Abstract
The Escherichia coli cold shock protein CsdA is a member of the DEAD box family of ATP-dependent RNA helicases, which share a core of nine conserved motifs. The DEAD (Asp-Glu-Ala-Asp) motif for which this family is named has been demonstrated to be essential for ATP hydrolysis. We show here that CsdA exhibits in vitro ATPase and helicase activities in the presence of short RNA duplexes with either 3′ or 5′ extensions at 15°C. In contrast to wild-type CsdA, a DQAD variant of CsdA (Glu-157→Gln) had no detectible helicase or ATPase activity at 15°C in vitro. A plasmid encoding the DQAD variant was also unable to suppress the impaired growth of the csdA null mutant at 15°C. Plasmid-encoded CsdAΔ444, which lacks most of the carboxy-terminal extension, enhanced the growth of a csdA null mutant at 25°C but not at 15°C; this truncated protein also has limited in vitro activity at 15°C. These results support the physiological function of CsdA as a DEAD box ATP-dependent RNA helicase at low temperature.
RNA helicases are involved in various cellular processes that require modulation of RNA structure, such as RNA splicing, ribosome biogenesis, translational initiation, mRNA degradation, and cell division (9, 23, 30). Driven by nucleoside triphosphate hydrolysis, these enzymes catalyze unwinding of RNA duplexes and disruption of RNA-protein interactions (9, 23, 30). Based upon the conservation of several motifs, RNA helicases are grouped into related families. Members of superfamily 2 (SF2) share eight conserved motifs and include the DExD/H helicase family, comprising the DEAD, DEAH, DExH, and DExD families (5, 34). The DEAD box family of ATP-dependent helicases, consisting of at least 500 eukaryotic and prokaryotic proteins, is the largest family (reviewed in reference 8). The prototype is eukaryotic initiation factor 4A (eIF4A), which exhibits helicase activity (21, 28).
Proteins in the DEAD box family contain a core of nine conserved sequence motifs, including the Q motif, which is unique to this subset of SF2 helicases (33), and the Asp-Glu-Ala-Asp (DEAD) motif that gives the family its name (21). The DEAD motif has been demonstrated to be essential for ATPase and/or RNA unwinding activity of several helicases, including the mammalian and yeast initiation factor 4A, the yeast protein Ded1p, and the Escherichia coli enzyme RhlB (3, 14, 26, 36). The recent crystal structure of the RNA-bound Drosophila melanogaster Vasa DEAD box helicase demonstrated that the DEAD sequence participates with residues of other conserved motifs to bind ATP (32). An intricate network of interactions between canonical helicase motifs serves to couple ATP binding and hydrolysis with RNA binding and unwinding activities in a manner consistent with roles for these motifs previously established by biochemical and genetic studies (8).
In addition to the core of conserved motifs, DEAD box proteins contain variable amino- and carboxy-terminal sequences. It has been suggested that these flanking sequences aid in the binding of substrates and cofactors or regulate the various activities of the enzyme (26, 41). However, a general role for these domains is tentative. Although the C-terminal domain of E. coli DEAD box helicase DpbA has been demonstrated to confer binding specificity to helix 92 (H92) of 23S rRNA (19), the C-terminal domains of several yeast DEAD box proteins have been reported to be dispensable for in vivo activity (34).
CsdA (cold-shock DEAD box protein A) is one of five DEAD box proteins in E. coli (17). The CsdA gene (deaD) was originally discovered as a multicopy suppressor of a temperature-sensitive mutant of the rpsB gene, which encodes the ribosomal protein S2 (35). Multicopy CsdA expression in the rpsB mutant resulted in reincorporation of ribosomal proteins S1 and S2 into the ribosome (24). CsdA is a cold-induced protein, as its expression is upregulated in response to a downward shift in growth temperature (16). Consistent with a specialized role for CsdA at low temperatures, deletion of the csdA gene has a negligible effect on growth at 37°C but impairs growth at low temperatures (7, 16). CsdA has been implicated in various cellular processes at low temperature, including 50S ribosomal biogenesis (7), association with an RNase E degradosome (18, 27, 29), degradation of CSP mRNA (39), and translation initiation (16, 22).
CsdA has been biochemically analyzed in vitro for enzymatic activities. CsdA-catalyzed ATP hydrolysis was observed in the presence of various RNA substrates (1). In addition, a truncated form of CsdA that lacks 185 amino acids from the C terminus catalyzed ATP-dependent unwinding of a 14-mer RNA duplex with 5′ or 3′ extensions, suggesting that CsdA functions as a bidirectional ATP-dependent RNA helicase (1).
While ATPase and helicase activities of CsdA are observed in vitro at 25°C, neither activity has been demonstrated at physiologically relevant lower temperatures. Furthermore, requirement for the conserved DEAD box motif for in vitro or in vivo CsdA function has not been determined. Here we report that both CsdA and CsdAΔ, which lacks most of the C-terminal extension, possess ATPase and helicase activities in vitro at 15°C. We also demonstrate that the C-terminal extension is required for in vivo function at 15°C, which is consistent with the observed limited in vitro activity of the truncated protein. Site-directed mutagenesis reveals that the DEAD motif is required for efficient in vitro ATPase and helicase activities and for in vivo function at 15°C, confirming the cold shock protein CsdA as a DEAD box ATP-dependent RNA helicase. While the natural substrate(s) of CsdA remains to be determined, our results suggest some processivity of helicase activity is required for efficient cellular function at 15°C and that CsdA's C-terminal extension may contribute to substrate recognition and/or processivity.
MATERIALS AND METHODS
Protein expression and purification. (i) CsdA.
Plasmid pECRSA contains the csdA gene cloned into pET11a (gift of Masayori Inouye, University of Medicine and Dentistry of New Jersey). Cultures of BL21 Star(DE3) cells (Invitrogen, Carlsbad, CA) transformed with pECRSA were grown to an A600 of ≈0.6 prior to induction of protein expression with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 1 h at 30°C. Cells were harvested by centrifugation and lysed by French press in 20 mM Tris-HCl (pH 7.6), 0.2 M KCl. Cleared cell lysate was loaded on a Bio-Rad MT-5 column containing UNO-S cation exchange resin (Hercules, CA). The protein was eluted with 20 mM Tris-HCl (pH 7.6) with a gradient of 0.2 M to 1 M KCl. CsdA eluted at approximately 0.4 M KCl. Fractions containing CsdA were identified by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Fractions were pooled and dialyzed against 20 mM Tris-HCl (pH 6.5) and 0.2 M KCl and run again on the same column at pH 6.5. Fractions containing CsdA were pooled and dialyzed against a storage buffer containing 20 mM Tris-HCl (pH 7.6), 0.4 M KCl, and 10% glycerol. The high salt allowed concentration of CsdA fractions to approximately 1 mg/ml without precipitation. Protein aliquots were flash-frozen in liquid nitrogen and stored at −80°C until use.
