, and vortexed, and the optical density (OD) of the solution was measured at 600 nm. Error bars represent the standard deviations from three independent experiments.
Contribution of IS256 to rapid biofilm switching in the ΔσB strain.
The insertion/excision activity of the IS256 transposable element in biofilm-essential genes has been described as a mechanism of biofilm phase variation (48). To determine whether the transposition activity of IS256 was involved in the rapid phenotypic switching observed in the ΔσB strain, we performed Southern blot analysis using a probe specific for IS256. Figure 3 shows that new DNA bands detected by the IS256 probe appeared as the ΔσB strain was serially passaged in the laboratory. After 7 days, the number of copies of IS256 increased dramatically, suggesting that the transposition activity of IS256 was derepressed in the absence of σB.
FIG. 3.
Analysis of the IS256 element in biofilm-negative variants of the ΔσB strain. Southern hybridization of EcoRI-SalI-digested chromosomal DNA with an IS256-specific DNA probe. Lanes 1 to 4, ΔσB biofilm-negative variants obtained from overnight cultures of experiment 1; lanes 5 to 8, ΔσB biofilm-negative variants obtained from overnight cultures of experiment 2; lanes 9 to 10, ΔσB biofilm-negative variants obtained after 7 days of subcultivation of the ΔσB mutant in TSB-gluc. WT, wild type.
This finding suggested that the insertion of new copies of IS256 into the icaADBC operon is a likely mechanism for the rapid biofilm-negative switching of the ΔσB strain. To explore this possibility, we characterized the icaADBC operons of several biofilm-negative variants by Southern blotting and by PCR. Southern blot experiments using a probe specific for the icaA gene revealed three different hybridization patterns (Fig. 4A and B). Group I included biofilm-negative variants (smooth colonies) with hybridization patterns identical to that of the parental biofilm-positive ΔσB strain. These variants were unable to produce the PIA/PNAG exopolysaccharide (Fig. 4C), although transcription of the icaADBC operon showed no difference from that of the wild-type strain, as determined by RT-PCR using an icaC-specific TaqMan probe (data not shown). Biofilm formation of these biofilm-negative variants was restored by complementation with a recombinant plasmid carrying the icaADBC operon (pSC18), suggesting that a point mutation in the ica operon might be responsible for biofilm deficiency. Group III included biofilm-negative variants exhibiting a DNA band with a lower electrophoretic mobility than those of the parental biofilm-positive ΔσB strain. Biofilm-negative variants of group II showed a hybridized band of higher electrophoretic mobility than was observed for bands in the parental strain. To investigate more precisely the reasons for the band shifts observed in biofilm mutants of groups II and III, the ica genes of several colonies were amplified by PCR and sequenced. The results revealed that the increased size of the ica restriction fragment of variants of group II was due to the insertion of the IS256 element at different positions of the icaC gene (Table 3). To determine the frequency at which IS256 insertions occurred in icaC, we performed PCR analysis of 110 biofilm-negative variants from the wild-type and ΔσB strains. The results revealed that 27 of the variants from the ΔσB strain contained an icaC::IS256 insertion. In contrast, only three of the biofilm-negative variants derived from the wild-type strain contained an IS256 insertion at the ica operon. The decreased size of the hybridized band of group III variants was due to a deletion of icaC and the DNA region downstream of the icaADBC operon because we were unable to amplify the icaC gene in these variants. As expected, dot blot experiments using specific anti-PNAG polyclonal antisera showed that all of the biofilm-negative smooth variants were unable to synthesize PIA/PNAG (Fig. 4C).
FIG. 4.
Analysis of the ica operon of biofilm-negative variants of the ΔσB strain. (A) Scheme of the ica operon EcoRI restriction sites. The icaA DNA probe was generated using primers icaA-1 and icaA-2. (B) Southern blot analysis of EcoRI-digested chromosomal DNA of several ΔσB biofilm-negative variants using the amplified icaA DNA probe. WT, wild type. (C) Dot blot analysis of PNAG accumulation in the 15981 wild-type strain, the ΔσB strain, and the ΔσB biofilm-negative variants that were classified into group I (no insertion of IS256 in the ica operon), group II (icaC::IS256), and group III (icaC deletion). Cell surface extracts of stationary-phase cultures were treated with proteinase K and spotted onto nitrocellulose filters. PNAG production was detected with anti-S. aureus PNAG antiserum. The transposon insertion mutation in the icaC gene was used as a negative control.
