Abstract
Pseudomonas syringae pv. phaseolicola is the causal agent of halo blight disease of beans (Phaseolus vulgaris L.), which is characterized by water-soaked lesions surrounded by a chlorotic halo resulting from the action of a non-host-specific toxin known as phaseolotoxin. This phytotoxin inhibits the enzyme ornithine carbamoyltransferase involved in arginine biosynthesis. Different evidence suggested that genes involved in phaseolotoxin production were clustered. Two genes had been previously identified in our laboratory within this cluster: argK, which is involved in the immunity of the bacterium to its own toxin, and amtA, which is involved in the synthesis of homoarginine. We sequenced the region around argK and amtA in P. syringae pv. phaseolicola NPS3121 to determine the limits of the putative phaseolotoxin gene cluster and to determine the transcriptional pattern of the genes comprising it. We report that the phaseolotoxin cluster (Pht cluster) is composed of 23 genes and is flanked by insertion sequences and transposases. The mutation of 14 of the genes within the cluster lead to a Tox− phenotype for 11 of them, while three mutants exhibited low levels of toxin production. The analysis of fusions of selected DNA fragments to uidA, Northern probing, and reverse transcription-PCR indicate the presence of five transcriptional units, two monocistronic and three polycistronic; one is internal to a larger operon. The site for transcription initiation has been determined for each promoter, and the putative promoter regions were identified. Preliminary results also indicate that the gene product of phtL is involved in the regulation of the synthesis of phaseolotoxin.
Pseudomonas syringae pv. phaseolicola is the causal agent of halo blight disease of beans (Phaseolus vulgaris L.), which is characterized by water-soaked lesions surrounded by a chlorotic halo (23). This halo results from the action of a non-host-specific toxin known as phaseolotoxin [Nδ(N′-sulfodiaminophosphinyl)-ornithyl-alanyl-homoarginine] (22, 25). Phaseolotoxin is a reversible inhibitor of the enzyme ornithine carbamoyltransferase, (OCTase) (EC 2.1.3.3) (7, 17) that catalyzes the formation of citrulline from ornithine and carbamoylphosphate in the sixth step of the arginine biosynthetic pathway. In planta phaseolotoxin is readily cleaved by peptidases to release Nδ(N′-sulfodiaminophosphinyl)-ornithine (PSOrn), the major toxic chemical species present in diseased leaf tissue (23).
The production of this phaseolotoxin is temperature dependent, being optimally produced at 18 to 20°C, while no detectable amounts of the toxin are present at 30°C (9, 21, 27). Phaseolotoxin toxin is active against not only plant OCTases but also bacterial and mammalian enzymes (6, 24, 38). This has led to the development of a rapid bioassay that measures the growth inhibition of a bacterial culture exposed to this toxin (38). However, P. syringae pv. phaseolicola is insensitive to the effect of its own toxin due to the presence of a phaseolotoxin-resistant OCTase (7, 37), which is produced under conditions leading to the synthesis of phaseolotoxin (13, 14) and is encoded by the argK gene (10, 26).
The argK gene had been previously found to be part of a gene cluster contained in a cosmid clone, pRCP17, which also included genes required for the synthesis of phaseolotoxin (28, 29). In an independent study, a second clone was isolated which contained a 25-kb insert and complemented several Tox− mutants. This clone, pHK120, was shown to contain eight transcriptional units that were designated phtA through phtH. Both clones pRCP17 and pHK120 were shown to be partially overlapping; however, pHK120 lacked argK but contained sequences missing in pRCP17 (43). A gene coding an amidinotransferase, amtA, and shown to be involved in the synthesis of the homoarginine residue of the tripeptide moiety of phaseolotoxin was also found to be linked to argK (12). All this evidence suggested that most or perhaps all of the genes required for the synthesis of phaseolotoxin were part of a large gene cluster.
In P. syringae pv. phaseolicola, the nucleotide sequence of argF, a gene coding for a phaseolotoxin-sensitive OCTase present under conditions not permissive for phaseolotoxin synthesis, has very low similarity with the sequence from argK; it is so low in fact that these genes do not cross-hybridize (26, 29). The sequence of argK also presents a G+C content of 49.4%, which is below that of argF (57.3%) and the hrp genes (56 to 58%). These characteristics suggest that in P. syringae pv. phaseolicola, argK did not evolve from the housekeeping gene argF, but rather seems to have been acquired from another microorganism through horizontal gene transfer. The close linkage of argK to some of the other genes involved in the synthesis of phaseolotoxin and its essential role in protection against this toxin also suggests that not only argK but also all of the genes involved in phaseolotoxin biosynthesis and immunity may have been acquired together (10).
We decided to sequence the entire region around the argK and amtA genes in P. syringae pv. phaseolicola NPS3121 in order to determine the limits of the putative phaseolotoxin gene cluster as well as to determine the transcriptional pattern of the genes comprising this cluster. The phaseolotoxin cluster (Pht cluster) is composed of 23 genes and is flanked by insertion sequences and transposases. Furthermore, we demonstrate that the mutation of 14 of the genes within the cluster results in a Tox− phenotype for 11 of them, while three mutants exhibited only low levels of toxin production. The 23 genes are organized in five transcriptional units, two monocistronic and three polycistronic, with one overlapping a larger operon. The promoter regions have been determined for each transcriptional unit, and preliminary evidence for a regulatory role for the gene product of phtL is discussed.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. syringae pv. phaseolicola NPS3121 and mutant derivatives were grown on King's B medium (32) or on M9 medium (33) at 18 or 28°C. Escherichia coli strains DH5α and JM103 were grown in Luria-Bertani medium at 37°C. For phaseolotoxin production, P. syringae pv. phaseolicola was grown in M9 medium at 18°C/48 h. When required, the following supplements were added: carbenicillin, 100 μg ml−1; kanamycin, 50 μg ml−1; rifampin, 50 μg ml−1; spectinomycin, 50 μg ml−1; and X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid), 30 μg ml−1.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Bacterial strains | ||
