Abstract
Large-scale chromosomal inversions (455 to 535 kbp) or deletions (266 to 320 kbp) were found to accompany spontaneous loss of beta-lactam resistance during drug-free passage of the multiresistant Staphylococcus haemolyticus clinical strain JCSC1435. Identification and sequencing of the rearranged chromosomal loci revealed that ISSha1 of S. haemolyticus is responsible for the chromosome rearrangements.
After the first report concerning the Haemophilus influenzae genome, a large number of microbial genomes were sequenced, and these genomes are available for comparison (2). A comparison of a pair of closely related chromosomes of members of the same species usually reveals that homologous genes, or orthologs, are arranged in the same order in the chromosome. However, noncolinear gene order is also found in certain strains of the same species, which is thought to be caused by chromosome rearrangement between the homologous sequences scattered along the chromosome (19). With this mechanism, it is presumed that genome rearrangements occur more frequently in the strains having many insertion sequences (IS) or prophages with closely related structures in their chromosomes. In fact, a comparison of three Bordetella species revealed that the rearrangements occurred more frequently in Bordetella pertussis (carrying 261 IS copies) than in Bordetella parapertussis and Bordetella bronchiseptica (112 and no IS copies, respectively) (12).
Of 11 staphylococcal strains belonging to four species whose entire genomes have been determined, Staphylococcus haemolyticus strain JCSC1435 has by far the greatest number of ISs in its chromosome (10, 18). This strain has as many as 82 copies of ISs in its chromosome, and notably 60 of these copies have been judged to be intact based on their nucleotide sequences. S. haemolyticus is notorious for its multidrug resistance phenotype and historical early acquisition of antimicrobial resistance to methicillin and glycopeptide antibiotics (3, 5). It has been reported that it is difficult to identify clinical strains as S. haemolyticus strains by traditional biochemical properties (17). These characteristics of S. haemolyticus may result from frequent structural alteration of the chromosome due to the recombination caused by the presence of abundant IS copies in the chromosome.
We have reported previously that large DNA segments that were up to 420 kbp long were deleted in ceftizoxime- or teicoplanin-susceptible variant strains of JCSC1435, presumably due to the IS-encoded transposase activity (18). We report here a different type of genome rearrangement, which also seems to contribute to the phenotypic instability or flexibility of S. haemolyticus.
In a previous study, all five of the IS-mediated genome deletions analyzed were localized in the first one-sixth of the chromosome, which, starting from oriC, roughly corresponded to the “oriC environ” (18). Following the previous study, we obtained 11 additional ceftizoxime-susceptible JCSC1435 derivatives by serial 11-day drug-free passages. Pulsed-field gel electrophoresis (PFGE) analysis showed that eight derivative strains had deletions in the oriC environ. However, the other three strains, UP01, UP03, and UP04, turned out to have more complicated rearrangements. They had the same pulsotype, which indicated that they had the same clonal origin. A detailed PFGE analysis of strain UP03 with three enzymes, SacII, SmaI, and BglI, revealed the presence of two chromosome rearrangements. One was an approximately 305-kbp deletion located in the oriC environ, and the other was a rearrangement localized on the opposite side of oriC. Furthermore, the deletion in the oriC environ was not a simple deletion, because we could not amplify a DNA fragment using a PCR primer set designed in a colinear manner on the chromosome map of JCSC1435, the parent strain. We then analyzed 10 and 18 other ceftizoxime-susceptible derivative strains obtained from two additional independent passage experiments. Each culture was started with an inoculum consisting of 100 μl of a JCSC1435 overnight culture, and the ceftizoxime-susceptible derivatives were obtained by replica plating after 9-day serial passages. All derivatives had altered SacII-restricted PFGE patterns, and we found that strains B5EC1 and C4EC2, obtained from the two independent experiments, had banding patterns that were similar, but not identical, to that of UP03 (Fig. 1a). SmaI- and BglI-restricted PFGE analysis further demonstrated that similar chromosome rearrangements had occurred in the three independent derivative strains (data not shown). We then used PCR scanning and Southern blot hybridization to determine the detailed chromosome structures of the three strains (Fig. 1).
