Abstract
The M13 phage assembles in the inner membrane of Escherichia coli. During maturation, about 2,700 copies of the major coat protein move from the membrane onto a single-stranded phage DNA molecule that extrudes out of the cell. The major coat protein is synthesized as a precursor, termed procoat protein, and inserts into the membrane via a Sec-independent pathway. It is processed by a leader peptidase from its leader (signal) peptide before it is assembled onto the phage DNA. The transmembrane regions of the procoat protein play an important role in all these processes. Using cysteine mutants with mutations in the transmembrane regions of the procoat and coat proteins, we investigated which of the residues are involved in multimer formation, interaction with the leader peptidase, and formation of M13 progeny particles. We found that most single cysteine residues do not interfere with the membrane insertion, processing, and assembly of the phage. Treatment of the cells with copper phenanthroline showed that the cysteine residues were readily engaged in dimer and multimer formation. This suggests that the coat proteins assemble into multimers before they proceed onto the nascent phage particles. In addition, we found that when a cysteine is located in the leader peptide at the −6 position, processing of the mutant procoat protein and of other exported proteins is affected. This inhibition of the leader peptidase results in death of the cell and shows that there are distinct amino acid residues in the M13 procoat protein involved at specific steps of the phage assembly process.
M13 bacteriophage infects a bacterial cell by binding to the tip of an F pilus. After binding, the pilus is retracted and disassembled in the inner membrane, leaving the phage DNA in the cytoplasm. The single-stranded circular DNA is rapidly converted into a double strand and replicated for continuous production of progeny phage. Phage-directed protein synthesis is dedicated mainly to expression of the five structural coat proteins, the products of genes III, VI, VII, VIII, and IX. All these coat proteins are inserted into the bacterial inner membrane, where the assembly of progeny particles occurs. The assembly of a particle starts with the formation of the coat cap structure consisting of the VII and IX gene products and is followed by assembly of the major coat protein, gpVIII. On the phage particle, units of five proteins cover the DNA in a shingle-like arrangement (4, 23). The integrity of the phage particle is based on strong protein-protein interactions and electrostatic protein-DNA interactions.
During the biogenesis of bacteriophage M13, the membrane-inserted major coat protein is processed to the mature form by the leader peptidase before it is assembled onto the extruding single-stranded phage DNA. The precursor protein, termed the procoat protein, spans the membrane twice, exposing a negatively charged region in the periplasm. The signal sequence is required for the membrane insertion process, although another filamentous phage has a coat protein without a signal sequence (29). The membrane insertion process is independent of SecA and the SecYEG translocase, but it requires the membrane insertase YidC and an electrochemical membrane potential (11, 31). The processed and membrane-integrated major coat protein consisting of 50 amino acid residues includes a 20-residue negatively charged region in the periplasm, a 19-residue membrane-spanning region, and an 11-residue positively charged C-terminal region in the cytoplasm.
The assembly of the progeny phage particle takes place at the inner membrane of Escherichia coli. The products of the I, III, VI, VII, VIII, IX, and XI genes are integrated into the inner membrane, whereas gpIV forms a complex consisting of 10 to 12 subunits in the outer membrane (16), allowing passage of the nascent phage particle through the outer membrane. The assembly of the phage is initiated by the VII and IX proteins that bind to the “morphogenic signal” of the newly replicated single-stranded DNA. During the phage assembly process, the coat protein (the VIII protein) participates by binding the positively charged C-terminal region to the single-stranded DNA (5, 7). The hydrophobic region of the coat protein is in an α-helical conformation in the membrane and in the phage particle, whereas the adjacent regions have to undergo a conformational change. Remarkably, the tilt of the helix axis of the coat protein in the membrane remains similar in the phage, where the protein is tilted with respect to the axis of the virion (20, 22).
One open question in the assembly process is how the coat protein binds onto the extruding single-stranded DNA molecule (21). It is possible that coat proteins form multimers in the membrane that bind in a concerted fashion on the DNA, such that lipid molecules are excluded. This implies that the coat proteins interact in the membrane with each other to form oligomeric complexes that are ready to bind onto the DNA molecule. Previously, Haigh and Webster (6) introduced single cysteine residues at positions 31, 35, 37, and 39 of the coat protein. The mutant with a mutation at position 31 was able to form dimers when it was extracted from phage, but it was monomeric when it was extracted from the cytoplasmic membrane. However, the mutant with a mutation at position 35 was found to be partially dimeric in the membrane, but the protein was not capable of assembling progeny particles. Since only a single cysteine had been introduced into the coat proteins, higher-order oligomers could not be detected. In addition, the membrane environment might be too reductive to observe existing protein-protein interactions.
We addressed this question by introducing cysteine residues at positions 30, 31, 32, and 33 into the transmembrane region of the coat protein. When only a single cysteine residue was present in the procoat protein, phage multiplication was normal for proteins with mutations at most positions. We found that the coat proteins were able to form dimers readily in the membrane when the cells were treated with copper phenanthroline (CuP). When the coat protein contained two cysteine residues, oligomeric forms were found, suggesting that the oligomeric forms accumulated in the membrane ready to assemble onto phage particles. We describe a model showing how the oligomeric coat proteins are loaded onto the extruding phage particles from the membrane.
MATERIALS AND METHODS
Plasmid construction.
Plasmid pJF119HE (3) was used to clone the VIII gene of M13 phage and was used as a template for site-specific mutagenesis (10). All mutations were verified by DNA sequencing.
Strains, phage, and growth conditions.