TABLE 3.
Localization of IS256 insertions in the icaC gene
| ΔσB biofilm-negative variant | Position | Sequencea |
|---|---|---|
| 8 | 381 | TTTATCGTTG*TTATCATGCA |
| 23 | 432 | ATTAACTATA*ACCTATTCAA |
| 29 | 417 | ATATCATTTT*TAAAATTAAC |
| 37 | 848 | TTCTCATTCT*TTATTTATTT |
| 53 | 933 | GTCTTTCTAG*CGATATCACT |
| 20 | 844 | TGCTTTCTCA*TTCTTTATTT |
| 22 | 405 | TTATTTTGAG*TTATATCATT |
| 24 | 906 | ACAAATATAT*TTGAGGATAA |
| 25 | 405 | TTATTTTGAG*TTATATCATT |
* represents the insertion point of IS256.
IS256-mediated biofilm phenotypic variation in the ΔσB strain is a reversible process.
To determine whether the ΔσB biofilm-negative variants could revert to biofilm-positive phenotypes, representative biofilm-negative derivatives were serially subcultured in TSB-gluc broth for 5 days as described in Materials and Methods. Rough black colonies were isolated on CRA after 4 days. Southern hybridization with an IS256-specific DNA probe of several biofilm-positive revertants revealed decreases in the numbers of copies of IS256 in the biofilm-positive revertants (Fig. 5A). In addition, rough colonies regained the capacity to produce a biofilm in microtiter plates (Fig. 5B) and to synthesize PIA/PNAG exopolysaccharide (Fig. 5C). Consistent with previous results by Ziebuhr et al. (48) for S. epidermidis, these data suggest that insertion of IS256 in the icaC gene of ΔσB biofilm-negative variants is a reversible process.
FIG. 5.

Analysis of the IS256 element, the biofilm formation capacity, and PIA/PNAG synthesis in ΔσB biofilm-positive revertants. (A) Southern hybridization of EcoRI-SalI-digested chromosomal DNA with an IS256-specific DNA probe. Lanes: 1, S. aureus 15981 ΔσB strain; 2, S. aureus 15981 ΔσB biofilm-negative variant; 3 to 5, S. aureus 15981 ΔσB biofilm-positive revertants. (B) Biofilm formation in microtiter dishes. (C) Dot blot analysis of PIA/PNAG accumulation.
IS256 insertions outside the ica operon.
In a previous study, Conlon et al. (9) reported that the sarA gene was another chromosomal hot spot for IS256 insertions, leading to the generation of biofilm-negative variants. Therefore, we decided to analyze the frequency of insertion of IS256 in the sarA gene in the ΔσB mutant. For this purpose, we took advantage of the fact that proteases are upregulated in the sarA mutant (7). A single colony each of the wild-type and ΔσB mutant strains was grown overnight in TSB-gluc. Then, an aliquot of each culture was plated daily on TSA-milk. The results of three independent experiments revealed that switching frequencies to protease-overproducing variants were 10 times higher for the ΔσB mutant than for the wild-type strain. PCR analysis of 34 of the ΔσB protease-overproducing variants revealed that 9 (26%) contained sarA::IS256 insertions but that none of the 9 protease-overproducing variants obtained from the wild-type strain contained sarA::IS256 insertions. Interestingly, all of the protease-overproducing variants containing sarA::IS256 insertions had a reduced capacity to produce biofilms, whereas some of the protease-overproducing variants obtained from the wild-type strain still retained the capacity to produce a biofilm (data not shown).
Although insertion of IS256 has been correlated only with biofilm phenotype switching, we wondered whether increased IS256 insertion activity might also play a role in the variation of other phenotypes. We analyzed the frequency of switching for enzymatic activities easily detectable on agar plates, such as coagulase, α-hemolysin, and lipase production. After 5 days of subcultivation, we were unable to detect any phenotypic variants in coagulase, hemolysin, or lipase production (Fig. 6). Taken together, these data suggest that insertion of IS256 into the sarA gene occurs at frequencies similar to that of the icaC gene, but insertion does not randomly occur in all genes of the staphylococcal genome.
FIG. 6.