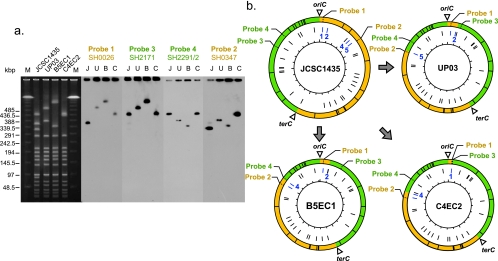
FIG. 1.
PFGE and Southern blot hybridization analysis of the three ceftizoxime-susceptible derivatives (UP03, B5EC1, and C4EC2). (a) SacII-restricted PFGE and Southern blot hybridization of S. haemolyticus JCSC1435 and three ceftizoxime-sensitive derivatives. Southern blot hybridization was carried out using a SacII-restricted PFGE gel with PCR products obtained with SH0026, SH2171, SH2291/2292, and SH0347 as probes. Lanes M, DNA size standard (lambda ladder); lanes J, JCSC1435; lanes U, UP03; lanes B, B5EC1; lanes C, C4EC2. (b) SacII restriction sites and locations of the open reading frames used as probes in the chromosomes of JCSC1435 and ceftizoxime-susceptible derivatives. The outer circle indicates the SacII restriction sites and the locations of the open reading frames used as probes (the clockwise and counterclockwise replication arms of JCSC1435 are yellow and green, respectively). The middle circle indicates the locations of ISSha1, and the inner circle indicates the nucleotide positions (in 100-kbp increments), starting from oriC and moving clockwise.
First, PCR scanning was performed in the chromosomal regions corresponding to the altered PFGE bands. With UP03, PCR produced no band in the region flanked by the two copies of the insertion sequence ISSha1-2 (from bp 88353 to 86865) and ISSha1-5 (bp 393322 to 394810) (the suffix for each ISSha1 copy was assigned in the clockwise direction, starting from oriC). This indicated that the 310-kbp region between the two ISSha1 copies was deleted. A similar strategy revealed that the regions between ISSha1-2 and ISSha1-4 (bp 356056 to 354570) of B5EC1 and between ISSha1-1 (bp 34466 to 32978) and ISSha1-4 of C4EC2 were deleted. In all the three cases, the mecA gene that encodes methicillin resistance was deleted along with the other regions. This was reasonable since all the strains were obtained with loss of beta-lactam resistance as the selective phenotype.
Next, DNA bands of PFGE gels were transferred to a nylon membrane, and Southern blot hybridization was carried out to identify the rearranged chromosome structures of the strains. PCR products that were about 1 kbp long (SH0026, SH0347, SH2171, and SH2291/SH2292) were amplified from the JCSC1435 chromosome and used as the hybridization probes (Fig. 1). Probes SH2171 and SH2291/SH2292 hybridized with the 430-kbp band of parent strain JCSC1435. SH0026 hybridized with the 350-kbp band and SH0347 hybridized with the 310-kbp band of JCSC1435. On the other hand, probes SH0026 and SH2171 hybridized to aberrant bands at 490 kbp (UP03), 570 kbp (B5EC1), and 405 kbp (C4EC2), and probes SH2291/2292 and SH0347 hybridized to other aberrant bands at 375 kbp (UP03), 335 kbp (B5EC1), and 445 kbp (C4EC2). Taken together, these observations indicated that large-scale inversions had occurred in these strains in the oriC region.