(ii) Glu-157→Gln CsdA.
The glutamic acid in the conserved DEAD box was changed to glutamine by PCR site-directed mutagenesis of pECRSA using the QuikChange method (Stratagene, La Jolla, CA). The mutation was confirmed by dideoxy sequencing. This DQAD variant was expressed in an E. coli strain with a disruption of the chromosomal copy of csdA (16). Novagen's λDE3 lysogenization kit (Madison, WI) was used to make this cell line suitable for pET vector expression according to the manufacturer's protocol. DQAD CsdA was expressed at 30°C and purified using the same method used for wild-type CsdA.
(iii) CsdAΔ.
The Pro-444 codon was converted to a stop codon, removing 185 amino acids from the C terminus of CsdA (1). CsdAΔ was cloned into pROEX-HT (Invitrogen, Carlsbad, CA), resulting in an N-terminally His6-tagged protein. This clone was a gift from Thierry Bizebard (Institut de Biologie Physico-Chimique, Paris, France). Following overexpression in BL21 Star(DE3) cells (Invitrogen, Carlsbad, CA), CsdAΔ was purified using a Ni-nitrilotriacetic acid affinity resin according to the manufacturer's protocol (QIAGEN, Valencia, CA). CsdAΔ eluted in 100 mM Tris-HCl, 0.5 M NaCl, and 0.5 M imidazole. The protein precipitated during attempts to lower the salt and imidazole concentrations; thus, the His6 tag was removed by direct treatment of the pooled elution fractions with TEV protease (Invitrogen, Carlsbad, CA). Removal of the affinity tag was confirmed by SDS-polyacrylamide gel electrophoresis, and CsdAΔ was flash-frozen at −80°C following addition of glycerol to 10%.
RNA substrates.
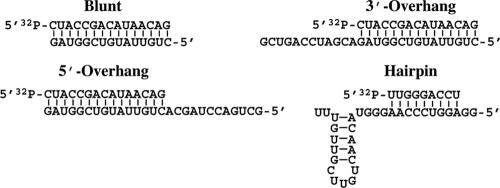
Substrates included a blunt-ended 15-bp duplex, the same duplex with an added 3′ overhang of 12 bases, and the duplex with an added 5′ overhang of 12 bases (Fig. 1). Sequences of the duplex substrates were arbitrary, designed to minimize secondary structure. The hairpin substrate was a modification of one provided by Olke C. Uhlenbeck (Northwestern University) that is the minimal 23S rRNA hairpin 92 substrate of the E. coli DEAD box helicase DbpA (10). We altered the stem portion of hairpin 92 to generate a non-rRNA substrate with similar structure and G-C content. The second, third, and fourth base pairs of the 32-mer were switched from G-C, U-A, and U-A in hairpin 92 to the C-G, A-U, and A-U pairs shown in Fig. 1. Such changes are expected to have minimal effect on hairpin stability. All substrates were synthesized using solid-phase phosphoramidite chemistry at the Wake Forest University DNA Synthesis Facility, deprotected as previously described (38), purified by denaturing gel electrophoresis, and recovered by electroelution using an Elutrap device (Whatman, Inc., Florham Park, NJ). The 15-mer strand of the blunt, 3′-overhang, and 5′-overhang substrates and the 9-mer strand of the hairpin substrate were 5′-end labeled with [γ-32P]ATP (Amersham) and T4 polynucleotide kinase (Promega, Madison, WI) as described in the manufacturer's protocol. Excess radiolabeled ATP was removed by phenol-chloroform (1:1) extraction followed by ethanol precipitation. The two strands of each substrate were annealed in equimolar amounts before each reaction by heating to 90°C followed by slow cooling to room temperature. As the mixtures cooled, MgCl2 was added to a final concentration of 1 mM at 65°C.
FIG. 1.
RNA substrates used in ATPase and helicase reactions. RNAs were synthesized by solid-phase phosphoramidite chemistry, deprotected, and gel purified (38). For each substrate, the indicated strand was radiolabeled at the 5′ end with [γ-32P]ATP using T4 polynucleotide kinase. The hairpin substrate is a variant of 23S rRNA hairpin 92 (10).
ATPase assay.
For the ATPase assay, reaction mixtures contained 20 mM Tris-HCl (pH 7.6), 5 mM MgCl2, 200 μM ATP, 0.1 mg/ml bovine serum albumin, 5 μM double-stranded RNA (dsRNA; duplex, 3′ overhang, 5′ overhang, or hairpin), and 5 μCi [α-32P]ATP (Amersham, Piscataway, NJ). All reaction mixtures were incubated at 15°C (unless otherwise noted) and initiated by the addition of 500 nM protein (CsdA, CsdAΔ, or DQAD). Aliquots (2 μl) were removed every 10 min from 0 to 60 min and quenched in 8 μl 40 mM EDTA. A 1-μl portion of the quenched reaction mixture was spotted on a polyethyleneimine-cellulose plate prewashed in water. The plate was developed using 0.75 M KH2PO4 (pH 3.4). Developed plates were imaged using Bio-Rad's molecular imager FX ProPlus, and radiation was quantified with the manufacturer's QuantityOne software (Bio-Rad, Hercules, CA). The percent of ATP hydrolyzed at each time point was determined, and the initial rates of hydrolysis were calculated based on the linear portion (first 30 min) of each assay.
Helicase assay.
For the helicase assay, reaction mixtures contained 20 mM Tris-HCl (pH 7.6), 5 mM MgCl2, 5 μM unlabeled 15-mer or 9-mer single-stranded RNA (as a trap for the displaced unlabeled strand), 0.1 mg/ml bovine serum albumin, and 500 nM protein (CsdA, CsdAΔ, or DQAD). All reaction mixtures were incubated at 15°C (unless otherwise noted) and initiated by the addition of 50 nM 32P-labeled dsRNA. Aliquots (10 μl) were removed every 15 min from 0 to 45 min (four total) from reaction mixtures which contained either 2 mM ATP or no ATP. Aliquots were quenched in 7 μl stop solution (40 mM EDTA, 2% SDS, bromophenol blue, and xylene cyanol) and kept on ice until the end of the assay. A 15-μl portion of each sample was then loaded onto a running 16% native polyacrylamide gel (19:1 acrylamide-bisacrylamide) in 1× TBE buffer (89 mM Tris-HCl pH 7.6, 89 mM boric acid, 2 mM EDTA). No significant reannealing of denatured RNA substrates was observed following incubation on ice (data not shown). Gels were dried, exposed, and imaged using Bio-Rad's molecular imager FX ProPlus.
In vivo complementation analysis.