Detection of phenotypic variants in coagulases, hemolysins, lipases, and proteases. After 5 days of subcultivation, a dilution of ΔσB biofilm-negative variant culture was plated on Baird Parker agar (Difco), TSA with 5% sheep blood (Difco), Spirit Blue agar (Difco), or TSA supplemented with skim milk (5%, wt/vol) to determine the number of phenotypic variants in coagulase, hemolysin, lipase, or protease production, respectively. The percentage of colonies that did not present phenotypic variation is represented in black. The percentage of colonies with phenotypic variation is shown in gray.
σB deficiency does not affect transposase expression.
To investigate the possibility that σB affects IS256 by regulating the expression of the transposase, we measured the transcription levels of the IS256 transposase in wild-type and ΔσB strains by RT-PCR. This analysis revealed no significant differences in transposase transcription levels between the wild-type and the ΔσB mutant strains (data not shown). These results suggest that σB influences the activity of IS256 by a mechanism other than repression of transcription of the transposase.
Recombinant IS256 exhibits high insertion activity in a ΔσB strain.
To determine whether the increased frequency of insertion of IS256 was due to a higher IS256 activity in the ΔσB mutant, we generated IS256r by insertion of an erythromycin resistance gene downstream of the transposase gene (Fig. 7A). IS256r was cloned into the thermosensitive pBT2 vector (5), generating pLA1. We also generated a derivative of IS256r in which the transposase gene was inactivated by site-directed mutagenesis of the conserved DDE motif. IS256r-ΔTnp was cloned into pBT2, generating pLA1-ΔTnp. To determine whether the transposase of IS256r was functional and capable of inducing IS256r excision and circularization, pLA1 and pLA1-ΔTnp were cloned in the recombination-deficient host E. coli XL1-Blue. For visualization of IS256r circles, extrachromosomal DNA from E. coli(pLA1) and E. coli(pLA1-ΔTnp) was isolated and used as the template in PCRs with IS256-specific, outward-directed primers (Fig. 7A). In this experiment, a specific 2.1-kb PCR product can be generated only if IS256 forms an extrachromosomal DNA circle. Figure 7B shows the appearance of a band of 2.1 kb only in the extrachromosomal DNA fraction from E. coli(pLA1), indicating that IS256r transposase was able to excise the insertion sequence and produce circular intermediaries.
FIG. 7.
Transposition analysis of the recombinant element IS256r. (A) Construction of the recombinant element IS256r. The natural chromosomal icaC::IS256 insertion was amplified and fused with an erythromycin resistance gene that included the left inverted repeat (IRL) of the IS256 element. IS256r was cloned into the pBT2 vector. IRR, right inverted repeat. (B) Agarose gel electrophoresis of PCR fragments amplified with circle-specific, outward-directed primers. (C) Percentages of chloramphenicol-sensitive colonies among the erythromycin-resistant colonies of S. aureus 15981 and its isogenic ΔσB mutant. Error bars represent the standard deviations from nine independent experiments. WT, wild type. (D) Southern hybridization of EcoRI-NcoI-digested chromosomal DNA with IS256 (left)- and erythromycin (right)-specific DNA probes. The arrows indicate new DNA bands hybridizing with the IS256 and erythromycin probes. Lanes 1 to 4, erythromycin-resistant/chloramphenicol-sensitive derivatives obtained from the ΔσB strain.
To determine whether the insertional activity of IS256r depends on the presence of σB, plasmid pLA1 was cloned in the S. aureus 15981 and ΔσB strains. The insertion frequency was quantified as the number of chromosomal insertion events of IS256r in each strain, as determined by the number of erythromycin-resistant/chloramphenicol-sensitive colonies (Fig. 7C). The colonies were expected to be chloramphenicol sensitive because we were screening for loss of the plasmid. The results showed a significant, 2.5-fold increase (P < 0.01) in the percentage of erythromycin-resistant/chloramphenicol-sensitive colonies obtained with the ΔσB strain compared to the percentage obtained with the wild-type strain. To confirm that the integration of IS256r in the chromosome was mediated by transposition activity and not by double crossover with chromosomal copies of IS256, Southern blotting was performed using IS256-specific and erythromycin-specific gene probes. The results revealed the appearance of new bands in the erythromycin-resistant/chloramphenicol-sensitive colony derivatives that hybridized with the IS256 and erythromycin gene probes, suggesting integration of IS256r into the chromosome by transposition (Fig. 7D). Overall, these results indicate that the transpositional activity of IS256r is increased in the absence of σB.