E. coli K38 (HfrC T2R relA1 pit-10 spoT1 tonA22 ompF627 phoA4 λ−) (18) was used as a host for M13 phage. To study complementation of plasmid-derived coat expression, a phage with an amber mutation at position 2 in gene VIII (M13am8-M2) was used. For some protein expression experiments strain BL-21 [hsd6 gal(λcIts857) ind1 Sam7 nin5 lacUV5-T7gene1] (32) was used. Media were prepared and bacterial manipulations were performed by using standard methods (19). When appropriate, ampicillin (final concentration, 200 μg/ml) was added to the medium.
For the complementation experiments, 103 or 105 M13am8-M2 phage particles were mixed with E. coli K38 cells and top agar and applied to a plate. In some cases (see below), 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added. The plates were incubated at 37°C overnight. The efficiency of plating (EOP) was calculated by comparison with the plaque formation by the wild-type phage. Multiple platings resulted in a standard deviation for each value of less than 20%.
For procoat protein expression, the plasmid-bearing cells were grown overnight at 37°C and back-diluted 1/100. At a density of 2 × 108 cells per ml, 1 mM IPTG was added, and the culture was grown for 4 h (or for the time indicated below) at 37°C. To analyze the procoat protein and coat protein, one portion of the culture was treated with 1 mM CuP for 10 min (or for the time indicated below). The 50 mM CuP stock solution was freshly prepared essentially as described previously (9) by mixing 225 mM 1,10-phenanthroline monohydrate in ethanol and 150 mM CuSO4 at a ratio of 2:1 (vol/vol). The reaction was stopped with 10 mM EDTA and 10 mM N-ethylmaleimide (NEM). The samples were precipitated with 10% trichloroacetic acid and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Membrane isolation.
One-liter cultures were grown as described above, cells were harvested by centrifugation, and the pellets were resuspended in 6 ml of 50 mM Na phosphate-1 mM EDTA. The cells were lysed by adding 0.2 mg/ml lysozyme for 1 h on ice following ultrasonication (three 15-s pulses). After low-speed centrifugation (12,000 × g for 10 min at 4°C) the membranes were pelleted by centrifugation at 350,000 × g for 10 min at 4°C and resuspended in 1 ml of 50 mM Na phosphate-1 mM EDTA. For oxidation, 1 mM CuP was added for different times at 37°C, and the reaction was stopped by adding 10 mM EDTA and 10 mM NEM on ice. Rereduction was performed by adding 0.1 M dithiothreitol (DTT) after the reaction was stopped. The samples were precipitated with 10% trichloroacetic acid and subjected to SDS-PAGE.
SDS-PAGE.
For reductive SDS-PAGE, the samples were incubated in sample buffer containing 0.1 M DTT at 95°C for 3 min; for nonreducing gels no DTT was added to the sample buffer, and the samples were incubated at 40°C for 30 min prior to electrophoresis. A 22% SDS-polyacrylamide gel was prepared as described previously (8). For Western blot analysis the proteins were transferred to nitrocellulose (Amersham) and incubated for 150 min at 4°C with serum to M13 coat protein (diluted 1:5,000 in 20 mM Tris-HCl[pH 7.5]-150 mM NaCl), followed by peroxidase-linked antibody to rabbit immunoglobulin G (1:5,000 dilution) for 90 min. The reaction was visualized by enhanced chemoluminescence detection (Amersham).
Pulse-labeling experiments and protein mapping.
E. coli K38 cells were grown overnight in minimal medium lacking methionine and diluted 1/100 into fresh minimal medium. After 2 h of growth the cells were induced with 1 mM IPTG and pulse-labeled with [35S]methionine (10 μCi/ml) for different times. To generate spheroplasts, the pulse-labeled cells were collected by centrifugation at 4°C and resuspended in 40% sucrose-33 mM Tris-HCl (pH 8.0). Lysozyme (5 μg/ml) and 1 mM EDTA (pH 8.0) were added, and the preparations were kept on ice for 20 min. In some cases, proteinase K was added (0.5 mg/ml) and the preparations were incubated on ice for 1 h. Immunoprecipitation was performed as previously described (12). The samples were acid precipitated and analyzed by SDS-PAGE.
RESULTS
Cysteine mutations in the M13 procoat protein allow normal phage multiplication.
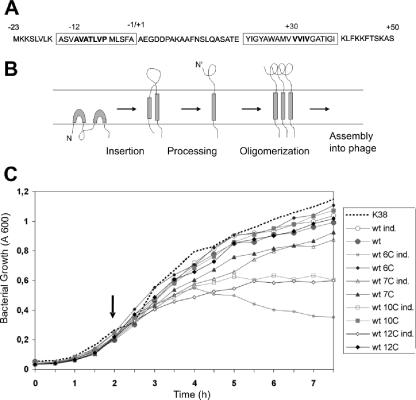
To study whether an intramolecular disulfide bond between the leader and the mature region of the M13 procoat protein affects membrane insertion, processing by the leader peptidase, or phage assembly, we introduced single TGC or TGT codons encoding cysteine residues at positions −6 to −12 in the leader sequence and at positions 30 to 33 in the mature sequence using site-directed mutagenesis (Fig. 1A). These codons were introduced into an IPTG-inducible plasmid in the cloned VIII gene encoding the procoat protein. E. coli K38 cells bearing the plasmids were then infected with M13 phage harboring an amber mutation in the VIII gene (M13am8-M2). The phage multiplied, forming plaques on plates, only when a functional plasmid-derived coat protein was expressed (Table 1). In the absence of induction or in the absence of the gene, less than 10−2 plaque per infecting particle was formed as a result of revertants of the amber mutation or low-level protein expression. When expression of the plasmid-encoded wild-type protein was induced, phage progeny were assembled, and the EOP was about 1. Similar efficiencies were observed (EOP, 0.6 to 0.8) for the mutants having procoat proteins with single cysteine residues at positions −7, −8, −9, −11, 30, 31, 32, and 33. Slightly lower EOP were observed for the mutants with proteins with a cysteine at position −10 or −12. Except for the mutant protein with a cysteine at position −6, all single-cysteine mutant proteins allowed productive phage multiplication.