| Escherichia coli | ||
| DH5α | 33 | |
| TOP10 | Commercial strain used for cloning | Invitrogen |
| JM103 | 42 | |
| P. syringae pv. phaseolicola | ||
| NPS3121 | Wild type, Tox+ | 28 |
| YNorf1P | Tcr; phtA::tet polar mutant of NPS3121 | This study |
| YNorf2P | Tcr; phtB::tet polar mutant of NPS3121 | This study |
| SAorf5P | Tcr; phtE::tet polar mutant of NPS3121 | This study |
| SAorf6P | Tcr; phtF::tet polar mutant of NPS3121 | This study |
| SAorf8P | Tcr; phtH::tet polar mutant of NPS3121 | This study |
| SAorfBP | Kmr; phtK::uidA-aph polar mutant of NPS3121 | This study |
| SAorf10P | Kmr; phtL::uidA-aph polar mutant of NPS3121 | This study |
| AT3 | Kmr; amtA::aph polar mutant of NPS3121 | 12 |
| SAorf17P | Kmr; phtT::aph polar mutant of NPS3121 | This study |
| UIKsir2 | Tcr; sir2::tet polar mutant of NPS3121 | This study |
| SADesNP | Kmr; desI::uidA-aph nonpolar mutant of NPS3121 | This study |
| SAorfBNP | Kmr; phtK::uidA-aph nonpolar mutant of NPS3121 | This study |
| SAorf10NP | Kmr; phtL::uidA-aph nonpolar mutant of NPS3121 | This study |
| SAorf11NP | Kmr; phtM::uidA-aph nonpolar mutant of NPS3121 | This study |
| UIKorf1 | Kmr; phtO::uidA-aph nonpolar mutant of NPS3121 | This study |
| UIKorf4 | Kmr; phtQ::uidA-aph nonpolar mutant of NPS3121 | This study |
| SAorf17NP | Kmr; phtT::aph nonpolar mutant of NPS3121 | This study |
| Plasmids | ||
| pUC19 | Apr; 2.69 kb; lacZ′ | 42 |
| pCR4Blunt-TOPO | Apr Kmr; 3.95 kb | Invitrogen |
| pRG960sd | Smr Spr; 17.0 kb; contains a promoterless SD-uidA gene | 40 |
| pWM6 | Kanamycin cassette | 20 |
| pKL66 | Smr Spr; InReKAa; PCR fragment in pRG960sd | This study |
| pYUDF1 | Smr Spr; InReKAb; PCR fragment in pRG960sd | This study |
| pYUDF2 | Smr Spr; InReAB; PCR fragment in pRG960sd | This study |
| pSELF1 | Smr Spr; InReCD; PCR fragment in pRG960sd | This study |
| pSELF2A | Smr Spr; InRedesI-phtJ; BamHI-SspI chromosomal fragment | This study |
| pSELF4 | Smr Spr; InReTU; PCR fragment in pRG960sd | This study |
Molecular biology techniques.
Routine techniques were performed as described previously (33). Plasmids and DNA from agarose gels were purified with QIAGEN columns and kits (Valencia, CA). Restriction enzymes were used according to instructions provided by the suppliers. Chromosomal DNA from P. syringae pv. phaseolicola was obtained as described previously (4). DNA fragments used as probes for Southern blot analysis were labeled with fluorescein with the ECF random primer labeling kit, and signal detection was performed with the ECF signal amplification system, both from Amersham Pharmacia Biotech (Piscataway, NJ).
Construction and screening of a genomic library of P. syringae pv. phaseolicola.
Chromosomal DNA from P. syringae pv. phaseolicola was obtained, and fragments of 15 to 20 kb were purified from Sau3A1 partially digested DNA, ligated into BamHI-cut EMBL3 vector, and packaged in vitro according to instructions of the supplier of commercial packaging extracts (Promega). The library was amplified in E. coli LE392, plated, and hybridized according to standard methods (33). Positive recombinant phages were amplified and replated.
DNA sequencing and analysis.
The DNA inserted in the recombinant phages was digested with SalI and BamHI and subcloned into pUC19 prior to sequencing. Universal and reverse primers were used for the primary reactions, and synthesized primers were used to sequence both strands. The DNA sequence was analyzed using DNAStar and Vector NTI software. Database comparisons were made using available BLAST programs (2).
Construction of P. syringae pv. phaseolicola mutants.
Restriction fragments or PCR-derived amplicons containing selected open reading frames (ORFs) from the phaseolotoxin gene cluster were cloned into pUC19 or pCR4Blunt-TOPO vectors. Mutants were obtained in these constructs by the interruption of the gene of interest with different antibiotic/uidA cassettes, followed by the replacement of the wild-type allele in the P. syringae pv. phaseolicola chromosome. Mutants with polar effects on downstream ORFs were obtained by using a 2.2-kb NruI tetracycline cassette and, in some cases, either a 3.8-kb SmaI fragment containing the uidA-aph cassette or a 1.38-kb SalI fragment containing only the aph cassette from pWM6 (20) inserted in an orientation opposite to the transcription of the ORFs. Nonpolar insertions were obtained by using the 3.8-kb SmaI fragment containing the uidA-aph cassette or the 1.38-kb SalI fragment containing only the aph cassette from pWM6 inserted in line with the transcription of the ORFs. All constructs were confirmed by restriction digests and introduced by electroporation into P. syringae pv. phaseolicola. Tetracycline or kanamycin resistance was used to select for double-recombination events. The fidelity of double recombination in all cases was confirmed by Southern blot hybridization or PCR analysis.
Construction of transcriptional uidA fusions and β-glucuronidase (GUS) assays.
Plasmid pRG960sd (40), which contains a promoterless uidA gene preceded by a Shine-Dalgarno sequence (SD-uidA) downstream of a multiple cloning site, was used to identify putative promoter sequences within intergenic regions (InRes) upstream of some of the ORFs from the phaseolotoxin cluster. These fragments were obtained as restriction fragments or by PCR using primers designed to include suitable restriction sites (data available on request) and were cloned into the BamHI-SmaI sites of pRG960sd to create pKL66, pYUDF1, and pSELF1. The orientations were determined by restriction digests, and constructs were mobilized into P. syringae pv. phaseolicola via electroporation. The intergenic region between argK and phtA (InReKA) (Fig. 1C) was cloned in both orientations to assess promoter activity towards argK and phtA.
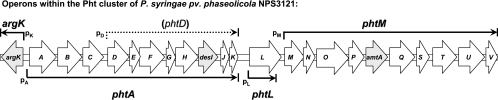
FIG. 1.