Plasmid pKX164 is a pACYC184 derivative containing the wild-type csdA gene (40). Plasmid pPJ41 encodes the Glu157→Gln substitution, which was generated by QuikChange mutagenesis of pKX164. The Pro-444 codon was converted to a stop codon by QuikChange mutagenesis of pKX164, generating plasmid pPJ60. Mutations were confirmed by DNA sequencing. E. coli K-12 strain WJW45 (4) and the csdA deletion strain WJW45ΔcsdA (7) were used for the in vivo complementation analysis. WJW45ΔcsdA contains an in-frame csdA deletion that results in the fusion between the first 6 and last 13 codons, with the corresponding removal of 97% of the coding region (7). Strains WJW45 and WJW45ΔcsdA were grown in LB broth containing 30 μg/ml chloramphenicol in the presence of the indicated plasmid, and growth was monitored by absorbance at 420 nm. Steady-state cultures at 37°C and 25°C were obtained from dilution of overnight cultures at the corresponding temperature. To obtain steady-state cultures at 15°C, the cultures were initially grown at 37°C to an optical density at 420 nm of 0.5 to 0.7 followed by a shift in growth temperature to 15°C. Following overnight incubation at 15°C, the cultures were diluted and steady-state growth was monitored for the time indicated. Complementation was assayed by the ability of a plasmid to increase the growth of the csdA null mutant at the indicated temperatures.
RESULTS
ATPase and helicase activities of CsdA and CsdAΔ.
E. coli DEAD box protein CsdA was previously demonstrated to catalyze RNA-dependent ATP hydrolysis (1). CsdAΔ, which contains all nine conserved motifs of canonical DEAD box helicases but lacks most of the C-terminal extension, was shown to catalyze ATP-dependent unwinding of a 14-mer RNA duplex with single-stranded extensions at either the 3′ or 5′ end (1). These activities were characterized in vitro at 25°C. Because a decrease in temperature can lead to increased stability of nucleic acid duplexes, which may indirectly affect the activity of weakly processive helicases, we examined the effect of low temperature on enzymatic activities of E. coli CsdA in vitro.
We initially monitored baseline ATP hydrolysis for the various purified proteins to ensure that any increase in ATP hydrolysis was due to the addition of the dsRNA substrates. RNA-independent ADP formation by CsdA, CsdAΔ, and the DQAD variant was negligible (Table 1).
TABLE 1.
Stimulation of ATPase activity of CsdA variantsa
| RNA | Rate of ATP hydrolysis (mol ATP hydrolyzed/min/mol of protein)
|
||
|---|---|---|---|
| CsdA | CsdAΔ | DQAD | |
| None | 0.26 | 0.09 | 0.05 |
| Blunt | 0.64 | 0.10 | 0.31 |
| 3′ overhang | 2.18 | 1.81 | 0.34 |
| 5′ overhang | 3.30 | 1.34 | 0.19 |
| Hairpin | 4.90 | 3.20 | 0.08 |
Activity is reported as moles of ATP hydrolyzed per minute per mole of protein and is the average of three determinations. Assays were performed with 5 μM RNA, 0.2 mM ATP, and 500 nM protein.
Four different double-stranded RNA substrates were used to stimulate ATPase activity in vitro at 15°C, including a blunt-ended duplex and duplexes with single-stranded 3′ or 5′ extensions (Fig. 1). The sequences of these substrates were arbitrarily chosen and designed to minimize possible secondary structure. A hairpin substrate, composed of a 9-bp duplex region with a hairpin-forming extension at the 3′ end, is a sequence variant of 23S rRNA hairpin 92, a minimal substrate of E. coli DEAD box helicase DbpA (Fig. 1) (10). While identical amounts of the RNA and protein were used for all assays, different levels of hydrolysis were observed for each RNA substrate. ATP hydrolysis was most efficient in the presence of the hairpin substrate (Fig. 2 and Table 1). The 5′-overhang duplex and 3′-overhang duplex also stimulated ATPase activity, although to a lesser extent, while the blunt substrate stimulated only minimal ATP hydrolysis (Fig. 2 and Table 1). The ability of CsdA to hydrolyze GTP was also tested using all RNA substrates. No GDP formation was observed in the presence of any substrate (data not shown).
FIG. 2.
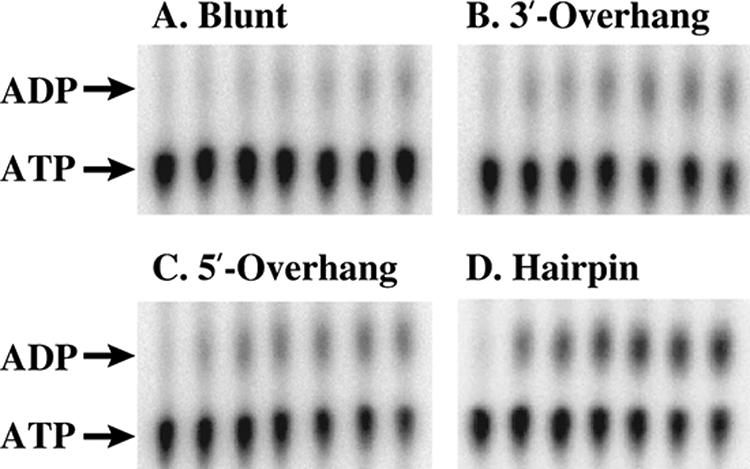
ATPase activity of CsdA. Representative thin-layer chromatography separations of ATPase reaction aliquots are shown for the blunt (A), 3′-overhang (B), 5′-overhang (C), and hairpin (D) substrate. Lanes 1 to 7 represent reaction progress at 10-minute intervals from 0 to 60 min.
As with full-length CsdA, CsdAΔ catalyzed ATP hydrolysis in the presence of the hairpin and overhanging duplex substrates and only minimal activity in the presence of the blunt-ended RNA duplex (Table 1). In general, RNA-stimulated ATPase activity of CsdAΔ was reduced compared to full-length CsdA for each substrate. For the blunt RNA duplex, ATPase activity of the truncated protein was reduced nearly to the level observed in the absence of RNA.
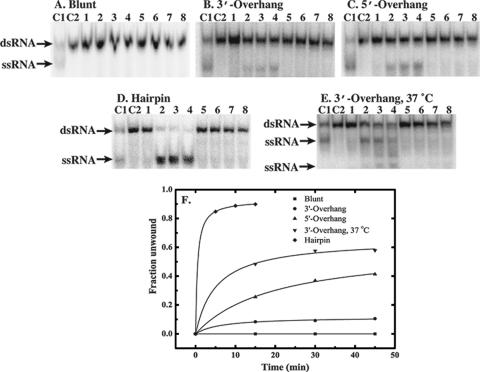
The same substrates were used to evaluate the helicase activity of CsdA, which correlated qualitatively with RNA-stimulated ATPase activity. CsdA was unable to unwind the blunt-ended duplex (Fig. 3A), which stimulated only minimal ATP hydrolysis (Fig. 2). CsdA was able to unwind the 3′-overhang duplex and the 5′-overhang duplex to similar extents (Fig. 3B and C). We also observed that CsdA exhibited a higher level of helicase activity for the 3′-overhang duplex at 37°C (Fig. 3D). Of the substrates tested, the highest level of helicase activity was observed using the hairpin substrate (Fig. 3E), which also stimulated the most ATP hydrolysis (Fig. 2). Approximately 85% of the hairpin was converted to its single-stranded form after only 5 min at 15°C (Fig. 3F). No helicase activity was observed for any substrate in the absence of ATP, indicating that unwinding of RNA is not simply a result of CsdA destabilizing the RNA or trapping the single strand upon thermal breathing of the nucleic acid (Fig. 3A to E, lanes 5 to 8 of each panel).