FIG. 1.
M13 procoat protein cysteine mutants. (A) Amino acid sequence of the M13 procoat protein. Bold type indicates the positions at which the amino acid was changed to a single cysteine residue. The leader sequence is numbered from −23 to −1, and the mature region is numbered from +1 to +50. The boxes indicate the membrane-spanning regions. (B) Membrane insertion and assembly pathway of the M13 procoat protein. (C) Expression of the cloned procoat protein from plasmids bearing single-cysteine mutants affects the growth of E. coli K38. Exponentially growing cultures were induced (ind.) (open symbols) at 2 h (arrow), and the optical density at 600 nm was monitored. Uninduced cultures (solid symbols) were used as controls. The results for wild-type procoat protein (wt), mutant −6C, mutant −7C, mutant −10C, and mutant −12C are shown. The results for a culture without a plasmid are indicated by the dotted line.
TABLE 1.
Single-cysteine mutants affecting the efficiency of plating of M13am8-M2 on E. coli K38
| Plasmid | Protein | IPTG | EOP |
|---|---|---|---|
| pJF119 | None | + | <0.001 |
| pJQ-8 | Wild type | − | 0.005 |
| pJQ-8 | Wild type | + | 1 |
| pJQ-8L6C | −6C | + | Lethal |
| pJQ-8L7C | −7C | + | 0.77 |
| pJQ-8L8C | −8C | + | 0.73 |
| pJQ-8L9C | −9C | + | 0.63 |
| pJQ-8L10C | −10C | + | 0.49 |
| pJQ-8L11C | −11C | + | 0.81 |
| pJQ-8L12C | −12C | + | 0.44 |
| pJQ-8M30C | 30C | + | 0.62 |
| pJQ-8M31C | 31C | + | 0.79 |
| pJQ-8M32C | 32C | + | 0.71 |
| pJQ-8M33C | 33C | + | 0.80 |
Bacterial growth inhibition after induction of cysteine mutants with mutations at positions −6, −10, and −12.
To investigate why the single-cysteine mutants having procoat proteins with cysteine residues at positions −6, −10, and −12 exhibited low complementation efficiencies for the amber VIII phage, we monitored the growth of the induced and noninduced bacterial cultures in the absence of phage (Fig. 1C). The growth of the wild type and all the other cysteine mutants was not affected when the coat protein was induced. However, when expression of the cysteine mutants with mutations at positions −6, −10, and −12 was induced, cell growth was clearly affected. Two hours after induction the mutants with mutations at positions −10 and −12 showed a bacteriostatic reaction, whereas with the cysteine mutant with a mutation at position −6 lysis was observed. This explains why in these cases we observed lower levels of phage production (Table 1).
Cysteine-containing mutant procoat proteins are inserted into the membrane.
To investigate whether the membrane insertion or processing by the leader peptidase of the single-cysteine mutants was affected, expression of the mutant coat proteins was induced for 2 h and analyzed by Western blotting (Fig. 2). Most of the single-cysteine mutant proteins were expressed well and processed efficiently; the only exception was the −6C mutant. It is well known that the proline at position −6 in the wild-type protein is important for recognition by the leader peptidase (15). We concluded that the single cysteine residues at all the other positions do not have an inhibitory effect on the membrane insertion and cleavage of the M13 coat protein.
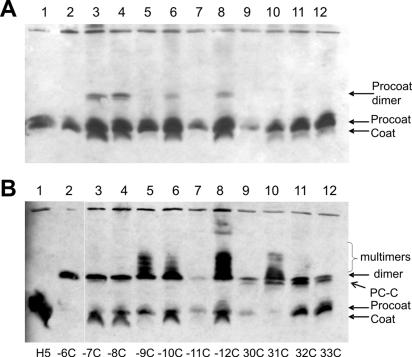
FIG. 2.
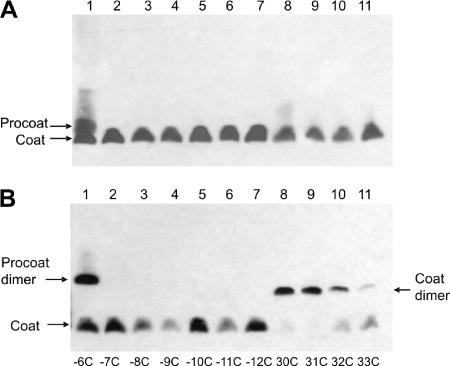
Processing and dimerization of M13 single-cysteine mutant procoat proteins. (A) E. coli BL-21 cells bearing a plasmid expressing a single-cysteine mutant procoat protein was induced for 4 h, analyzed on a nonreducing SDS-PAGE gel, and visualized by Western blotting. (B) After 4 h of induction, the samples were oxidized with 1 mM copper phenanthroline and analyzed as described above. The cysteine mutant proteins with mutations at positions −6 (lane 1), −7 (lane 2), −8 (lane 3), −9 (lane 4), −10 (lane 5), −11 (lane 6), −12 (lane 7), 30 (lane 8), 31 (lane 9), 32 (lane 10), and 33 (lane 11) were analyzed.