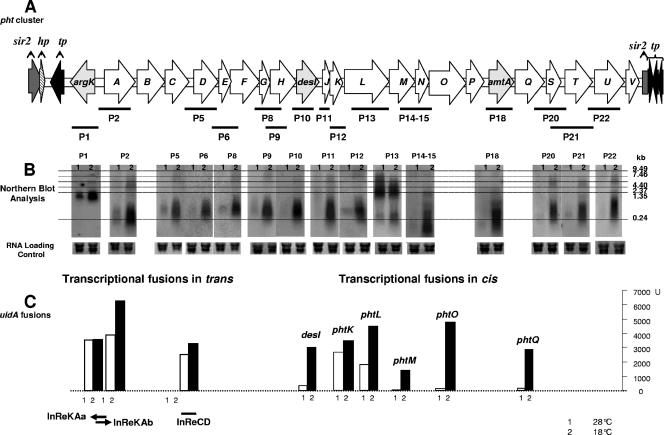
Pht cluster of P. syringae pv. phaseolicola NPS3121 and expression patterns. (A) Graphic representation of the Pht cluster. Each arrow represents each gene, with the direction of the arrow indicating the direction of transcription. The probes (P1, P2, etc.) used for the Northern blot analysis are graphically represented by the bars under the genes. (B) Northern blot results with each of the corresponding probes. Assessment of transcription activity as β-glucuronidase activity or mRNA-DNA hybridization. (C) Transcriptional fusions in trans: β-glucuronidase activity from the InRe cloned into the promoter trap vector pRG960sd. The cloned regions are represented graphically under the corresponding value bars. Transcriptional fusions in cis shows β-glucuronidase activity from pht::uidA constructs. The fused genes are indicated above the corresponding value bars. The small numbers under the bars or on top of the Northern blots represent the temperatures at which expression was assayed: 1 indicates 28°C and 2 indicates 18°C.
To measure GUS activity, P. syringae pv. phaseolicola strains carrying uidA fusions were grown to an optical density at 600 nm of 0.8 in M9 medium at 18 or 28°C. Bacteria were harvested and resuspended in GUS extraction buffer (50 mM NaHPO4, [pH 7.0], 10 mM β-mercaptoethanol, 10 mM disodium EDTA, 0.1% [wt/vol] sodium lauryl sarcosine, 0.1% [wt/vol] Triton X-100). GUS activity was determined by measuring the accumulation of 4-methylumbelliferone in a reaction mixture consisting of the bacterial lysate supplemented with 1 mM 4-methylumbelliferyl glucuronide. To terminate the reaction, 50 μl of the reaction mixture was combined with 1,950 μl of 0.2 mM Na2CO3. 4-Methylumbelliferone was determined fluorometrically (15) by using a VersaFluor fluorometer (Bio-Rad Laboratories, CA). GUS activity is reported as specific activity (1 pmol of methylumbelliferone formed per minute per milligram of protein). Protein in the bacterial lysates was estimated by the procedures of Bradford (3). Lysates of wild-type P. syringae pv. phaseolicola, which exhibited no detectable GUS activity, were used as controls in these experiments.
Phaseolotoxin bioassays.
Phaseolotoxin production by P. syringae pv. phaseolicola and mutants was assayed by the E. coli growth inhibition assay (38) as previously described (12, 35). In every case, plates containing arginine were used as controls to confirm that growth inhibition was due to phaseolotoxin.
RNA extraction and Northern blot analysis.
RNA was isolated from cultures of P. syringae pv. phaseolicola grown in M9 medium at 18 or 28°C when they reached an optical density at 600 nm of 0.8. Total RNA was extracted from cells by using TRIzol reagent as recommended by the manufacturer (Invitrogen). Genomic DNA was removed by digestion with RNase-free DNase (Roche). Samples of total RNA (20 μg) were denatured by treatment with formamide and separated by electrophoresis using 1.3% denaturing agarose gels. The RNA was transferred to Hybond N+ nylon membranes (Amersham) and cross-linked by exposure to UV radiation. The hybridization of the nylon membranes was performed using Church buffer (33) and different DNA probes for each ORF. The probes were labeled with [α-32P]dCTP by using the Rediprime random primer labeling kit (Amersham). The hybridization was carried out overnight at 65°C. The membranes were washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate for 3 min at room temperature, followed by a wash with 1× SSC-0.1% sodium dodecyl sulfate for 3 min at 65°C. The membranes were exposed in a Storm 860 apparatus, and the analysis was made using ImageQuant 1.1 software.
Reverse transcription-PCR analysis.
DNA-free RNA was obtained from cultures grown in M9 medium for 48 h at 18 and 28°C. The RNA was checked for integrity in an agarose gel and used for reverse transcription (RT) and PCR using the SuperScript one-step kit (Invitrogen). A list of the primers used in this analysis is available on request. Controls used for each set of primers were (i) PCR without the reverse transcription step to verify the absence of DNA, (ii) RT-PCRs performed without RNA templates to detect any contaminating DNA/RNA, (iii) PCRs performed using chromosomal DNA as a template to ensure primer fidelity, and (iv) amplification of a portion of the 23S rRNA operon using suitable primers as an internal control of the reaction. A portion of the 23S rRNA operon was amplified using suitable primers. The RT reaction was performed at 50°C for 40 min, followed by PCR amplification at 94°C for 2 min for 1 cycle; 94°C for 35 s, 58°C for 30 s, and 72°C for 2.0 min for 25 cycles; and 72°C for 15 min for 1 cycle.
Mapping the site of transcription initiation.
The site of transcription initiation for each operon was obtained by sequencing the 5′ end of the different mRNA transcripts using a 5′ circular first-strand cDNA-mediated RACE (rapid amplification of cDNA ends) method (5′ cRACE) (18) as follows. First, the synthesis of single-stranded cDNA was performed. Total DNA-free RNA was obtained from cultures grown in M9 medium for 48 h at 18°C. One microgram of this RNA was used as a template to synthesize the first-strand cDNA of the mRNA by using a cDNA synthesis kit (Invitrogen) with a gene-specific oligonucleotide primer designed to anneal within the coding region of the gene. Second, a poly(A) tail was added to the 3′ end of the single-stranded cDNA. After hydrolysis of the template mRNA with 0.5 N NaOH (final concentration) at 37°C for 10 min, the single-stranded cDNA was precipitated with ethanol and redissolved in 10 μl of a reaction mixture containing 25% polyethylene glycol (molecular weight, ∼8,000), 10 mM Tris-HCl (pH 8.0), 1 mM hexamine cobalt chloride, 10 mM MgCl2, 0.01 mM ATP, 1 μg/ml bovine serum albumin, 10 U T4 RNA ligase (New England Biolabs), and a commercial 30mer poly(A) oligonucleotide previously phosphorylated with T4 polynucleotide kinase (New England Biolabs). This ligation mixture was incubated at 22°C for 16 h. Third, the first PCR amplification was performed. An aliquot of the previous ligation mixture was directly used as a template for the first PCR amplification. The PCR was set up in a total volume of 50 μl containing 0.5 μl of the cDNA ligation mixture, a poly(T) oligonucleotide, and a gene-specific primer. The PCR was carried out for 30 cycles with the following conditions: 94°C for 30 s denaturation, 58°C for 30 s annealing, and 72°C for a 1.5-min extension. Last, nested PCR was performed. The resulting PCR product was diluted 1,000-fold with sterile H2O, and a 2-μl aliquot was used as template for a second nested PCR amplification with a 30mer commercial poly(T) oligonucleotide and a gene-specific nested primer under the same conditions. The amplification products were cloned into a TOPO-TA cloning vector (Invitrogen) and sequenced.