Effect of culture media and temperature on the IS256 activity of the ΔσB strain.
To test whether environmental conditions affect the insertional activity of IS256 in the ΔσB strain, we determined the number of biofilm phenotype variants of the 15981 and ΔσB strains on CRA plates and on TSA-milk after serial passage in different media (TSB, TSB-gluc, and TSB-NaCl [0.5 M]) and at different temperatures (30°C and 37°C). Interestingly, the presence of 0.5 M NaCl (or 0.5 M KCl [data not shown]) in the growth medium decreased the appearance of smooth and protease-overproducing variants in the ΔσB mutant to the levels in the wild type (Fig. 8). In contrast, a high frequency of biofilm phenotype variants was observed for the ΔσB mutant under the remaining conditions tested. From these results, we concluded that osmotic stress was able to compensate for the biofilm phenotype instability of the ΔσB mutant, suggesting that the activity of IS256 was also regulated by environmental conditions. It is important to notice that the frequency of biofilm phenotypic variation slightly increased in the wild-type strain at 30°C (20%), strongly suggesting that temperature plays a role in the biofilm phenotypic variation process.
FIG. 8.
Effect of culture media and temperature on IS256 activity in the ΔσB strain. (A) Percentages of rough colonies (black) and smooth colonies (gray) of wild-type and ΔσB strains on CRA at days 1 and 5 of continuous subcultivation under different conditions (at 30°C or 37°C) and in TSB, TSB-gluc (TSBg), or TSB-NaCl. (B) Percentages of protease-overproducing colonies (black) and non-protease-overproducing colonies (gray) of wild-type and ΔσB strains on TSA-milk at days 1 and 5 of subcultivation under different conditions as described for panel A. Error bars represent the standard deviations from three independent experiments.
Two-component systems (TCS) are the principal signal transducers involved in environmental sensing in S. aureus. Seventeen TCS have been annotated to occur in the S. aureus genome. Among them, yycFG is essential for cell viability, and SA0016-SA0017 is present only in methicillin-resistant strains. Thus, we investigated whether 15 isogenic mutations in TCS of the 15981 strain have effects on biofilm phenotypic variation. The results showed no changes in the frequency of appearance of smooth colonies under our experimental conditions (data not shown), indicating that the signals due to salt stress that induce IS256 transposition activity are perceived by unknown mechanisms that are separate from TCS. Such signals might act directly to regulate the stability of the transposase transcript or transposase activity itself.
DISCUSSION
Biofilm formation in S. aureus is a tightly regulated process involving many different factors. In this regard, it has been reported that (i) IcaR is a strong negative regulator of icaADBC transcription, and in its absence, PIA/PNAG production and biofilm formation are enhanced (24); (ii) TcaR, a putative transcriptional regulator of the teicoplanin-associated locus, represses icaADBC transcription without affecting PIA/PNAG production (24); and (iii) SarA, a global virulence regulator, positively regulates icaADBC operon transcription and PIA/PNAG accumulation (4, 45). In parallel to this refined regulatory procedure, it has been shown that biofilm development in S. aureus and S. epidermidis is subject to an “all-or-none” control mechanism based on the integration/excision of the IS256 insertion element in the icaADBC operon (9, 28, 48). IS256 is a common element in the genomes of staphylococci and enterococci and is often present in multiple copies either flanking the Tn4001 transposon or independent of it. Several studies have indicated that IS256 elements are significantly more frequent in S. epidermidis strains of clinical origin than in commensal isolates, and the use of IS256 as a genetic marker has been proposed (14, 19, 30, 42). Host cells have adopted different mechanisms for limiting the deleterious effects that uncontrolled transposition of insertion elements might cause (for a review, see Nagy and Chandler [38]). In the case of IS256, nothing was known about the mechanisms used by staphylococci to control IS256 insertion/excision activity.