The cysteine mutant proteins were monomers on a nonreducing gel (Fig. 2A). Treatment of the cells with copper phenanthroline changed the membrane to an oxidative milieu. This allowed efficient disulfide bond formation between two cysteine residues located close to each other in the mature regions, which led to dimeric structures (Fig. 2B). The efficiency of disulfide formation was highest for the cysteine residues at positions 30 and 31, suggesting that these residues are exposed and interact with each other. Cysteine residues at positions 32 and 33 produced dimers with efficiencies of only 64 and 18%, respectively. As expected, only the mature coat proteins with cysteine residues could oligomerize, whereas the proteins that had a cysteine in the signal sequence and no cysteine in the mature region (Fig. 2B, lanes 2 to 7) remained monomers. The −6C mutant, however, which was only partially cleaved, produced dimers with the precursor form (Fig. 2B, lane 1). We suspected that the membrane-inserted procoat proteins might oligomerize when their cleavage is inhibited. However, when the procoat protein is cleaved normally by the leader peptidase, formation of cysteine dimers is prevented by rapid processing to coat proteins.
Procoat protein −6C inhibits the leader peptidase.
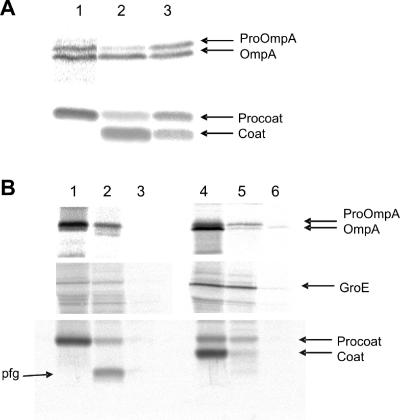
Since the growth of cells expressing the cysteine mutant proteins with mutations at positions −6, −10, and −12, was inhibited, we analyzed at which stage of the pathway the procoat protein was blocked. E. coli K38 bearing the appropriate plasmid was grown to the exponential phase, and expression of the mutant protein was induced with 1 mM IPTG. The cells grew normally without a plasmid or with a plasmid without induction. In comparison, cells expressing procoat protein −6C stopped growing 2 h after induction and started to lyse 30 min later (Fig. 1C). Similarly, growth of the cells expressing cysteine mutant proteins with mutations at positions −10C and −12C stopped 2 h after induction; however, no lysis was observed. At this time, cells expressing the mutant proteins were tested for procoat protein processing by the leader peptidase and translocation across the membrane. The processing of proOmpA was used as an indicator of leader peptidase function. The cells were labeled with [35S]methionine for 1 min, and the processing to OmpA was analyzed. Whereas cells expressing the wild-type protein (not shown) and the −7C mutant showed normal processing of proOmpA and the procoat protein (Fig. 3A, lane 2), processing of proOmpA in the cells expressing the mutants with mutations at positions −6C and −12C was affected (lanes 1 and 3). Also, the processing of the −12C procoat protein (lane 3) was inhibited. Likewise, processing of the −6C procoat protein was completely blocked (lane 1). The observed defect in processing might have been caused by enzymatic inhibition of the leader peptidase or by inhibition of membrane translocation. To test this hypothesis, we analyzed the membrane translocation of the −6C procoat protein with protease mapping (Fig. 3B, left panels). An experiment in which proteinase K was added to the periplasmic side of the inner membrane showed that most of the accumulated procoat protein was accessible and was therefore translocated (Fig. 3B, lane 2). This was also observed for most of the proOmpA protein (Fig. 3B, upper panel, lane 2) that had accumulated. After proteolysis of the −6C procoat protein, a 3-kDa fragment was immunoprecipitated, suggesting that the uncleaved procoat protein was partially protected from the added protease. This fragment was not observed in cells expressing the −7C procoat protein that was processed normally (lanes 4 to 6). Based on the molecular weight and the antigenic region of the coat (residues 2 to 8), the fragment most likely contained the region between residues −23 and 10. This suggests that the mutant protein remained bound to the leader peptidase and was protected in the region from residue −23 to residue 10, which included the antigenic region. This indicated that the activity of the leader peptidase was affected in these cells. Intriguingly, a double mutant combining the −6C mutation with the noncleavable H5 mutation at position −3, which inhibited the binding to the leader peptidase, had no lytic effect on the bacterial growth (data not shown).
FIG. 3.
Expression of procoat protein −6C inhibits the leader peptidase. E. coli K38 cells bearing a plasmid expressing procoat protein −6C, −7C, or −12C were grown in minimal M9 medium and induced for 2 h with 1 mM IPTG. [35S]methionine was added for 1 min, and the sample was immediately chilled by mixing it with ice-cold medium. (A) The cells were acid precipitated and analyzed to determine their processing of proOmpA (upper panel) and procoat protein (lower panel) by immunoprecipitation, SDS-PAGE, and phosphorimaging. The results for cells expressing procoat protein −6C (lane 1), procoat protein −7C (lane 2), and procoat protein −12C (lane 3) are shown. The positions of proOmpA, OmpA, procoat protein, and coat protein are indicated by arrows. (B) To analyze the membrane translocation of the proteins, the cells were converted to spheroplasts, and 1 mg/ml proteinase K was added to the outside of the cells for 1 h on ice (lanes 2 and 5). The results for cells expressing procoat protein −6C (lanes 1 to 3) and procoat protein −7C (lanes 4 to 6) are shown. Cells without proteinase K (lanes 1 and 4) and cells treated with TritonX-100 and proteinase K (lanes 3 and 6) were used as controls. The cytoplasmic GroE protein was analyzed to show that the spheroplasts remained intact during proteolysis.