RESULTS
Sequence of the phaseolotoxin gene cluster of P. syringae pv. phaseolicola NPS3121.
By the initial use of probes derived from argK and amtA, we were able to identify several overlapping λ clones which allowed us to sequence the regions around both of these genes. With this strategy, we were able to sequence and assemble around 25 kb before we had access to the preliminary release of the complete sequence of P. syringae pv. phaseolicola 1448A (GenBank accession no. CP000058 to CP000060). Once we had this information, the gaps in the sequence of P. syringae pv. phaseolicola NPS3121 were covered by sequencing amplicons obtained through PCR generated with primers designed from both the partial sequence of NPS3121 and the reported sequence from 1448A. A total of 30,245 bp were sequenced on both strands (GenBank DQ141263).
When we aligned the sequence we obtained for NPS3121 to the corresponding region reported for strain 1448A, we found only 35 nucleotide differences, all of them occurring after position 14694 in the sequence of NPS3121. Among these changes, there were 15 bases not found in 1448A, including a stretch of 9 contiguous nucleotides, 4 bases missing in NPS3121 with respect to 1448A, and 16 single-point changes corresponding to 6 transitions and 10 transversions. Of these, the most significant change corresponds to the 9-nucleotide stretch found in NPS3121 between positions 18748 and 18756 since this introduces a stop codon in the region corresponding to locus PSPPH4306 of 1448A, and as a consequence, in strain NPS3121, we found two separate and distinct ORFs (phtM and phtN) at this locus.
The computational analysis of the 30,245-bp region of P. syringae pv. phaseolicola NPS3121 suggested the presence of 27 ORFs in this region (Fig. 1A). Of these, three ORFs at the 5′ end correspond to a sir2 homolog, a “hypothetical protein,” and an ISPsy25 transposase. At the 3′ end, between positions 28997 and 29324, we found a 328-bp region whose translation showed 59% identity to Sir2 from P. syringae pv. tomato DC3000, 55% to that from Pseudomonas fluorescens Pf-5, and 54% to that from Pseudomonas entomophila L48 among others. This truncated region of similarity is linked through a 127-bp region to a region with homology to ISPsy5/ISPpu13 transposases. Between these ends, we defined 23 ORFs, each one preceded by a putative ribosome-binding sequence. In this work, we refer to this group of ORFs as the phaseolotoxin (Pht) cluster as previously proposed (11). A BLASTX analysis of the Pht cluster returned the same information reported for strain 1448A (16). The unambiguous genes identified at the moment are argK (coding for the phaseolotoxin-resistant ornithyl carbamoyl transferase [10, 26]), amtA (coding for an l-arginine:lysine amidinotransferase [12]), and the des gene (desI) (coding for a fatty acid desaturase [11]).
Phenotype of mutants.
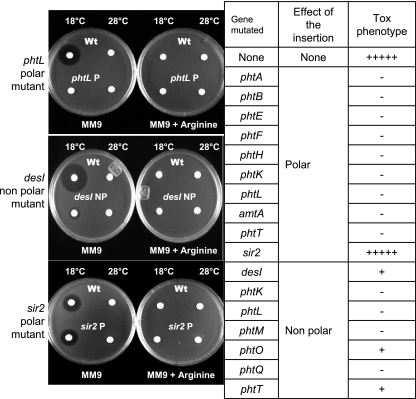
Mutants with polar effects on downstream genes were obtained for phtA, -B, -E, -F, -H, -K, -L, -T, amtA, and sir2, and these were unable to produce phaseolotoxin as shown by the growth inhibition assay, the only exception being the mutant on the sir2 gene located outside the Pht cluster which produces phaseolotoxin at a level comparable to that of the wild-type strain. Nonpolar mutants were obtained for desI and the pht genes phtK, -L, -M, -O, -Q, and -T. In this case, mutants in phtK, -L, -M, and -Q showed a clear Tox− phenotype, whereas desI, phtO, and phtT were still capable of producing phaseolotoxin, although at a very low level (Fig. 2).
FIG. 2.
Phenotype of mutants of P. syringae pv. phaseolicola NPS3121. The table lists the genes mutated, type of insertion, and phenotypic evaluation compared qualitatively to the wild-type (Wt) strain in terms of phaseolotoxin production using the E. coli growth inhibition assay. The photographs illustrate examples of the growth inhibition assays. −, no phaseolotoxin produced; +, low phaseolotoxin levels; +++++, wild-type phaseolotoxin production.
Expression pattern and putative promoter regions.
In order to assess the expression pattern of the Pht cluster with respect to temperature, we performed Northern blot analysis using restriction fragments or PCR-amplified regions from 15 genes of the Pht cluster (Fig. 1A) as probes. The hybridization results (Fig. 1B) show that all of the 15 genes are transcribed at high levels at 18°C and that most of the ORFs show some basal level of expression at 28°C, including argK, as previously reported (10). However, although 28°C does not allow for the biosynthesis of phaseolotoxin, we could still clearly see a high level of transcription of phtL at this temperature.
In order to identify the promoters responsible for the observed expression, InRes of the Pht cluster were cloned into a vector containing the promoterless uidA gene preceded by a SD sequence. These constructs were electroporated into P. syringae pv. phaseolicola to assess promoter activity in trans. Also, the nonpolar insertions in desI and the pht genes phtK, -L, -M, -O, and -Q, were obtained with a construct comprising the SD-uidA gene followed by the aph gene, allowing for the promoter of the interrupted gene/operon to drive uidA expression in cis. Results from these experiments are depicted graphically in Fig. 1C; nevertheless, we are aware that the levels of expression do not necessarily correlate to levels in vivo since the reporter in trans occurs in multiple copies (40), whereas in the fusions in cis, a direct or indirect effect on the levels of transcription due to the mutation itself could not be excluded.
Expression was unambiguously detected for the InRe between argK and phtA, both in the direction of argK (InReKAa←) and divergently into phtA (InReKAb→), suggesting the presence of two distinct promoters. Promoter activity was also detected within the InRe between phtC and phtD (InReCD), but not within the 68-bp intergenic region between phtA and phtB (InReAB), indicating that phtABC comprise an operon and that phtD is the first gene of a different transcriptional unit. The 141-bp region between desI and phtJ and the 129-bp region between phtT and phtU were also unable to drive uidA expression, indicating the lack of promoter sequences (data not shown).
With respect to the uidA fusion in cis, activity was detected within desI. Considering that the largest InRe found within the phtD-desI fragment is only 13 bp (within phtD and phtE) and the lack of promoter activity in the InRe between desI and phtJ, the result strongly suggests that phtD, -E, -F, -G, -H, desI, and phtJ are part of a single operon, which is also supported by the expression pattern (observed through Northern blot analysis) that suggests a similar pattern for both desI and phtJ. The expression of phtK as assayed with Northern blotting suggests that it could be part of the same operon as that for phtJ.