FIG. 3.
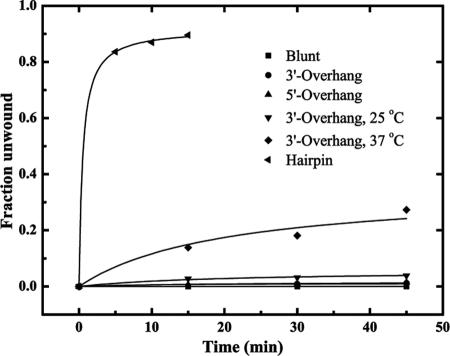
Helicase activity of CsdA. Annealed substrates were incubated with CsdA, quenched in gel loading buffer, and separated by nondenaturing acrylamide gel electrophoresis followed by phosphorimager analysis. Controls are indicated by C1 (substrate denatured by incubation at 95°C for 5 min) and C2 (annealed substrate). Lanes 1 to 4 are from reactions with 2 mM ATP, and lanes 5 to 8 lacked ATP. CsdA was incubated with the blunt (A), 3′-overhang (B), 5′-overhang (C), or hairpin (D) substrate at 15°C. (E) CsdA was incubated with 3′-overhang substrate at 37°C. Reaction aliquots were removed at 5-minute intervals (t = 0, 5, 10, or 15 min) for the hairpin substrate and at 15-minute intervals (t = 0, 15, 30, or 45 min) for the other substrates. (F) Quantitation of CsdA helicase activity. The fractional conversion of each annealed substrate to single-stranded product in the presence of 2 mM ATP was quantified. A fraction of 1.0 corresponds to 100% conversion to single-stranded product. The assay temperature was 15°C unless otherwise indicated.
Only the hairpin substrate was unwound to a detectable level by CsdAΔ (Fig. 4); this activity was ATP dependent (data not shown). Although other substrates did not trigger the truncated enzyme's helicase activity, this defect was not simply a decrease in overall activity of CsdAΔ, as unwinding of the hairpin substrate reached a level similar to that produced by full-length CsdA (∼85% conversion to single-stranded form). The activity observed with the hairpin substrate, which features a smaller, less stable double-stranded region than the other duplex substrates, suggests that CsdA is a weakly processive helicase in vitro. Furthermore, the C-terminal extension of CsdA may increase the processivity of the enzyme, allowing unwinding of substrates containing longer helical portions.
FIG. 4.
Helicase activity of CsdAΔ. Annealed substrates were incubated with CsdAΔ in the presence of 2 mM ATP and subjected to nondenaturing acrylamide gel electrophoresis as in Fig. 3. Gels were analyzed by phosphorimaging, and the fractional conversion to single-stranded product was quantified. A fraction of 1.0 corresponds to 100% conversion to single-stranded product. The assay temperature was 15°C unless otherwise indicated.
In vivo analysis of CsdAΔ.
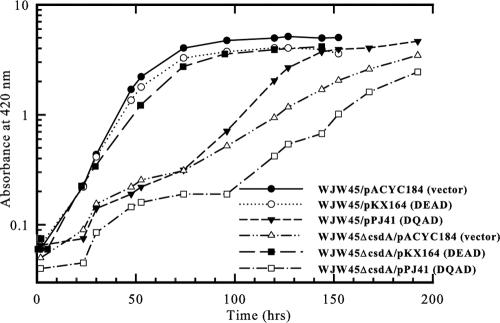
In the absence of a chromosomally encoded copy of csdA, E. coli displays impaired growth at low temperature, consistent with an adaptive role for CsdA in bacterial physiology at low temperature (7, 16). In the presence of the control vector pACYC184, the parent strain WJW45 grew with a doubling time of 1.3 h at 25°C (Fig. 5A) and 8.0 h at 15°C (Fig. 5B). In contrast, WJW45ΔcsdA/pACYC184 grew with a doubling time of 2.4 h at 25°C (Fig. 5A) and 30 h at 15°C (Fig. 5B). The csdA deletion resulted in a 1.8-fold increase in the doubling time at 25°C compared to a 3.75-fold increase in the doubling time at 15°C. Therefore, E. coli has a higher requirement of CsdA for optimal growth at 15°C than at 25°C. The presence of plasmid pKX164, encoding full-length CsdA, complemented the growth of the mutant, resulting in doubling times of 1.3 h at 25°C (Fig. 5A) and 10 h at 15°C (Fig. 5B).
FIG. 5.