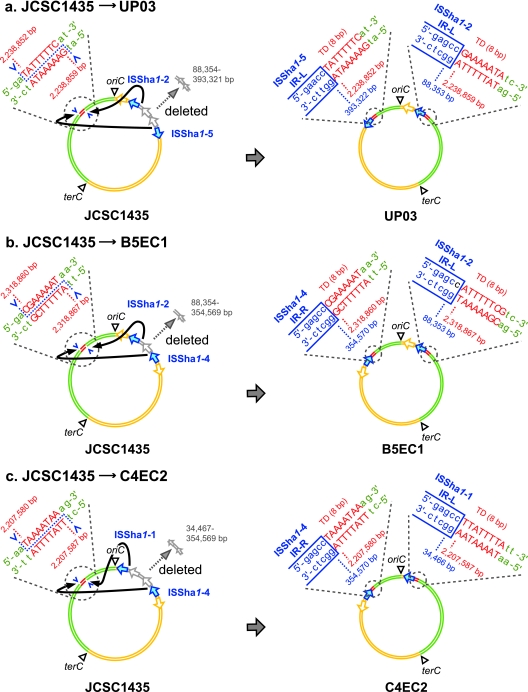
Finally, we determined exact genome rearrangement sites by nucleotide sequencing (Fig. 2). The large-scale inversion involved two ISSha1 copies located in oriC environ and an AT-rich octamer present on the other side of oriC. In addition, the regions between the two copies of ISSha1 were deleted. For example, in UP03, ISSha1-2 stayed at the same position relative to oriC, whereas ISSha1-5 moved to the other side of oriC from bp 393321 to bp 2238852 (Fig. 2a). A total of 304,968 bp between the two copies of ISSha1 was deleted, leaving two identical octamer sequences at nucleotide positions 2238852 and 2238859, which were located next to the IS copies ISSha1-2 and ISSha1-5, respectively. Similar inversions/deletions were found in the B5EC1 and C4EC2 chromosomes (Fig. 2b and c). The octamer sequences next to ISSha1 corresponded to one of the “direct repeats” (DRs) that are known to be generated as a result of target duplication in the integration of the insertion sequence ISSha1. The characteristic 26-bp inverted repeats (IRs) of ISSha1 were found to abut the newly generated octamers, as is the case with all the ISSha1 copies in JCSC1435. ISSha1 belongs to the ISL3 family, and each member of this family has a unique transposase (tnp) and a 15- to 39-bp IR and is known to produce 8-bp DRs during the integration event (11). Thus, our data clearly indicated that the rearrangements with a newly generated octamer sequence were caused by the integration of ISSha1. The deletion associated with this rearrangement, therefore, seemed to be coupled with the precise excision and integration of the ISSha1 copies. More specifically, it can be explained by the excision and self-integration of a composite transposon. Figure 3 illustrates this hypothesis. A composite transposon with a large chromosomal region flanked by two ISSha1 copies excises itself (with cleavage sites at the outside edges of the IRs) and subsequently integrates itself into the self DNA, generating the octamer DR at the target site of integration. As a result, the chromosome is left with a big inversion and deletion.
FIG. 2.
Genome rearrangements observed in three derivatives of S. haemolyticus JCSC1435. In addition to deletion between two copies of ISSha1, inversion has occurred between the two ISs and the 8-bp site in opposite replication arms. The blue arrows indicate ISSha1 involved in the genome rearrangements. The octamer sequences are indicated by uppercase red letters. They are considered to be target duplications (TD) accompanying the integration of ISSha1. The nucleotide positions of the derivatives are based on the nucleotide sequence of JCSC1435 (accession no. AP006716).
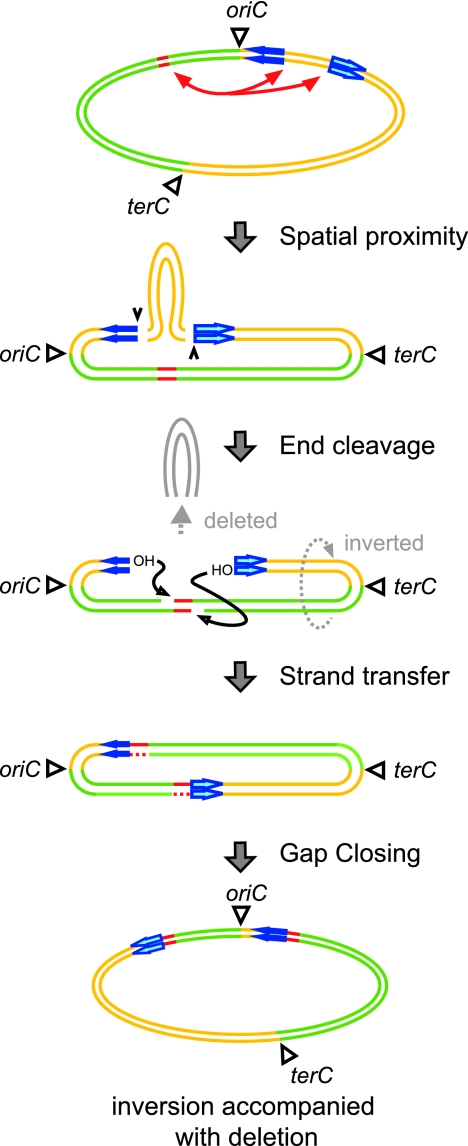
FIG. 3.