With respect to genes downstream of phtL, we observed GUS activity from the uidA fusion to phtM, phtO, and phtQ. The largest intergenic space found in the region from phtM to phtQ is 116 bp between phtP and amtA. However, our previous primer extension analyses of the region upstream of amtA consistently suggested that this gene was transcribed as part of a larger transcript (data not shown), therefore it seems reasonable to assume that phtM, -N, -O, -P, amtA, and phtQ are all part of a single operon. To analyze the operon structure downstream of phtQ, confirm the structure suggested by the expression experiments, and solve the uncertainties remaining, we conducted reverse transcription-PCR experiments.
Reverse transcription-PCR analysis.
We conducted experiments in which we tried to amplify specific fragments derived from cDNA of contiguous ORFs to investigate whether these were being transcribed from the same mRNA. In every case, we included as a control the amplification within a single pht ORF besides the amplification of the 23S rRNA operon. The results are shown graphically in Fig. 3A. Primers spanning argK and phtA did not produce any amplicon, whereas amplification from argK produced the expected fragment in the same reaction, which confirms that argK and phtA belong to different transcription units.
FIG. 3.
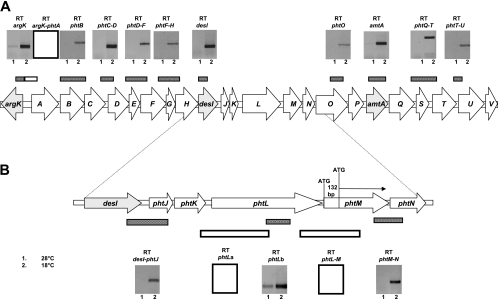
Reverse transcription-PCR of the Pht cluster of P. syringae pv. phaseolicola NPS3121. (A) Each amplification result is shown and labeled with the amplicon name. Also, each amplicon obtained is illustrated graphically as bars above the Pht cluster. (B) Detail of the analysis of the desI-phtN region. Negative results are shown for clarity as empty boxes or empty bars. The small numbers under the result boxes represent the temperature at which expression was assayed.
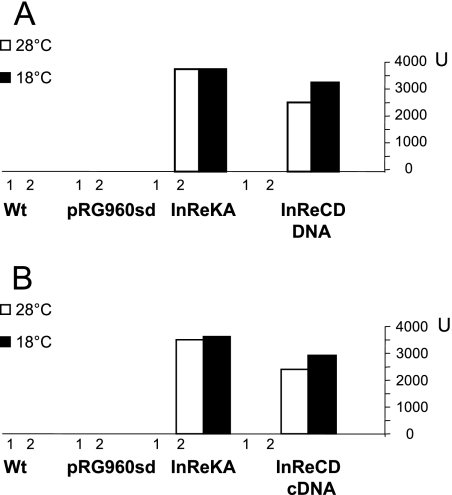
Surprisingly, amplification was also obtained using primers spanning phtC to phtD, indicating that these genes are cotranscribed, despite the observation that the intergenic region between these genes harbors a promoter sequence that, when placed in direction to phtD, could drive the expression of a promoterless uidA gene (InReCD [Fig. 1C]). To investigate this further, we performed RT-PCR from mRNA extracted from cultures grown at 18 and 28°C and ensured that this mRNA was free of contaminating DNA by incorporating controls to which we added RNase. The control reactions showed no amplification (data not shown), indicating that the reaction was free of contaminating DNA, whereas the expected cDNA band of 671 bp was obtained from the intergenic region between phtC and phtD. A sample of this cDNA was treated with EcoRV, and two fragments of 362 and 309 bp were obtained, confirming the identity of the amplified region. The 671-bp cDNA fragment was then cloned into the promoter probe plasmid pRG960sd in the orientation present in the wild-type sequence, towards phtD, and the construct was electroporated into P. syringae pv. phaseolicola NPS3121. The results obtained were almost identical to those previously obtained with the equivalent construction using InReCD (pSELF1), in which the same 671-bp fragment was amplified from chromosomal DNA (Fig. 4).
FIG. 4.
Promoter activity from the phtC to phtD intergenic region. (A) A 671-bp DNA fragment from the intergenic region between phtC and phtD was obtained through PCR using chromosomal DNA as a template and cloned upstream of a promoterless uidA gene in pRG960sd. The construct was electroporated into wild-type (Wt) P. syringae pv. phaseolicola, and promoter activity assayed as β-glucuronidase at both 18 and 28°C. (B) The same experiment was repeated but obtained was the 671-bp fragment as a cDNA amplicon produced through reverse transcription of total RNA from a culture of P. syringae pv. phaseolicola grown at 18°C in M9 medium. As controls, β-glucuronidase activity was measured from the Wt P. syringae pv. phaseolicola and P. syringae pv. phaseolicola (pRG960sd).
Amplification products were also obtained from primers spanning phtD to phtF, phtF to phtH, and internal to desI, supporting the conclusions obtained through the uidA fusion data that all these genes belonged to a single transcription unit (Fig. 3A). With respect to desI and phtJ, a primer pair was used to amplify this region by RT-PCR, which started within desI and included most of phtJ, indicating the cotranscription of these genes (Fig. 3B). However, primers starting within the 3′ of phtK and ending within phtL failed to produce amplification products (RT-phtLa) (Fig. 3B), whereas an internal fragment of phtL did produce an amplicon (RT-phtLb) (Fig. 3B). These results indicated that phtK and phtL do not belong to the same operon. Therefore, we concluded that phtK is part of the operon that begins at phtD.
There are 14 bp between the stop codon of phtL and the predicted ATG of phtM; however, no amplification product was obtained using primers spanning both genes (Fig. 3B). The expression of phtL was detected at temperatures permissive, 18°C, and nonpermissive, 28°C, for phaseolotoxin production. This pattern of expression was observed in Northern blot experiments (P13) (Fig. 1B), with the uidA fusion in cis (phtL) (Fig. 1C), and in the RT-PCR experiments (phtLb) (Fig. 3B). This expression pattern contrasts with that of phtM, which in similar experiments, is always negligible at 28°C (Fig. 1B and C and 3B). This result indicates that phtL and phtM cannot be transcribed from the same mRNA. Therefore, to explain the apparent lack of space to accommodate the promoter driving the expression of phtM, we assumed that the coding sequence could begin 51 or 132 bp downstream of the postulated ATG, at a second or third ATG codon in the same reading frame, leaving up to 146 bp to accommodate the promoter for phtM and having phtL as a monocistronic transcriptional unit.