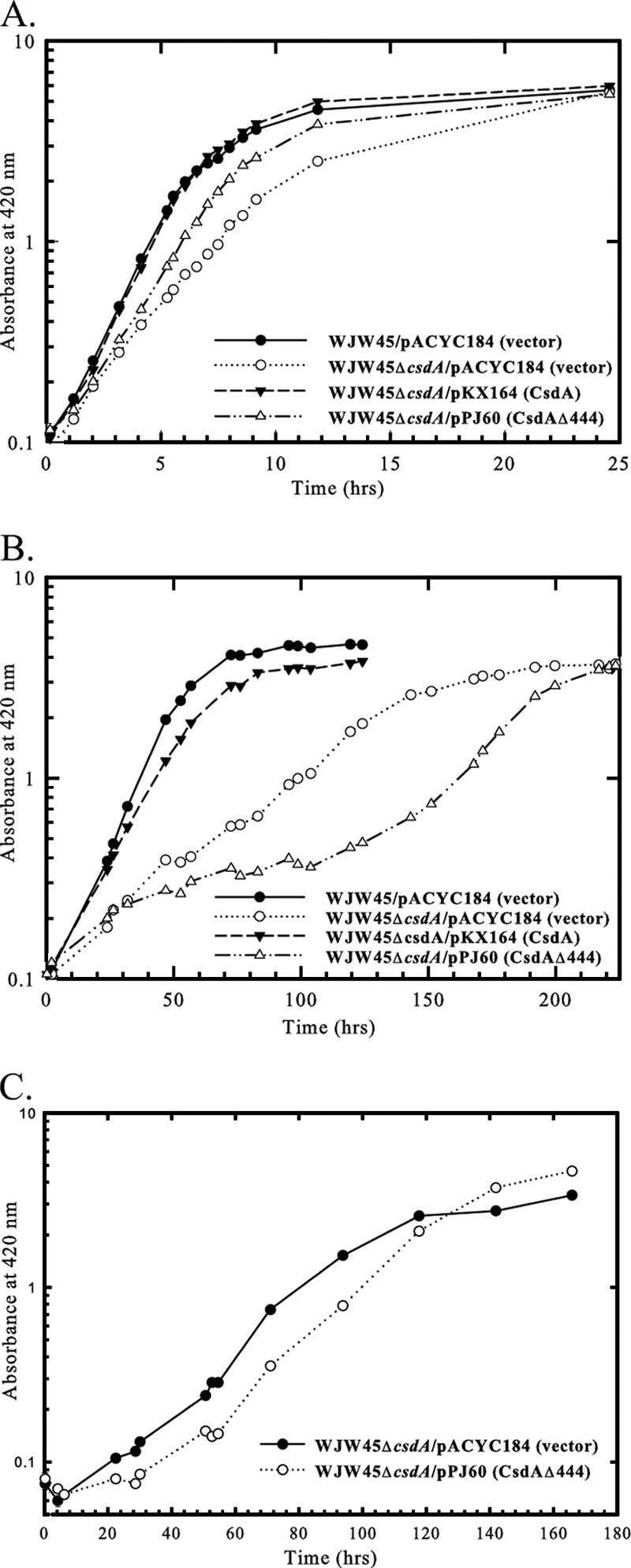
In vivo complementation analysis of the csdA null mutant with plasmid-encoded wild-type CsdA and CsdAΔ444. Strains WJW45 and WJW45ΔcsdA were grown in LB broth containing 30 μg/ml chloramphenicol at 25°C (A) and 15°C (B) in the presence of the vector pACYC184, plasmid encoding wild-type CsdA (pKX164), or plasmid encoding CsdAΔ444 (pPJ60). (C) Growth of subcultured strain WJW45ΔcsdA at 15°C in the presence of the vector pACYC184 and plasmid encoding CsdAΔ444 (pPJ60).
The enzymatic activity observed for CsdAΔ indicates that at 15°C, the C-terminal variable region is dispensable for in vitro unwinding of the hairpin substrate, containing a 9-mer duplex region, but not the 15-mer duplex (either with or without a single-stranded extension). However, at 25°C, CsdAΔ has been demonstrated to catalyze in vitro unwinding of a 14-mer substrate with 5′ or 3′ single-stranded extensions (1). To determine whether the C-terminal extension is necessary for functional activity at 25°C and 15°C in vivo, the csdA null strain was grown in the presence of plasmid pPJ60, encoding CsdAΔ444. CsdAΔ444 lacks 185 amino acids from the protein's C terminus. The presence of plasmid-encoded CsdAΔ444 partially complemented the growth of the csdA null mutant at 25°C, as evidenced by the doubling time of 1.7 h observed in the presence of pPJ60 compared to the 2.4-h doubling time of the csdA null mutant observed in the presence of vector pACYC184 (Fig. 5A).
In contrast to the enhanced growth observed at 25°C, the presence of plasmid-encoded CsdAΔ444 was unable to stimulate growth of the csdA null mutant at 15°C (Fig. 5B). After 32 h at 15°C, the presence of pPJ60 inhibited growth of the csdA null mutant, as indicated by the doubling time of 92 h. However, after 125 h at 15°C, the doubling time of the csdA null mutant with plasmid-encoded CsdAΔ444 decreased to 27 h, which is similar to the 30-h doubling time of the csdA null mutant with vector pACYC184 (Fig. 5B). This latter phase of growth may be due to the accumulation of compensatory mutations on the plasmid or chromosome that bypass or alleviate the CsdAΔ444 block, as redilution of stationary-phase cultures of WJW45ΔcsdA/pPJ60 and WJW45ΔcsdA/pACYC184 at 15°C resulted in a similar rate of growth (Fig. 5C). These observations indicate that the C-terminal extension contributes to optimal functional activity at 25°C but is a necessary requirement for enzyme function at 15°C.
In vitro and in vivo analysis of the DQAD variant.
It has been shown by site-directed mutagenesis that the DEAD sequence (motif II) is essential for the in vitro ATPase and helicase activities of eIF4A, as substitution of the glutamic acid with glutamine dramatically reduced enzyme function (26). We made the equivalent substitution in CsdA (Glu-157→Gln), resulting in the DQAD variant. This single mutation essentially abolished ATP hydrolysis for all substrates (Table 1).
Helicase activity of the DQAD variant was also either abolished or severely reduced. No helicase activity was observed using duplex substrates in the presence or absence of ATP. A very low level of helicase activity, less than 8% conversion to single-stranded form, was observed using the hairpin substrate (Fig. 6).
FIG. 6.
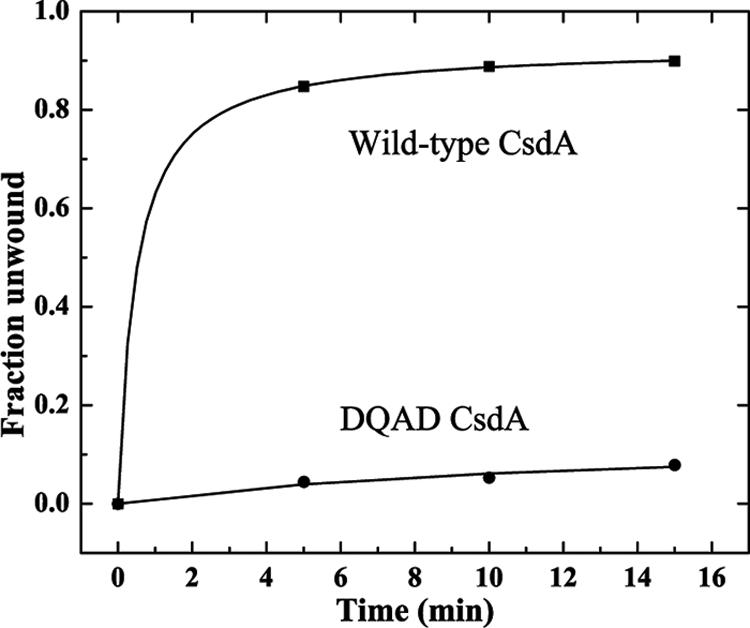
Unwinding of the hairpin substrate by wild-type and DQAD CsdA. Annealed hairpin substrate was incubated at 15°C with either wild-type CsdA or the DQAD variant in the presence of 2 mM ATP and subjected to nondenaturing gel electrophoresis as in Fig. 3. Gels were analyzed by phosphorimaging, and the fractional conversion to single-stranded product was quantified. A fraction of 1.0 corresponds to 100% conversion to single-stranded product.