The results of our study revealed that the absence of the global stress response regulator σB increased the frequency of smooth colonies, completely reversing the balance of smooth/rough bacteria in the culture over a 7-day period. The molecular mechanism responsible for the change to smooth, biofilm-negative colonies in the σB mutant strain might involve either the activation of a new genetic mechanism of phenotypic variation or the increased transposition activity of the IS256 element. Genetic analysis of biofilm-negative variants of the σB strain showed that insertion of IS256 at different positions in the icaC gene occurs in 30% of the biofilm-negative variants. This percentage coincided with the frequencies already described for wild-type strains of S. epidermidis, strongly suggesting that similar mechanisms of biofilm phenotypic variation take place in the wild-type and σB strains. To demonstrate that the transposition of IS256 increases in the absence of σB, the insertion frequency of IS256r, containing an erythromycin resistance gene, was determined. The results showed that the chromosomal insertion frequency of IS256r from a plasmid was almost three times higher in the σB mutant than in the parental strain. According to RT-PCR experiments, the absence of σB was not associated with an increased transcription of the transposase. Therefore, it appears that σB regulates the expression of an unknown element that, either alone or in concert with additional factors, represses the translation of IS256 transposase mRNA or the transposase's enzymatic activity. Further studies are required to determine the molecular mechanism by which σB regulates IS256 transposase activity.
The mechanisms of target recognition by the IS256 transposase have not been characterized, but preferential insertion of IS256 in the icaC, rsbU, and sarA genes has previously been reported to occur in Staphylococcus epidermidis (9, 48). In agreement with these results, we observed that the increased activity of IS256 in the absence of σB did not influence target specificity, and the same preferential insertion of IS256 in the icaC and sarA genes was observed among the ΔσB-derived, biofilm-negative variants. rsbU, a positive activator of σB, was previously reported to accumulate 11% of the IS256 insertions in biofilm-negative colonies (9). It has been shown that the absence of rsbU causes a reduction in σB activity; therefore, insertional inactivation of rsbU is expected to mimic σB mutation (40). The evolutionary selection of rsbU as a preferential IS256 insertion site might represent a positive-feedback mechanism for increasing transposition activity under specific environmental conditions. This hypothesis is also supported by the finding that σB-deficient S. aureus strains occur frequently in nature (27). Indeed, we have observed that natural IS256-containing σB-negative isolates of S. aureus very often contained a very high copy number of the IS256 element, suggesting that IS256 activity is increased in naturally σB-deficient S. aureus strains (our unpublished results). We have not been able to determine the frequency of insertion of IS256 into the rsbU gene because rsbU-deficient variants of S. aureus, in contrast to S. epidermidis, are effective biofilm producers that display rough colony morphology in CRA. An intriguing result relating to IS256 target recognition is the enormous number of copies of IS256 present in biofilm-negative variants that, in principle, should seriously compromise bacterial survival.
The high frequency of emergence of biofilm-negative variants in the σB mutant might also explain previous reports conferring opposite roles to σB in S. aureus biofilm formation (41, 45). It was first reported that the disruption of σB in the clinical strain MA12 impaired the biofilm formation capacity of the bacteria (41). Then, we reported that the deletion of σB from two genetically unrelated S. aureus strains did not affect biofilm formation capacity (45). It is likely that a biofilm-negative variant could be unconsciously selected during the mutagenesis procedure of σB in strain MA12 that would distort the observed phenotype. Similar conjectures might be applied to other studies that analyzed the effects of σB deficiency in bacterial biology. For example, one study investigated the effect of nutrient limitation on S. epidermidis biofilms (23). Biofilms of strain S. epidermidis 8400 with deletion of both rsbU and σB disintegrated within 2 to 3 days in TSB without glucose, whereas biofilms of mutants in negative regulators of σB (RsbW) displayed high stability over at least 6 days. Based on these results, those authors concluded that the stability of established biofilms under conditions of nutrient limitation depends on σB activity (23). However, it is possible that the increased activity of IS256 in the σB-deficient S. epidermidis 8400 strain might favor the appearance of biofilm-negative variants that caused the biofilm disintegration phenotype. To date, most of the studies investigating the role of σB in S. aureus have been performed using IS256-free S. aureus 8325-4 and COL strains; therefore, phenotypes would not be affected by this mobile element (8, 17, 18, 32). Nevertheless, it is important to keep in mind the potential influence of mobile elements when regulatory gene deficiencies are analyzed in natural bacterial isolates because transposon-mediated phase variation might underlie contradictory phenotypes associated with particular bacterial strains.