In agreement with the fusion data, amplification was obtained from phtMN, phtO, amtA, phtQRST, and phtTU, indicating that most likely all of the genes from phtM to at least phtU comprise a single transcriptional unit (Fig. 3A).
Taken together, all these results suggest that there are five transcriptional units (Fig. 5), two of which are monocistronic, argK and phtL. Additionally, there is a promoter driving the expression of a large operon that includes 11 genes, from phtA to phtK, with an internal promoter immediately downstream of phtC capable of driving expression of phtD to phtK, and a further transcriptional unit that includes 10 genes, phtM to phtV.
FIG. 5.
Proposed operon structure of the Pht cluster of P. syringae pv. phaseolicola. The Pht cluster contains 23 genes arranged into five main transcriptional units, including two monocistronic (argK and phtL) and three polycistronic operons, one comprising 11 genes from phtD to phtK and a second large polycistronic operon comprising 10 genes from phtM to phtV. There is a secondary promoter capable of driving the expression of genes from phtD to phtK. The polycistronic operons are named after the first gene of the operon.
Mapping the site of transcription initiation for each operon predicted.
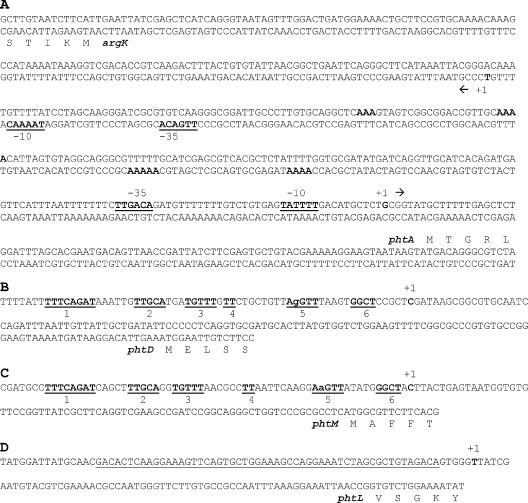
To determine the exact site of transcription of the four putative promoters, PA, PD, PL, and PM, the 5′ ends of the corresponding mRNAs were obtained through 5′ cRACE. Ten clones containing the cDNA from the 5′ end of the mRNA from each putative transcriptional unit were randomly selected among a large number of clones obtained, and these were found to have similar restriction patterns. Five clones from each 5′ cRACE experiment were then sequenced, and the results indicated the same site of transcription initiation in all cases. The results are shown in Fig. 6. The promoter for argK has been previously determined (10) and shown to be a Pribnow-type (σ70) promoter with appropriate −10 and −35 regions (41). The initiation of transcription that was determined for phtA also shows a well-conserved −10 and −35 region characteristic of a Pribnow-type promoter, and these divergent promoters show two A-rich regions on both strands that could be implicated in polymerase binding (Fig. 6A) (1).
FIG. 6.
Promoter regions of the Pht cluster. (A) Nucleotide sequence of the argK-phtA region showing the proposed −10 and −35 promoter region for argK (10) and for phtA as determined in this work. The double-stranded sequence is shown to facilitate the identification of relevant sequences on both strands as the promoters are divergent. A-rich regions that may be involved in polymerase binding are shown in bold type. (B and C) Putative promoter regions of phtD (B) and phtM (C). Six DNA regions common to both promoters are shown in bold type and underlined. (D) Promoter region of phtL. Underlined type indicates the putative RNA polymerase binding site. In all cases, the position of the first nucleotide to be transcribed is labeled as +1 and shown in bold type.
The promoter regions for phtD, phtM, and phtL do not show similarity to any of the known eubacterial consensus sequences identified as targets of known σ factors (41) or of the HrpL σ factor (8). However the promoter region of phtD and phtM share at least six obvious conserved regions within 60 bp upstream of the site of transcription initiation, suggesting that both promoters are under a common mechanism of transcriptional regulation. None of the promoters identified show, within the predicted polymerase binding region, the TRR elements previously postulated to be involved in thermoregulation of phaseolotoxin synthesis (30, 31), although these TRR sequences could possibly be located outside this region to exert their effects.
Transcriptional analysis of a phtL mutant.
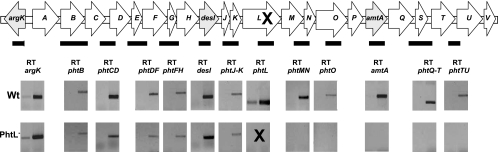
It was interesting to note that the expression of phtL occurred at 28°C at a level significantly above that observed for the other pht genes. The observation that transcripts of phtL were present at 28°C, a temperature nonpermissive for phaseolotoxin synthesis, suggested that the gene product of phtL could have a role in regulation, such as being a temperature sensor and/or signal transducer. Consistent with this role, the protein from this gene should be present at 28°C and a mutation in this gene should result in a Tox− phenotype because perhaps the temperature change would not be detected or this information would not be relayed to the cognate effector. To investigate this further, we analyzed the transcription pattern of the Pht cluster in a phtL insertion mutant through RT-PCR. Consistent with the idea that the PhtL product may be involved in regulation, all of the transcripts of the phtMV operon were clearly absent at 18°C, contrasting with the situation observed in the wild-type strain (Fig. 7).
FIG. 7.
Phenotype of a phtL-null mutant. The Pht cluster is shown diagrammatically, and the bar under the cluster represents the amplicons obtained through RT-PCR to assess the transcription activity from different pht genes. Results of the RT-PCR amplifications for the wild-type strain are shown on top of those obtained for the phtL-null mutant (indicated by an X). Negative results are shown as empty gels for clarity. Numbers indicate the temperature at which bacteria was grown for mRNA extraction.
DISCUSSION
We have sequenced a 30,245-bp region from the chromosome of P. syringae pv. phaseolicola NPS3121 which includes argK, whose product is a phaseolotoxin-resistant OCTase (10, 26); amtA, which codes for a l-arginine:lysine amidinotransferase involved in the synthesis of ornithine and homoarginine, both precursors in the synthesis of phaseolotoxin (12); and the desI gene coding for a fatty acid desaturase (11). A comparison of this sequence to the corresponding region on the chromosome of P. syringae pv. phaseolicola 1448A resulted in only 35 nucleotide differences. Of these, the most significant difference is the presence of a 9-bp sequence in NPS3121 which is missing within locus PSPPH4306 of 1448A. This introduces a stop codon, and therefore, NPS3121 presents two ORFs at this locus instead of one. We resequenced the region for both strains with the same result, indicating that, in fact, locus PSPPH4306 codes for a larger polypeptide that represents a fused protein of the two ORFs found in NPS3121, as instances of fused genes are not uncommon in bacterial genomes (36). The presence of insertion sequences and related transposases at both ends of the Pht cluster, together with the differences in G+C content, 58% for the chromosome of 1448A and 51.9% for the Pht cluster, agrees with the proposal that this entire region has been acquired by horizontal transfer (16, 34, 35). Also, the observation that the mutation of the sir2 homolog located outside the transposase/insertion sequence border at the argK end of the cluster does not affect phaseolotoxin production may indicate the boundary of the transferred region. Whether the presence of a complete sir2 gene at one end of the Pht cluster and a truncated sir2 gene at the other end adjacent to transposase sequences represents a specific target for the insertion of the cluster remains to be determined.