Model for the ISSha1-mediated deletion/inversion and simple deletion. The diagram shows the model for ISSha1-mediated deletion/inversion. The blue and purple arrows indicate ISSha1. The deletion (designated End cleavage) is thought to occur as a result of precise excision of a huge composite transposon made up of a large S. haemolyticus chromosome flanked by two copies of ISSha1 at both extremities. Subsequent to the excision, the transposon is integrated into itself at a preferred site (8-bp AT-rich sequence). The resultant genome is characteristic in its big part of the chromosome inverted relative to the oriC region. The integration makes the target site duplication that is identifiable (as “inverted” repeats) in the chromosomes of derivative strains adjacent to the two ISSha1 copies involved in the integration.
ISs are known to cause inversion or deletion through a homologous recombination mechanism (4). However, the members of many IS families are reported to form chromosomal deletions mediated by their transposases, where recombination takes place precisely at the end of the IS element (1, 7, 8, 13). In staphylococci, we previously described IS431 transposase-mediated large-scale deletions in a methicillin-resistant Staphylococcus aureus chromosome (20). All deletions start precisely at the edge of a unique IS431 copy but end at various chromosomal loci in the chromosome. S. haemolyticus possesses IS431 copies. ISs also generate deletions in S. haemolyticus strains (6) but were found only occasionally in JCSC1435 (18).
IS10/Tn10 also causes deletions with a similar mechanism (15). Tn10 is a composite transposon demarcated by two IS10 copies at both ends. It has been reported recently that an experimentally inserted Tn10 copy in the bacteriophage lambda genome or the Salmonella enterica serovar Typhimurium chromosome causes two types of genome rearrangements similar to those observed in this study (7-9, 14, 16). The first type is a transposase-promoted deletion that removes a single contiguous DNA segment starting precisely at one internal terminus of IS10 of the transposon. The second type is a more complex rearrangement involving both an inversion and a specific deletion of Tn10. The observed DNA rearrangements occurred almost always at the internal termini of IS10 copies. The ISSha1-mediated simple deletion and inversions/deletions described here appear to be caused by a mechanism similar to the IS10/Tn10 mechanism (Fig. 3).
ISSha1 may be the most frequent cause of genome rearrangements in S. haemolyticus (it is by far the most abundant IS in JCSC1435). In order to investigate the distribution of ISSha1 among staphylococci, PCR detection of ISSha1 was carried out using 74 S. haemolyticus clinical strains, 75 clinical strains of coagulase-negative staphylococci belonging to other species, and 28 Staphylococcus type strains, including S. haemolyticus type strain ATCC 29970. ISSha1 was harbored by all 74 clinical isolates of S. haemolyticus tested. No staphylococcal type strain except S. haemolyticus ATCC 29970 carried ISSha1. When clinically isolated coagulase-negative staphylococcal strains were examined, only three species other than S. haemolyticus carried the IS element; these species were Staphylococcus capitis (1/7 strains), Staphylococcus epidermidis (4/42 strains), and Staphylococcus warneri (1/1 strain). Strains of six other species did not carry ISSha1; these species were Staphylococcus auricularis (four strains), Staphylococcus caprae (six strains), Staphylococcus hominis (five strains), Staphylococcus hyicus (one strain), Staphylococcus lungdunensis (one strain), and Staphylococcus sciuri (eight strains). This strongly suggests that carriage of ISSha1 is one of the significant features of S. haemolyticus.