Growth conditions modulate the transposition activities of mobile elements by affecting the expression of transposition-related genes and/or their activities and/or of host factors that regulate transposition activity (38). In particular, stress situations such as carbon starvation, extreme temperature, and UV light have been correlated with the enhanced transposition of bacterial mobile elements (10, 15, 22, 31, 33, 39). These findings support the idea that bacteria derepress the movement of mobile elements as an adaptive response to stress and to permit new traits to evolve (6, 46). In contrast to this idea, we observed that σB, a global regulator that enables adaptation to environmental stresses, is a repressor of IS256 activity. In this situation, environmental stress conditions would activate σB activity and decrease the generation of biofilm-negative variants. Accordingly, salt stress (NaCl or KCl), which has been shown to induce biofilm formation (41), represses IS256 activity in the absence of σB, indicating that the sensor for osmotic stress influences IS256 activity downstream of σB.
Direct consequences of the change in biofilm switching frequency and the enrichment of smooth colonies in the bacterial population are the progressive degradation of an established biofilm and the release of biofilm-negative variants that can colonize new niches. An enzymatic activity (dispersin B) able to degrade the PIA/PNAG exopolysaccharide matrix to facilitate bacterial detachment from biofilms has been described for Actinobacillus actinomycetemcomitans (25, 26). In the absence of a dispersin homologue in the S. aureus genome, it appears that the generation of an appropriate ratio of biofilm-negative variants through the insertion/excision activity of IS256 plays an important role in controlling the transition from a biofilm to a planktonic lifestyle.
Acknowledgments
Marta Vergara-Irigaray is a predoctoral fellow (FPU) from the Ministerio de Educación y Ciencia, Spain. Nekane Merino is a predoctoral fellow from the Basque Government, Spain. This work was supported by the BIO2005-08399 grant from the Spanish Ministerio de Educación y Ciencia and grant LSHM-CT-2006-019064 from the European Union.
We thank Susanne Hennig, Wilma Ziebuhr, Alejandro Toledo-Arana, Josep Casadesus, and Cristina Solano for critical reading of the manuscript.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Arnaud, M., A. Chastanet, and M. Débarbouillé. 2004. A new vector for efficient allelic replacement in naturally nontransformable low GC% gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrizubieta, M. J., A. Toledo-Arana, B. Amorena, J. R. Penades, and I. Lasa. 2004. Calcium inhibits Bap-dependent multicellular behavior in Staphylococcus aureus. J. Bacteriol. 186:7490-7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1990. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 4.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruckner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 6.Capy, P., G. Gasperi, C. Biemont, and C. Bazin. 2000. Stress and transposable elements: co-evolution or useful parasites? Heredity 85(Part 2):101-106. [DOI] [PubMed] [Google Scholar]
- 7.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2004. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J. Bacteriol. 186:6208-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coros, A. M., E. Twiss, N. P. Tavakoli, and K. M. Derbyshire. 2005. Genetic evidence that GTP is required for transposition of IS903 and Tn552 in Escherichia coli. J. Bacteriol. 187:4598-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 12.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deplano, A., M. Vaneechoutte, G. Verschraegen, and M. J. Struelens. 1997. Typing of Staphylococcus aureus and Staphylococcus epidermidis strains by PCR analysis of inter-IS256 spacer length polymorphisms. J. Clin. Microbiol. 35:2580-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichenbaum, Z., and Z. Livneh. 1998. UV light induces IS10 transposition in Escherichia coli. Genetics 149:1173-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman, D. J., F. R. Falkiner, and C. T. Keane. 1989. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 42:872-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gertz, S., S. Engelmann, R. Schmid, A. K. Ziebandt, K. Tischer, C. Scharf, J. Hacker, and M. Hecker. 2000. Characterization of the σB regulon in Staphylococcus aureus. J. Bacteriol. 182:6983-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu, J., H. Li, M. Li, C. Vuong, M. Otto, Y. Wen, and Q. Gao. 2005. Bacterial insertion sequence IS256 as a potential molecular marker to discriminate invasive strains from commensal strains of Staphylococcus epidermidis. J. Hosp. Infect. 61:342-348. [DOI] [PubMed] [Google Scholar]