The fact that insertion mutants in 11 different genes of the cluster, from phtA to phtT, result in Tox− phenotypes clearly indicates that all genes within this region encode proteins that are required at any of the different stages of phaseolotoxin production, such as synthesis, transport, and regulation. With respect to the transcriptional organization of the Pht cluster, argK has been reported to be a monocistronic transcriptional unit (10, 26) and our results show that there are four other regions where distinct promoter activity was observed by the use of fusions to a promoterless uidA gene either in cis or in trans. The initiation of the transcription for each promoter has been determined, indicating that the 23 genes of the Pht cluster are organized in five transcriptional units, two being monocistronic, argK and phtL, with a large complex polycistronic operon comprising 11 genes, from phtA to phtK, harboring an internal promoter upstream of phtD and another large operon comprising genes from phtM to phtV.
It is interesting to note that the largest operon, comprising 11 genes from phtA to phtK, presents an internal promoter, a feature observed in operons harboring genes for the production of secondary metabolites, such as antibiotics (5, 39), or for the expression of gene regulators, such as the ntrBC two-component regulatory system involved in regulating different aspects of nitrogen metabolism in gram-negative bacteria (19).
We have shown that the promoter driving the expression of phtABC is a Pribnow-type promoter very similar to the divergent promoter of argK and that both share an interpromoter region with distinct sequence features that suggest coordinate regulation. It is interesting to note that while trying to obtain evidence for a regulatory role for the phtL gene product, we observed that a null phtL mutant completely abolishes the transcription of the phtM operon, consistent with a regulatory role for the phtL gene product, with no effect on any of the other transcriptional units (Fig. 7). However, our findings indicate that both the phtD and pthM operons are transcribed from very similar promoters, suggesting a common mechanism of regulation. The fact that transcripts from the phtD operon are present in the phtL mutant may indicate that transcription is proceeding from the phtA promoter generating a transcript from phtA to phtK and overriding the effect of the phtL mutation. This in turn suggests that PhtL does not have an effect on the regulation of the argK or phtA promoters.
Therefore, we propose the Pht cluster to be composed of five transcriptional units, which we have named after the gene immediately downstream of the promoter region and presented in the structure depicted in Fig. 5.
We are presently investigating how these promoters are being regulated and the particular sequence of the phtL promoter as well as which type of σ factor is involved in recognition of the phtD, -M, and -L promoters. Also, we are currently investigating the precise role of the phtL gene product. The identity and role of most of the gene products of the Pht cluster remain to be determined as it is still uncertain which genes participate in regulation and in the biosynthesis of the inorganic moiety and the tripeptide composing phaseolotoxin.
As we have shown conclusively that all the genes within the Pht cluster (with the sole exception of phtV) are expressed and have a role in phaseolotoxin production, we propose the use of the nomenclature used in this article for further reference to these genes until more information is obtained for the proteins they encode and specific names can be assigned.
Acknowledgments
The work reported was funded by grants from CONACYT to A. Alvarez-Morales (Research grant) and S. Aguilera (graduate student scholarship) and from the Ministerio de Educación y Ciencia (AGL2004-03143) to J. Murillo.
Footnotes
Published ahead of print on 19 January 2007.
REFERENCES
- 1.Aiyar, S. E., R. L. Gourse, and W. Ross. 1998. Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase α subunit. Proc. Natl. Acad. Sci. USA 95:14652-14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Chen, W.-P., and T.-T. Kuo. 1993. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 21:2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Distler, J., A. Ebert, K. Mansouri, K. Pissowotzki, M. Stockmann, and W. Piepersberg. 1987. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 15:8041-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferguson, A. R., and J. S. Johnston. 1980. Phaseolotoxin: chlorosis, ornithine accumulation and inhibition of ornithine carbamoyltransferase in different plants. Physiol. Plant Pathol. 16:269-275. [Google Scholar]
- 7.Ferguson, A. R., J. S. Johnston, and R. E. Mitchell. 1980. Resistance of Pseudomonas syringae pv. phaseolicola to its own toxin, phaseolotoxin. FEMS Microbiol. Lett. 7:123-125. [Google Scholar]
- 8.Fouts, D. E., R. B. Abramovitch, J. R. Alfano, A. M. Baldo, C. R. Buell, S. Cartinhour, A. K. Chatterjee, M. D. Ascenzo, M. L. Gwinn, S. G. Lazarowits, N.-C. Lin, G. B. Martin, A. H. Rehm, D. J. Schneider, K. V. Dijk, X. Tang, and A. Collmer. 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. USA 99:2275-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goss, R. W. 1970. The relation of temperature to common and halo blight of beans. Phytopathology 30:258-264. [Google Scholar]
- 10.Hatziloukas, E., and N. J. Panopoulos. 1992. Origin, structure and regulation of argK, encoding the phaseolotoxin-resistant carbamoyltransferase in Pseudomonas syringae pv. phaseolicola, and functional expression of argK in transgenic tobacco. J. Bacteriol. 174:5895-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatziloukas, E., N. J. Panopoulos, S. Delis, D. E. Prosen, and N. W. Schaad. 1995. An open reading frame in the approximately 28-kb tox-argK gene cluster encodes a polypeptide with homology to fatty acid desaturases. Gene 166:83-87. [DOI] [PubMed] [Google Scholar]
- 12.Hernández-Guzmán, G., and A. Alvarez-Morales. 2001. Isolation and characterization of the gene coding for the amidinotransferase involved in the biosynthesis of phaseolotoxin in Pseudomonas syringae pv. phaseolicola. Mol. Plant-Microbe Interact. 14:545-554. [DOI] [PubMed] [Google Scholar]
- 13.Jahn, O., J. Sauerstein, and G. Reuter. 1987. Characterization of two ornithine carbamoyltransferases from Pseudomonas syringae pv. phaseolicola, the producer of phaseolotoxin. Arch. Microbiol. 147:174-178. [DOI] [PubMed] [Google Scholar]