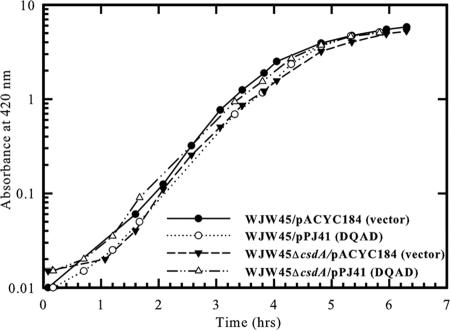
We also investigated the requirement of the DEAD motif for in vivo functional activity of CsdA. The effect of the Glu-157→Gln mutation on growth at low temperature was assayed by the ability of a plasmid encoding the DQAD variant to complement the growth defect of the csdA null mutant at 15°C. Unlike the plasmid encoding wild-type CsdA (pKX164), the plasmid encoding the DQAD variant of CsdA (pPJ41) did not stimulate growth of the null mutant (Fig. 7). The doubling time of the csdA null mutant in the presence of vector pACYC184 or pPJ41 was 32 h or 40 h, respectively, compared to 8- and 10-h doubling times for parent WJW45/pACYC184 and WJW45ΔcsdA/pKX164, respectively. Therefore, the DEAD motif is essential for in vivo and in vitro function of CsdA, supporting the cellular requirement of CsdA as a DEAD box ATP-dependent RNA helicase at low temperature.
FIG. 7.
In vivo complementation analysis of the csdA null mutant with plasmid-encoded wild-type or DQAD CsdA at 15°C. Strains WJW45 and WJW45ΔcsdA were grown in LB broth containing 30 μg/ml chloramphenicol in the presence of vector pACYC184, plasmid encoding wild-type CsdA (pKX164), or plasmid encoding the DQAD variant of CsdA (pPJ41).
The Glu-157→Gln substitution is a dominant negative mutation, as the presence of the plasmid encoding the DQAD variant inhibited growth of cells containing chromosomally encoded CsdA. Strain WJW45/pPJ41 initially grew with a doubling time of 32 h, which is the doubling time of WJW45ΔcsdA/pACYC184 (Fig. 7). In contrast, the presence of plasmid encoding wild-type CsdA (pKX164) in WJW45 had negligible effect on growth (Fig. 7). The reduced growth of the parent strain WJW45 in the presence of pPJ41 suggests that the plasmid-encoded DQAD variant prevented the function of wild-type CsdA, possibly by competing for binding to substrates. However, unlike WJW45ΔcsdA/pACYC184 or WJW45ΔcsdA/pPJ41, the doubling time of WJW45/pPJ41 decreased to 17 h after 76 h (Fig. 7). While full recovery of cell growth was not achieved, the improvement in growth rate suggests an increase in the enzymatic functioning of chromosomally encoded CsdA.
The Glu-157→Gln mutation is dominant negative only at low temperature. In contrast to 15°C, the presence of the plasmid encoding the DQAD variant had a negligible effect on growth of strain WJW45 at 37°C. The doubling time of WJW45 in the presence of pPJ41 was 27 min, compared to a doubling time of 25 min for WJW45 in the presence of vector pACYC184 (Fig. 8). Furthermore, the presence of the plasmid-encoded DQAD variant had a negligible effect on growth of the csdA null mutant at 37°C. The doubling time of WJW45ΔcsdA in the presence of pACYC184 or pPJ41 was 25 min or 27 min, respectively (Fig. 8). Therefore, the observed conditional dominant negative phenotype is consistent with the increased requirement of CsdA for cellular growth at low temperature.
FIG. 8.
Effect of plasmid-encoded DQAD CsdA on cell growth at 37°C. Strains WJW45 and WJW45ΔcsdA were grown in LB broth containing 30 μg/ml chloramphenicol in the presence of vector pACYC184 and plasmid encoding the DQAD variant of CsdA (pPJ41).
DISCUSSION
Temperature can modulate enzyme activity in vitro and in vivo. It has been widely observed that enzymes of psychrophilic and thermophilic organisms are specifically adapted to function at their respective optimal temperatures (11, 12, 15). Here we demonstrate that CsdA is functionally active at low temperature in vitro, exhibiting RNA-stimulated ATP hydrolysis and RNA unwinding activities at 15°C. These data are consistent with a physiological function of cold shock protein CsdA as an RNA helicase at low temperature. Bacillus subtilis, cyanobacteria, the Antarctic archaeon Methanococcoides burtonii, Hordeum vulgare, Pisum sativum, and Arabidopsis plants also increase the synthesis of a DEAD box protein upon a shift to low temperature (6, 13, 20, 25, 31, 37). These observations suggest an evolutionarily conserved requirement for helicase function to optimize cellular growth at low temperatures.
The DEAD motif of the prototype helicase eIF4A is essential for ATP hydrolysis (26). In particular, the glutamic acid is expected to participate in hydrolysis of bound ATP, as evidenced by its proximity to the presumptive attacking water (32). As was previously demonstrated for eIF4A (26), we found that substituting glutamine for glutamic acid to generate the DQAD variant of CsdA resulted in a significant loss of in vitro ATPase and helicase activities. Furthermore, a plasmid encoding the DQAD variant did not suppress the impaired growth of a csdA null mutant at low temperature. The presence of plasmid-encoded DQAD CsdA even inhibited growth of cells containing chromosomally encoded wild-type CsdA. The Glu-157→Gln mutation is dominant negative only at low temperature, as the presence of the plasmid encoding the DQAD variant in the parent strain had a negligible effect on growth at 37°C. The conditional dominant negative phenotype is consistent with the increased requirement of CsdA for cellular growth at low temperature. Therefore, the DEAD motif of CsdA is essential for functional activity both in vitro and in vivo at low temperature, confirming the physiological role of cold shock protein CsdA as a DEAD box ATP-dependent RNA helicase. In addition to the conserved requirement of DEAD motif for functional activity, CsdA shares 54% amino acid identity and 87% sequence similarity with eIF4A (22). It was demonstrated that CsdA increases translation in vitro of mRNA templates with secondary structures, further suggesting that the two DEAD box proteins share similar functions (22).
In vitro biochemical characterization of CsdA at 15°C revealed that only substrates containing a single-stranded overhang stimulated significant ATP hydrolysis and ATP-dependent helicase activities of CsdA. This is consistent with the previous observation that a single-stranded overhang may function as a platform to facilitate efficient loading of CsdA on the substrate and subsequent RNA unwinding (1). Furthermore, we found that CsdA exhibited RNA helicase activity towards substrates with either a 3′ or a 5′ overhang, confirming the previous observation that CsdA can function as a bidirectional helicase (1). The ATPase and helicase activities were not substantially different for the 3′- or 5′-overhang substrates. In the presence of the various RNA substrates, we also observed a qualitative correlation of ATPase activity with helicase activity. Of the substrates tested, the highest level of both ATP hydrolysis and RNA unwinding occurred with the hairpin RNA substrate derived from hairpin 92 of 23S rRNA. Hairpin 92 contains a sequence known to specifically stimulate DbpA helicase activity (10). To determine if CsdA has the same sequence-specific recognition of hairpin, we altered nucleotides in the hairpin 92 sequence to generate the hairpin substrate used here. CsdA retained its ability to efficiently unwind either hairpin 92 (data not shown) or our hairpin substrate (Fig. 3), suggesting a lack of sequence specificity for binding to CsdA.