- 20.Hall-Stoodley, L., and P. Stoodley. 2005. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13:7-10. [DOI] [PubMed] [Google Scholar]
- 21.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 22.Ilves, H., R. Horak, and M. Kivisaar. 2001. Involvement of σS in starvation-induced transposition of Pseudomonas putida transposon Tn4652. J. Bacteriol. 183:5445-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jager, S., D. Mack, H. Rohde, M. A. Horstkotte, and J. K. Knobloch. 2005. Disintegration of Staphylococcus epidermidis biofilms under glucose-limiting conditions depends on the activity of the alternative sigma factor σB. Appl. Environ. Microbiol. 71:5577-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jefferson, K. K., D. B. Pier, D. A. Goldmann, and G. B. Pier. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan, J. B., C. Ragunath, K. Velliyagounder, D. H. Fine, and N. Ramasubbu. 2004. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 48:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan, J. B., K. Velliyagounder, C. Ragunath, H. Rohde, D. Mack, J. K. Knobloch, and N. Ramasubbu. 2004. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 186:8213-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsson-Kanth, A., K. Tegmark-Wisell, S. Arvidson, and J. Oscarsson. 2006. Natural human isolates of Staphylococcus aureus selected for high production of proteases and alpha-hemolysin are sigma(B) deficient. Int. J. Med. Microbiol. 296:229-236. [DOI] [PubMed] [Google Scholar]
- 28.Kiem, S., W. S. Oh, K. R. Peck, N. Y. Lee, J. Y. Lee, J. H. Song, E. S. Hwang, E. C. Kim, C. Y. Cha, and K. W. Choe. 2004. Phase variation of biofilm formation in Staphylococcus aureus by IS 256 insertion and its impact on the capacity adhering to polyurethane surface. J. Korean Med. Sci. 19:779-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozitskaya, S., S. H. Cho, K. Dietrich, R. Marre, K. Naber, and W. Ziebuhr. 2004. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 72:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kretschmer, P. J., and S. N. Cohen. 1979. Effect of temperature on translocation frequency of the Tn3 element. J. Bacteriol. 139:515-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamrani, S., C. Ranquet, M. J. Gama, H. Nakai, J. A. Shapiro, A. Toussaint, and G. Maenhaut-Michel. 1999. Starvation-induced Mucts62-mediated coding sequence fusion: a role for ClpXP, Lon, RpoS and Crp. Mol. Microbiol. 32:327-343. [DOI] [PubMed] [Google Scholar]
- 34.Loessner, I., K. Dietrich, D. Dittrich, J. Hacker, and W. Ziebuhr. 2002. Transposase-dependent formation of circular IS256 derivatives in Staphylococcus epidermidis and Staphylococcus aureus. J. Bacteriol. 184:4709-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maira-Litran, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark III, D. A. Goldmann, and G. B. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenney, D., K. Pouliot, V. Wang, V. Murthy, M. Ulrich, G. Döring, J. C. Lee, D. A. Goldmann, and G. B. Pier. 1999. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science 284:1523-1527. [DOI] [PubMed] [Google Scholar]
- 38.Nagy, Z., and M. Chandler. 2004. Regulation of transposition in bacteria. Res. Microbiol. 155:387-398. [DOI] [PubMed] [Google Scholar]
- 39.Ohtsubo, Y., H. Genka, H. Komatsu, Y. Nagata, and M. Tsuda. 2005. High-temperature-induced transposition of insertion elements in Burkholderia multivorans ATCC 17616. Appl. Environ. Microbiol. 71:1822-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palma, M., and A. L. Cheung. 2001. σB activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 69:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohde, H., M. Kalitzky, N. Kroger, S. Scherpe, M. A. Horstkotte, J. K. Knobloch, A. R. Zander, and D. Mack. 2004. Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. J. Clin. Microbiol. 42:5614-5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 44.Tormo, M. A., M. Marti, J. Valle, A. C. Manna, A. L. Cheung, I. Lasa, and J. R. Penades. 2005. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J. Bacteriol. 187:2348-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 46.Wessler, S. R. 1996. Turned on by stress. Plant retrotransposons. Curr. Biol. 6:959-961. [DOI] [PubMed] [Google Scholar]
- 47.Ziebuhr, W. 2001. Staphylococcus aureus and Staphylococcus epidermidis: emerging pathogens in nosocomial infections. Contrib. Microbiol. 8:102-107. [DOI] [PubMed] [Google Scholar]
- 48.Ziebuhr, W., V. Krimmer, S. Rachid, I. Lossner, F. Gotz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]