- 14.Jahn, O., J. Sauerstein, and G. Reuter. 1985. Detection of two ornithine carbamoyltransferases in a phaseolotoxin-producin strain of Pseudomonas syringae pv. phaseolicola. J. Basic Microbiol. 25:543-546. [DOI] [PubMed] [Google Scholar]
- 15.Jefferson, R. A. 1987. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5:387-405. [Google Scholar]
- 16.Joardar, V., M. Lindeberg, R. W. Jackson, J. Selengut, R. Dodson, L. M. Brinkac, S. C. Daugherty, R. DeBoy, D. A. S., M. G. Giglio, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. Sullivan, Crabtree, T. Creasy, T. Davidsen, D. H. Haft, N. Zafar, L. Zhou, R. Halpin, T. Holley, H. Khouri, T. Feldblyum, O. White, C. M. Fraser, A. K. Chatterjee, S. Cartinhour, D. J. Schneider, J. Mansfield, A. Collmer, and C. R. Buell. 2005. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187:6488-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langley, D. B., M. D. Templeton, B. A. Fields, R. E. Mitchell, and C. A. Collyer. 2000. Mechanism of inactivation of ornithine transcarbamoylase by Nδ-(N′-sulfodiaminophosphinyl)-l-ornithine, a true transition state analogue? J. Biol. Chem. 275:20012-20019. [DOI] [PubMed] [Google Scholar]
- 18.Maruyama, I. N., T. L. Rakow, and H. I. Maruyama. 1995. cRACE: a simple method for identification of the 5′ end of mRNAs. Nucleic Acids Res. 23:3796-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merrick, M. J., and R. A. Edwards. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59:604-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metcalf, W. W., and B. L. Wanner. 1993. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene 129:17-25. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell, R. E. 1978. Halo blight of beans: toxin production by several Pseudomonas phaseolicola isolates. Physiol. Plant Pathol. 13:37-49. [Google Scholar]
- 22.Mitchell, R. E. 1976. Isolation and structure of a chlorosis-inducing toxin of Pseudomonas phaseolicola. Phytochemistry 15:1941-1947. [Google Scholar]
- 23.Mitchell, R. E., and R. L. Bieleski. 1977. Involvement of phaseolotoxin in halo blight of beans. Transport and conversion to functional toxin. Plant Physiol. 60:723-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell, R. E., J. S. Johnston, and A. R. Ferguson. 1981. Phaseolotoxin and other phosphosulphanyl compounds: biological effects. Physiol. Plant Pathol. 19:227-235. [Google Scholar]
- 25.Moore, R. E., W. P. Niemczura, O. C. H. Kwok, and S. S. Patil. 1984. Inhibitors of ornithine carbamoyltransferase from Pseudomonas syringae pv. phaseolicola. Tetrahedron Lett. 25:3931-3934. [Google Scholar]
- 26.Mosqueda, G., G. Van den Broeck, O. Saucedo, A.-M. Bailey, A. Alvarez-Morales, and L. Herrera-Estrella. 1990. Isolation and characterization of the gene from Pseudomonas syringae pv. phaseolicola encoding the phaseolotoxin-insensitive ornithine carbamoyltransferase. Mol. Gen. Genet. 222:461-466. [DOI] [PubMed] [Google Scholar]
- 27.Nüske, J., and W. Fritsche. 1989. Phaseolotoxin production by Pseudomonas syringae pv. phaseolicola: the influence of temperature. J. Basic Microbiol. 29:441-447. [DOI] [PubMed] [Google Scholar]
- 28.Peet, R. C., P. B. Lindgren, D. K. Willis, and N. J. Panopoulos. 1986. Identification and cloning of genes involved in phaseolotoxin production by Pseudomonas syringae pv. “phaseolicola.” J. Bacteriol. 166:1096-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peet, R. C., and N. J. Panopoulos. 1987. Ornithine carbamoyltransferase genes and phaseolotoxin immunity in Pseudomonas syringae pv. phaseolicola. EMBO J. 6:3585-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowley, K. B., D. E. Clements, M. Mandel, T. Humphreys, and S. S. Patil. 1993. Multiple copies of a DNA sequence from Pseudomonas syringae pathovar phaseolicola abolish thermoregulation of phaseolotoxin production. Mol. Microbiol. 8:625-635. [DOI] [PubMed] [Google Scholar]
- 31.Rowley, K. B., R. Xu, and S. S. Patil. 2000. Molecular analysis of thermoregulation of phaseolotoxin-resistant ornithine carbamoyltransferase (argK) from Pseudomonas syringae pv. phaseolicola. Mol. Plant-Microbe Interact. 13:1071-1080. [DOI] [PubMed] [Google Scholar]
- 32.Saettler, A. W., N. W. Schaad, and D. A. Roth (ed.). 1989. Detection of bacteria in seed and other planting material. APS Press, St. Paul, MN.
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Sawada, H., S. Kanaya, M. Tsuda, F. Suzuki, K. Azegami, and N. Saitou. 2002. A phylogenomic study of the OCTase genes in Pseudomonas syringae pathovars: the horizontal transfer of the argK-tox cluster and the evolutionary history of OCTase genes on their genomes. J. Mol. Evol. 54:437-457. [DOI] [PubMed] [Google Scholar]
- 35.Sawada, H., F. Suzuki, I. Matsuda, and N. Saitou. 1999. Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. J. Mol. Evol. 49:6244. [DOI] [PubMed] [Google Scholar]
- 36.Serres, M. H., and M. Riley. 2005. Gene fusions and gene duplications: relevance to genomic annotation and functional analysis. BMC Genomics 6:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staskawicz, B. J., and N. J. Panopoulos. 1980. Phaseolotoxin transport in Escherichia coli and Salmonella typhimurium via the oligopeptide permease. J. Bacteriol. 142:474-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staskawicz, B. J., and N. J. Panopoulos. 1979. A rapid and sensitive assay for phaseolotoxin. Phytopathology 69:663-666. [Google Scholar]
- 39.Tieleman, L. N., G. P. Van Wezel, M. J. Bibb, and B. Kraal. 1997. Growth phase-dependent transcription of the Streptomyces ramocissimus tuf1 gene occurs from two promoters. J. Bacteriol. 179:3619-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van den Eede, G., R. Deblaere, K. Goethals, M. Van Montagu, and M. Holsters. 1992. Broad host range and promoter selection vectors for bacteria that interact with plants. Mol. Plant-Microbe Interact. 5:228-234. [DOI] [PubMed] [Google Scholar]
- 41.Wösten, M. M. S. M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127-150. [DOI] [PubMed] [Google Scholar]
- 42.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, Y., K. B. Rowley, and S. S. Patil. 1993. Genetic organization of a cluster of genes involved in the production of phaseolotoxin, a toxin produced by Pseudomonas syringae pv. phaseolicola. J. Bacteriol. 175:6451-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]