Oligomer formation for a coat protein with two cysteine residues.
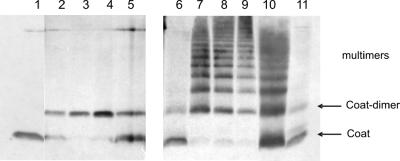
When two cysteine residues were placed into the membrane-spanning region of the procoat protein, oligomeric forms were observed (Fig. 4). The cysteine residues at positions 30 and 31 were combined in a double mutant, and the protein was analyzed using a nonreducing gel system. The coat proteins with the two cysteine residues were present mainly in the monomeric form (Fig. 4, lane 6). When the cells were treated with copper phenanthroline (lanes 7 to 9), oligomeric forms were readily observed, whereas for the single-cysteine mutant 30C, only dimers appeared (lanes 2 to 4). We concluded from these results that the coat proteins accumulate in the membrane in oligomeric clusters and that residues 30 and 31 are able to stabilize intermolecular contacts.
FIG. 4.
Oligomerization of the M13 coat protein in the membrane. E. coli BL-21 cells bearing a plasmid expressing procoat protein 30C (lanes 1 to 5) or procoat protein 30C/31C (lanes 6 to 11) were induced for 3 h (lanes 1 and 6) or were induced for 3 h and treated with 1 mM copper phenanthroline for 1 min (lanes 2 and 7), 10 min (lanes 3 and 8), or 30 min (lanes 4 and 9). For controls, the copper phenanthroline-treated sample was rereduced by adding 100 mM DTT (lanes 5 and 10) or was pretreated with 10 mM NEM and 10 mM EDTA before treatment with copper phenanthroline for 10 min (lane 11). The samples were acid precipitated and analyzed by nonreducing SDS-PAGE and Western blotting.
Cysteine procoat protein mutants can form dimers in the membrane.
To determine whether the precursor form, the procoat protein, can also form dimers, each cysteine mutant was combined with the H5 mutation, which prevented processing of the protein to its mature form. The H5 mutation replaces the serine at position −3 with a phenylalanine that interferes with leader peptidase recognition (13). Each of the mutant plasmids was transformed into E. coli BL-21, the cells were grown at 37°C to the exponential phase, and the expression of coat protein was induced for 4 h. The cells were acid precipitated and analyzed by Western blotting (Fig. 5A). To determine the exact position of the dimeric form, we genetically constructed a mutant procoat protein that included a twin fused procoat protein with 146 amino acid residues (not shown). A minor amount of H5 procoat proteins that had cysteine residues at positions −7, −8, −10, and −12 was found at the dimeric position (Fig. 5A, lanes 3, 4, 6, and 8). Intriguingly, the same mutants showed some processing to the coat protein. The procoat proteins with a cysteine residue in the mature region did not spontaneously form dimers (lanes 9 to 12).
FIG. 5.
Dimerization of H5 mutant procoat proteins. E. coli BL-21 cells bearing a plasmid expressing a single-cysteine mutant H5 procoat protein was induced for 4 h (A) or induced for 4 h and treated with 1 mM copper phenanthroline for 10 min (B). The cells were acid precipitated and analyzed by nonreducing SDS-PAGE and Western blotting. PC-C was the dimer of a coat protein and a procoat protein.
When the samples were analyzed under oxidizing conditions, most of the protein was shifted to the dimeric position (Fig. 5B). Whereas the control, the H5 procoat protein without a cysteine residue, did not shift (lane 1), all the cysteine mutants, including the −6C mutant, were present as dimers. Intriguingly, in addition to the dimer band the −9C, −10C, −12C, and 31C mutants produced higher-molecular-weight bands that most likely represented contacts with other proteins or procoat protein multimers. Since a single cysteine in a protein can participate in only one disulfide bond and thereby give rise to only one dimer (consisting of two procoat proteins), the additional bands must have been due to other interactions that might have been stabilized by disulfide bonds.
For some of the single-cysteine mutants with mutations in the mature region a portion shifted to a slightly lower position than the dimer (Fig. 5B, lanes 9 to 12). These protein bands might have represented disulfides between a procoat protein and a coat molecule (PC-C), which was possible if the cysteine residue was in the mature region. In these cases, the H5 mutation had not fully blocked the processing to coat protein.