The CsdAΔ variant, which retains all sequence motifs of canonical DEAD box helicases but lacks 185 amino acids from the enzyme's C terminus, unwound only the hairpin substrate containing a 9-mer duplex. The activity toward the hairpin substrate was as robust as for full-length CsdA, reaching approximately 85% conversion to single-stranded oligonucleotides within 5 min (Fig. 3F and 4). Although ATPase activity of CsdAΔ was stimulated in the presence of substrates containing a 15-mer duplex, the truncated protein did not measurably unwind these substrates at 15°C (Fig. 6). To identify any residual activity toward these substrates, we incubated the 3′-overhang substrate at 25 or 37°C; CsdAΔ was unable to significantly unwind this duplex even at these temperatures (Fig. 4). Although a previous study did demonstrate unwinding of 14-mer RNA duplexes with 3′ or 5′ overhangs by CsdAΔ (1), it should be noted that our duplex substrates contain different sequences and are marginally longer (by a single base pair). This may indicate either that the substrates used by Bizebard and coworkers (1) more closely resemble a natural substrate of CsdA or that substrates used in both studies are near the stability boundary for unwinding by CsdA. The reduced helicase activity of CsdA we observed towards substrates containing 15-mer duplex regions in comparison to the 9-mer hairpin substrate indicates that strand displacement of longer regions of double-stranded RNA is not efficient in vitro. The in vivo substrate or substrates of CsdA remain unknown, and while the isolated protein seems able to unwind a short duplex in vitro, additional protein factors may participate in the cellular activity of CsdA toward longer duplexes. The crystal structure of RNA-bound Drosophila Vasa revealed contacts encompassing seven nucleotides of the U10 oligonucleotide, with a distinct bend in the RNA strand resulting from steric conflict with residues of conserved motif 1b (32). A complementary oligo(A) strand could be modeled on either side of the bend, indicating that helicase activity may arise from the protein's destabilization of a duplex structure with or without a single-stranded overhang (32).
The robust unwinding of the hairpin substrate by either CsdA or CsdAΔ is not due to inherent instability of the hairpin at 15°C. Unwinding is ATP dependent (Fig. 3 and data not shown). We evaluated the structure of the hairpin at 15, 25, and 37°C using circular dichroism spectroscopy, and the spectra displayed the positive peak around 265 to 270 nm and negative peak around 240 nm characteristic of structured RNA (data not shown) (2). Furthermore, we determined the melting temperature and enthalpy of denaturation for UV-monitored thermal melting of the hairpin substrate (data not shown). The hairpin exhibited a melting temperature of 67.8°C and a vant'Hoff denaturation enthalpy of 31.9 kcal/mol, indicating that the hairpin is thermally stable in the absence of protein. Further studies will investigate the activity of CsdA toward a greater variety of substrates, including sequence variants of the hairpin described here. Complete kinetic characterization of the ATPase and helicase activities of CsdA will also be carried out.
Consistent with the observed limited in vitro activity of the CsdAΔ variant at 15°C, we found that a plasmid encoding CsdAΔ444 (lacking 185 amino acids from the protein's C terminus) did not suppress the impaired growth of the csdA null mutant at 15°C but further inhibited cellular growth. The subsequent faster growth of cultures upon redilution at 15°C indicates that improved growth is likely due to compensatory mutations on the plasmid or chromosome that bypass or alleviate the CsdAΔ444 block. In contrast to the impaired growth observed at 15°C, the presence of plasmid-encoded CsdAΔ444 partially complemented the growth of the csdA null mutant at 25°C. This is consistent with the previous finding that CsdAΔ catalyzed ATP-dependent unwinding of a 14-mer RNA duplex with 5′ or 3′ extensions at 25°C (1). Therefore, the C-terminal extension is necessary for optimal enzyme function in vivo at 25°C but is indispensable for function at 15°C. Relative differences in the doubling times of the parent and csdA null mutant indicate that E. coli has an increased functional requirement for CsdA at 15°C compared to 25°C. Furthermore, CsdA is multifunctional, having been associated with various cellular processes from ribosomal biogenesis to mRNA decay (7, 18, 22, 27, 29, 39). Therefore, the C-terminal portion may function in vivo to increase the activity or processivity of CsdA, particularly at low temperatures that demand a stringent requirement of CsdA for varied cellular processes and growth. Study of the mutations that result in bypass or alleviation of the CsdAΔ444 block may lead to a further understanding of the processes that become limiting in the absence of the CsdA and/or the requirement of the C-terminal extension for function of CsdA.
Acknowledgments
This work was supported by grant number MD00232 from the National Center for Minority Heath and Health Disparities at Winston-Salem State University.
We thank Isabella Iost, Laboratoire de Genetique Moleculaire, for generous gifts of bacterial strains WJW45 and WJW45ΔcsdA. R.W.A. thanks Michael Budiman for valuable technical assistance and helpful discussions.
Footnotes
Published ahead of print on 26 January 2007.