JCSC1435 is highly resistant to teicoplanin (MIC, 64 mg/liter). The extreme flexibility of the S. haemolyticus genome demonstrated in this study may also be associated with the acquisition of resistance. Studies of JCSC1435 taking advantage of its frequent chromosome rearrangement are under way.
Acknowledgments
This work was supported by a Grant-in-Aid for 21st Century COE Research, by a Grant-in-Aid for Scientific Research on Priority Areas (grant 13226114), and by research fellowships of the Japan Society for the Promotion of Science for Young Scientists (grant 18-53122) from The Ministry of Education, Science, Sports, Culture and Technology of Japan.
Footnotes
Published ahead of print on 19 January 2007.
REFERENCES
- 1.Chan, R. K., D. Botstein, T. Watanabe, and Y. Ogata. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology 50:883-898. [DOI] [PubMed] [Google Scholar]
- 2.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 3.Froggatt, J. W., J. L. Johnston, D. W. Galetto, and G. L. Archer. 1989. Antimicrobial resistance in nosocomial isolates of Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 33:460-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray, Y. H. 2000. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 16:461-468. [DOI] [PubMed] [Google Scholar]
- 5.Hiramatsu, K. 1998. Vancomycin resistance in staphylococci. Drug Resist. Updates 1:135-150. [DOI] [PubMed] [Google Scholar]
- 6.Katayama, Y., T. Ito, and K. Hiramatsu. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 45:1955-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleckner, N., D. F. Barker, D. G. Ross, and D. Botstein. 1978. Properties of the translocatable tetracycline-resistance element Tn10 in Escherichia coli and bacteriophage lambda. Genetics 90:427-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleckner, N., K. Reichardt, and D. Botstein. 1979. Inversions and deletions of the Salmonella chromosome generated by the translocatable tetracycline resistance element Tn10. J. Mol. Biol. 127:89-115. [DOI] [PubMed] [Google Scholar]
- 9.Krug, P. J., A. Z. Gileski, R. J. Code, A. Torjussen, and M. B. Schmid. 1994. Endpoint bias in large Tn10-catalyzed inversions in Salmonella typhimurium. Genetics 136:747-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroda, M., A. Yamashita, H. Hirakawa, M. Kumano, K. Morikawa, M. Higashide, A. Maruyama, Y. Inose, K. Matoba, H. Toh, S. Kuhara, M. Hattori, and T. Ohta. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. USA 102:13272-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 13.Reif, H. J., and H. Saedler. 1975. IS1 is involved in deletion formation in the gal region of E. coli K12. Mol. Gen. Genet. 137:17-28. [DOI] [PubMed] [Google Scholar]
- 14.Roberts, D. E., D. Ascherman, and N. Kleckner. 1991. IS10 promotes adjacent deletions at low frequency. Genetics 128:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross, D. G., J. Swan, and N. Kleckner. 1979. Physical structures of Tn10-promoted deletions and inversions: role of 1400 bp inverted repetitions. Cell 16:721-731. [DOI] [PubMed] [Google Scholar]
- 16.Shen, M. M., E. A. Raleigh, and N. Kleckner. 1987. Physical analysis of Tn10- and IS10-promoted transpositions and rearrangements. Genetics 116:359-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shittu, A., J. Lin, D. Morrison, and D. Kolawole. 2004. Isolation and molecular characterization of multiresistant Staphylococcus sciuri and Staphylococcus haemolyticus associated with skin and soft-tissue infections. J. Med. Microbiol. 53:51-55. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tillier, E. R., and R. A. Collins. 2000. Genome rearrangement by replication-directed translocation. Nat. Genet. 26:195-197. [DOI] [PubMed] [Google Scholar]
- 20.Wada, A., Y. Katayama, K. Hiramatsu, and T. Yokota. 1991. Southern hybridization analysis of the mecA deletion from methicillin-resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 176:1319-1325. [DOI] [PubMed] [Google Scholar]