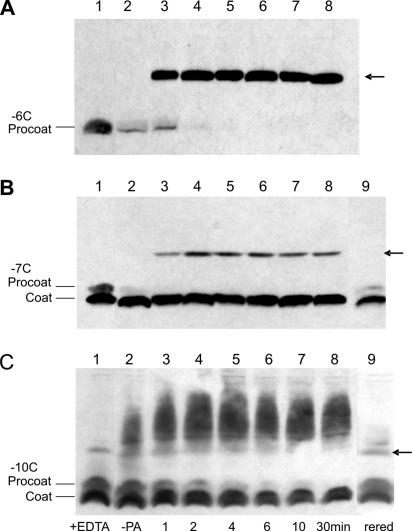
We then analyzed whether the dimers and multimers could also be found in isolated membranes. To do this, exponentially growing cells were induced for 4 h. The cells were harvested and treated with lysozyme and by sonication. The membranes were isolated by differential centrifugation. When the membranes with mutant procoat protein −6C were treated with 1 mM copper phenanthroline, dimers appeared in a time-dependent manner (Fig. 6A). Similarly, the −7C mutant showed efficient procoat protein dimer formation (Fig. 6B). In this case, a portion of the protein was cleaved to the coat protein form, possibly due to the long expression time (4 h) and the membrane isolation procedure. The resulting coat protein was not converted to a dimer and remained a monomer. With the −8C mutant, the −9C mutant (not shown), and the −10C mutant (Fig. 6C) there were some dimers in the untreated samples. This finding is similar to what we observed previously (Fig. 5A). Treatment of these mutants with copper phenanthroline resulted in the appearance of higher-molecular-weight bands and the simultaneous disappearance of the monomeric and dimeric procoat protein bands. When the treated membranes were rereduced, the higher-molecular-weight bands disappeared and the intensity of the monomeric procoat protein band increased (Fig. 6C, lane 9). In all cases, a portion of the mature coat protein remained at the monomer position since all these mutants had cysteine in the leader sequence.
FIG. 6.
Oxidation kinetics with copper phenanthroline. The formation of procoat protein dimers with different times of phenanthroline exposure was studied. E. coli BL-21 cells bearing plasmids coding for procoat proteins H5/−6C (A), H5/−7C (B), and H5/−10C (C) were induced for 4 h. The cell membrane fraction was collected and exposed to 1 mM copper phenanthroline for the times indicated at the bottom of panel C. The reaction was stopped by addition of 10 mM EDTA and 10 mM NEM. The samples were acid precipitated and analyzed by nonreducing SDS-PAGE and Western blotting. Lane 1, samples pretreated with 100 mM DTT; lane 2, untreated samples (no CuP) (−PA); lanes 3 to 8, copper phenanthroline-treated samples; lane 9, same as lane 8 but samples were rereduced (rered) with 100 mM DTT. The arrows indicate the position of the procoat protein dimer.
Procoat proteins with two cysteine residues affect phage multiplication.
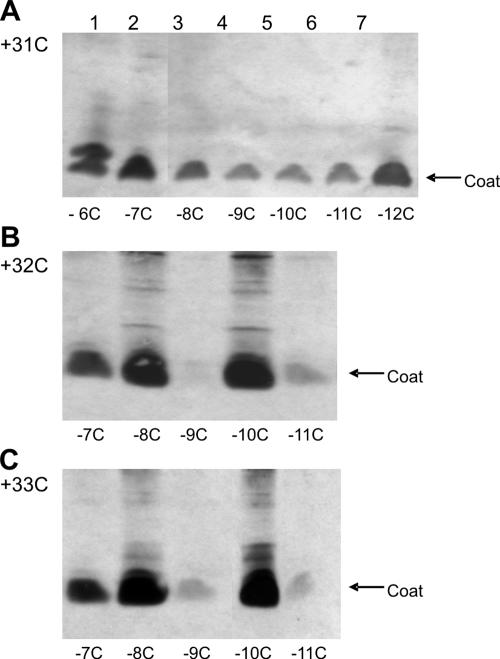
The single-cysteine mutants that supported phage assembly were combined with a second cysteine mutation (Table 2). To determine whether a possible intramolecular disulfide between the leader peptide and the mature hydrophobic region could interfere with phage multiplication, the mutants were analyzed in order to determine their abilities to support plaque formation in an M13am8-M2 infection. As shown in Table 2, most of the double-cysteine mutants allowed normal phage growth. However, the mutant proteins with a cysteine residue at position 32 and a second cysteine residue at position −7, −9, or −11 and the mutants with cysteine residues at positions 33 and −9 or −11 strongly inhibited phage growth and did not allow plaque formation. The double-cysteine mutants were tested to determine their expression and processing using SDS-polyacrylamide gels (Fig. 7). Most of the mutants migrated at the position of the coat protein, showing that the cleavage by the leader peptidase was not generally inhibited by two cysteine residues. We found that the amount of coat protein corresponded to the efficiency of phage propagation (Table 2). For the mutant proteins with one cysteine at position 32 or 33 and the second cysteine at position −9 or −11 only a weak coat protein band was visible, suggesting that these proteins were unstable. We verified in a pulse-chase experiment that these mutant proteins were processed to a mature form but were degraded (data not shown). Since the single-cysteine mutants were stable, the conformation of the double mutants might be different and defective for phage assembly. We concluded that the instability of the mutant proteins is related to the observed inhibition of phage assembly.
TABLE 2.
Double-cysteine mutants affecting the efficiency of plating of M13am8-M2 on E. coli K38
| First mutation | Second mutation | EOP |
|---|---|---|
| 30C | −7C | 0.81 |
| −8C | 0.56 | |
| −9C | 0.17 | |
| −10C | 0.43 | |
| −11C | 0.37 | |
| 31C | −7C | 0.80 |
| −8C | 0.72 | |
| −9C | 0.57 | |
| −10C | 0.65 | |
| −11C | 0.77 | |
| 32C | −7C | 0.04 |
| −8C | 0.77 | |
| −9C | <0.001 | |
| −10C | 0.21 | |
| −11C | 0.01 | |
| 33C | −7C | 0.41 |
| −8C | 0.90 | |
| −9C | 0.01 | |
| −10C | 0.77 | |
| −11C | 0.06 |
FIG. 7.