REFERENCES
- 1.Bizebard, T., I. Ferlenghi, I. Iost, and M. Dreyfus. 2004. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry 43:7857-7866. [DOI] [PubMed] [Google Scholar]
- 2.Blum, A. D., O. C. Uhlenbeck, and I. Tinoco, Jr. 1972. Circular dichroism study of nine species of transfer ribonucleic acid. Biochemistry 11:3248-3256. [DOI] [PubMed] [Google Scholar]
- 3.Blum, S., S. R. Schmid, A. Pause, P. Buser, P. Linder, N. Sonenberg, and H. Trachsel. 1992. ATP hydrolysis by initiation factor 4A is required for translation initiation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89:7664-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britton, R. A., B. S. Powell, S. Dasgupta, Q. Sun, W. Margolin, J. R. Lupski, and D. L. Court. 1998. Cell cycle arrest in Era GTPase mutants: a potential growth rate-regulated checkpoint in Escherichia coli. Mol. Microbiol. 27:739-750. [DOI] [PubMed] [Google Scholar]
- 5.Caruthers, J. M., and D. B. McKay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12:123-133. [DOI] [PubMed] [Google Scholar]
- 6.Chamot, D., W. C. Magee, E. Yu, and G. W. Owttrim. 1999. A cold shock-induced cyanobacterial RNA helicase. J. Bacteriol. 181:1728-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charollais, J., M. Dreyfus, and I. Iost. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 32:2751-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordin, O., J. Banroques, N. K. Tanner, and P. Linder. 2006. The DEAD-box protein family of RNA helicases. Gene 367:17-37. [DOI] [PubMed] [Google Scholar]
- 9.de la Cruz, J., D. Kressler, and P. Linder. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 10.Diges, C. M., and O. C. Uhlenbeck. 2001. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 20:5503-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerday, C., M. Aittaleb, M. Bentahir, J. P. Chessa, P. Claverie, T. Collins, S. D'Amico, J. Dumont, G. Garsoux, D. Georlette, A. Hoyoux, T. Lonhienne, M. A. Meuwis, and G. Feller. 2000. Cold-adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol. 18:103-107. [DOI] [PubMed] [Google Scholar]
- 12.Gianese, G., F. Bossa, and S. Pascarella. 2002. Comparative structural analysis of psychrophilic and meso- and thermophilic enzymes. Proteins 47:236-249. [DOI] [PubMed] [Google Scholar]
- 13.Hunger, K., C. L. Beckering, F. Wiegeshoff, P. L. Graumann, and M. A. Marahiel. 2006. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J. Bacteriol. 188:240-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iost, I., M. Dreyfus, and P. Linder. 1999. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 274:17677-17683. [DOI] [PubMed] [Google Scholar]
- 15.Jaenicke, R., and G. Bohm. 1998. The stability of proteins in extreme environments. Curr. Opin. Struct. Biol. 8:738-748. [DOI] [PubMed] [Google Scholar]
- 16.Jones, P. G., M. Mitta, Y. Kim, W. Jiang, and M. Inouye. 1996. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:76-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalman, M., H. Murphy, and M. Cashel. 1991. rhlB, a new Escherichia coli K-12 gene with an RNA helicase-like protein sequence motif, one of at least five such possible genes in a prokaryote. New Biol. 3:886-895. [PubMed] [Google Scholar]
- 18.Khemici, V., I. Toesca, L. Poljak, N. F. Vanzo, and A. J. Carpousis. 2004. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol. Microbiol. 54:1422-1430. [DOI] [PubMed] [Google Scholar]
- 19.Kossen, K., F. V. Karginov, and O. C. Uhlenbeck. 2002. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol. 324:625-636. [DOI] [PubMed] [Google Scholar]
- 20.Lim, J., T. Thomas, and R. Cavicchioli. 2000. Low temperature regulated DEAD-box RNA helicase from the Antarctic archaeon, Methanococcoides burtonii. J. Mol. Biol. 297:553-567. [DOI] [PubMed] [Google Scholar]
- 21.Linder, P., P. F. Lasko, M. Ashburner, P. Leroy, P. J. Nielsen, K. Nishi, J. Schnier, and P. P. Slonimski. 1989. Birth of the D-E-A-D box. Nature 337:121-122. [DOI] [PubMed] [Google Scholar]
- 22.Lu, J., H. Aoki, and M. C. Ganoza. 1999. Molecular characterization of a prokaryotic translation factor homologous to the eukaryotic initiation factor eIF4A. International J. Biochem. Cell Biol. 31:215-229. [DOI] [PubMed] [Google Scholar]
- 23.Luking, A., U. Stahl, and U. Schmidt. 1998. The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol. 33:259-296. [DOI] [PubMed] [Google Scholar]
- 24.Moll, I., S. Grill, A. Grundling, and U. Blasi. 2002. Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol. Microbiol. 44:1387-1396. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura, T., Y. Muramoto, S. Yokota, A. Ueda, and T. Takabe. 2004. Structural and transcriptional characterization of a salt-responsive gene encoding putative ATP-dependent RNA helicase in barley. Plant Sci. 167: 63-70. [Google Scholar]
- 26.Pause, A., and N. Sonenberg. 1992. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 11:2643-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prud'homme-Genereux, A., R. K. Beran, I. Iost, C. S. Ramey, G. A. Mackie, and R. W. Simons. 2004. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome’. Mol. Microbiol. 54:1409-1421. [DOI] [PubMed] [Google Scholar]
- 28.Ray, B. K., T. G. Lawson, J. C. Kramer, M. H. Cladaras, J. A. Grifo, R. D. Abramson, W. C. Merrick, and R. E. Thach. 1985. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J. Biol. Chem. 260:7651-7658. [PubMed] [Google Scholar]
- 29.Raynal, L. C., and A. J. Carpousis. 1999. Poly(A) polymerase I of Escherichia coli: characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol. Microbiol. 32:765-775. [DOI] [PubMed] [Google Scholar]
- 30.Schmid, S. R., and P. Linder. 1992. D-E-A-D protein family of putative RNA helicases. Mol. Microbiol. 6:283-291. [DOI] [PubMed] [Google Scholar]
- 31.Seki, M., M. Narusaka, H. Abe, M. Kasuga, K. Yamaguchi-Shinozaki, P. Carninci, Y. Hayashizaki, and K. Shinozaki. 2001. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13:61-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sengoku, T., O. Nureki, A. Nakamura, S. Kobayashi, and S. Yokoyama. 2006. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125:287-300. [DOI] [PubMed] [Google Scholar]
- 33.Tanner, N. K., O. Cordin, J. Banroques, M. Doere, and P. Linder. 2003. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell 11:127-138. [DOI] [PubMed] [Google Scholar]
- 34.Tanner, N. K., and P. Linder. 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8:251-262. [DOI] [PubMed] [Google Scholar]
- 35.Toone, W. M., K. E. Rudd, and J. D. Friesen. 1991. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J. Bacteriol. 173:3291-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanzo, N. F., Y. S. Li, B. Py, E. Blum, C. F. Higgins, L. C. Raynal, H. M. Krisch, and A. J. Carpousis. 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12:2770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vashisht, A. A., A. Pradhan, R. Tuteja, and N. Tuteja. 2005. Cold- and salinity stress-induced bipolar pea DNA helicase 47 is involved in protein synthesis and stimulated by phosphorylation with protein kinase C. Plant J. 44:76-87. [DOI] [PubMed] [Google Scholar]
- 38.Wincott, F., A. DiRenzo, C. Shaffer, S. Grimm, D. Tracz, C. Workman, D. Sweedler, C. Gonzalez, S. Scaringe, and N. Usman. 1995. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 23:2677-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamanaka, K., and M. Inouye. 2001. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J. Bacteriol. 183:2808-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamanaka, K., T. Ogura, E. V. Koonin, H. Niki, and S. Hiraga. 1994. Multicopy suppressors, mssA and mssB, of an smbA mutation of Escherichia coli. Mol. Gen. Genet. 243:9-16. [DOI] [PubMed] [Google Scholar]
- 41.Yang, L., J. Yang, Y. Huang, and Z. R. Liu. 2004. Phosphorylation of p68 RNA helicase regulates RNA binding by the C-terminal domain of the protein. Biochem. Biophys. Res. Commun. 314:622-630. [DOI] [PubMed] [Google Scholar]