Expression and processing of procoat protein double-cysteine mutants. E. coli K38 cells bearing plasmids expressing double mutants with one cysteine residue at position 31 (A), 32 (B), or 33 (C) and a second cysteine residue at the positions indicated below the lanes were induced for 3 h, acid precipitated, and analyzed by SDS-PAGE and Western blotting.
Procoat proteins with two cysteine residues form multimers.
The double-cysteine mutants were combined with the H5 mutation to see whether the precursor proteins could form intramolecular disulfides and multimeric forms in the membrane, as observed for the coat protein (Fig. 4). It is thought that intramolecular disulfides migrate faster than the monomer (35). When the procoat protein was analyzed on a Western blot, most of the mutant procoat proteins had the same mobility (Fig. 8A). However, for mutants H5/32C-8C, H5/32C-9C, H5/32C-10C, H5/33C-8C, and H5/33C-10C some of the protein exhibited increased mobility, possibly representing some cleaved coat protein. The amount, however, was less than 10% of the amount of the procoat protein, and most of the protein was at the normal position. These mutant proteins were also partially present in dimeric forms. We concluded that these mutant procoat proteins engage in intermolecular contacts and not in intramolecular contacts under normal growth conditions.
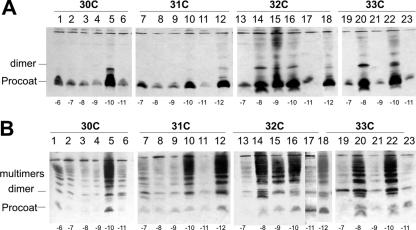
FIG. 8.
Oligomerization of H5 procoat protein double-cysteine mutants. E. coli BL-21 cells bearing plasmids expressing procoat protein H5 with two additional cysteine residues at the positions indicated below were induced for 4 h (A) or were induced for 4 h and incubated with 1 mM copper phenanthroline for 10 min (B). The samples were acid precipitated and analyzed by nonreducing SDS-PAGE and Western blotting with anti-coat protein. One cysteine residue was at position 30 (lanes 1 to 6), 31 (lanes 7 to 12), 32 (lanes 13 to 18), or 33 (lanes 19 to 23). The second cysteine was at position −6 (lane 1), −7 (lanes 2, 7, 13, and 19), −8 (lanes 3, 8, 14, and 20), −9 (lanes 4, 9, 15, and 21), −10 (lanes 5, 10, 16, and 22), −11 (lanes 6, 11, 17, and 23), or −12 (lanes 12 and 18).
The same cells were treated with copper phenanthroline to investigate whether all the procoat proteins can efficiently interact with each other under oxidative conditions. All double-cysteine mutants produced multimeric forms (Fig. 8B). This suggests that the procoat proteins were in close contact with each other, which allowed multiple disulfide bonding within the membrane under oxidizing conditions.
DISCUSSION
The assembly of the filamentous phage at the cytoplasmic membrane is a fascinating process that is still not well understood. Transmembrane coat proteins are loaded most likely as oligomers onto the single-stranded DNA and seal the virion into a tightly folded structure. To examine whether oligomers of the coat protein are formed in the membrane of E. coli prior to virus assembly, we generated a collection of mutant coat proteins that contained one or two cysteine residues in their hydrophobic domains. We anticipated that a cysteine residue in the leader sequence would result in the formation of procoat protein dimers and could block the processing step. Likewise, a cysteine in the mature region could result in the formation of coat protein dimers that could be blocked during assembly of the phage (6). Surprisingly, in general, the cysteine residues did not inhibit multiplication of the phage (Table 1). Analysis of the procoat and coat proteins localized in the cytoplasmic membrane by SDS-PAGE revealed that only a few mutant procoat proteins were present as dimers (Fig. 5A). This suggests that the membrane environment is rather reductive and does not allow disulfide bridges to form. When we treated the cells with copper phenanthroline, a hydrophobic oxidizing reagent, procoat protein dimers (Fig. 5B) and coat protein dimers were readily formed (Fig. 2B). Since this reaction occurred instantly (Fig. 6), we concluded that both the membrane-inserted procoat protein and coat protein are organized in clusters of multimers. Indeed, multimers of the procoat protein were found with mutants containing two cysteine residues (Fig. 8B), and multimers of coat protein were found when two cysteine residues were present in the mature region (Fig. 4).
Copper phenanthroline has been used previously to demonstrate close contact between two transmembrane helices. The Tar chemoreceptor of E. coli was shown to dimerize by its TM1 region but not by its TM2 region (17). Intramolecular disulfides were found for the Trg chemoreceptor, and the interacting faces of TM1 and TM2 matched on a helical wheel (14). The maximum distance between two helical axes is 7 to 12 Å, which allows the formation of a disulfide bond (1). When the disulfides are formed instantly, the two partner helices must be in close contact. When the formation of disulfides takes a longer time, the molecules may require movement within the membrane, or the two cysteine residues are shielded to react with copper phenanthroline.
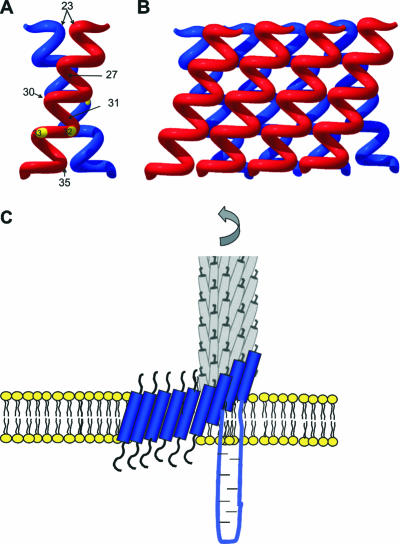
It has been shown previously that a mutant coat protein with a cysteine at position 35 forms a disulfide when it is integrated into the membrane (6). We found that the residues at positions 30 and 31 interact efficiently (Fig. 2B), whereas the residues at positions 32 and 33 are less reactive. This is consistent with the protein-interactive region that was proposed by Deber et al. (2). Such an interactive region suggests that the coat proteins in the membrane are organized in an ordered fashion and are not freely mobile. Intermolecular interaction of the hydrophobic regions is also implied by the presence of a GXXXG motif that is present at positions 34 to 38 in the transmembrane region of coat protein. The GXXXG motif has been found to promote helix-helix interactions in glycophorin A (24, 30). Taken together, the proposed contacts between two coat proteins can be summarized in a model, in which the two proteins are tilted. There are indications that such a tilted conformation of coat protein occurs in a lipid bilayer, and it has been shown that the protein is tilted about 20o with respect to the membrane normal (26). When two interacting coat proteins are tilted against each other at an angle of 40o, residues 23, 27, 31, and 35 are in close contact (Fig. 9A). The tilted helices could then interact with two additional neighboring helices, resulting in additional close contacts (Fig. 9B). As a consequence, a multimeric protein sheet structure involving more than two proteins is formed.
FIG. 9.
Model for the assembly of coat oligomers onto the nascent phage particle. (A) Two coat proteins interact, joining residues 27, 30, 31, and 35. (B) Multimeric sheets are formed. (C) The coat proteins retain their tilt and their interprotein contacts from the membrane to the viral particle.
Remarkably, in the phage particle, the coat proteins are also tilted at an angle of about 20o compared to the phage axis (22, 26). However, in the phage the coat proteins form an interdigitated helical array, such that one protein comes into contact with 10 neighboring subunits. All the coat proteins are tilted at the same angle. As a result, many residues (including residues 30, 31, and 35) are not exposed to the outside of the phage (22). Since the tilted coat helix is found in the membrane-anchored protein, as well as in the phage particle, we concluded that little structural rearrangement is necessary during the phage assembly process. Thus, the protein can keep its basic structural arrangement from the host membrane to the free phage particle so that the assembly process may occur by binding multimeric sheets of coat proteins out of the membrane (Fig. 9C). This assembly model contrasts with a previously proposed mechanism indicating that there is a major topological change from the membrane to the phage (21).
Whereas the formation of multimers of coat proteins makes sense for the phage assembly process, we were surprised to see that the procoat protein was also found in oligomeric forms (Fig. 8). Since all mutant procoat proteins were capable of initiating the formation of multimers, we concluded that the proteins can rotate in the membrane. However, we found that at positions −7, −8, −10, and −12 spontaneous dimers were formed (Fig. 5A), suggesting that there might be a preferred region of interaction between two procoat proteins.
We did not find a major fraction of intramolecular contacts in the mutant procoat proteins. In the normal pathway, cleavage by the leader peptidase occurs rapidly, and only cysteine residues in the mature region were able to form disulfides (Fig. 2). These dimers were found mainly when we treated the cells with the hydrophobic oxidant copper phenanthroline (Fig. 2B). In untreated cells, the intramembrane milieu was reductive, and only minor amounts of dimers were observed (Fig. 5A and 8A). This also explains why the cysteine mutant proteins generally had no effect on the multiplication of the phage (Tables 1 and 2). When the phage production was reduced, the reasons involved something other than interference by disulfide formation. For the single-cysteine mutants, expression of mutants −6C and −12C had lethal and static effects on bacterial growth, respectively. Interestingly, when the same mutations were combined with H5, a mutation that prevents cleavage (−3F), no growth defect was observed (data not shown). Topological analysis with proteinase K showed that the Sec-independent procoat proteins and also Sec-dependent proOmpA accumulated in a translocated form (Fig. 3B), suggesting that the processing of the precursor proteins by the leader peptidase, but not the translocation across the membrane, was inhibited. We suspect that the cysteine mutants remain bound to the leader peptidase and block the enzyme. Binding to the leader peptidase is prevented with the H5 mutation, which explains why expression of the H5 cysteine mutant proteins did not have a lethal effect.
In contrast, we did not find intermolecular disulfides of the −6C procoat protein with the leader peptidase. This finding is plausible, since the leader peptidase has no cysteine residues in the center of its two transmembrane segments (27). The formation of protein disulfides in the membrane has also been used to analyze the interactions between two different proteins. The interaction between SecE and SecY was studied by cysteine scanning (33), as was the interaction between SecG and SecY (25). Intramolecular contacts between two transmembrane regions were found in the Tar receptor protein (28), the lactose permease (34), and the leader peptidase (35).
Acknowledgments
We thank Sandra Facey, Dorothee Kiefer, and Ross Dalbey for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (grant SFB 495).
Footnotes
Published ahead of print on 19 January 2007.
REFERENCES
- 1.Chothia, C., M. Levitt, and D. Richardson. 1981. Helix to helix packing in proteins. J. Mol. Biol. 145:215-250. [DOI] [PubMed] [Google Scholar]
- 2.Deber, C. M., A. R. Khan, Z. Li, C. Joensson, M. Glibowicka, and J. Wang. 1993. Val-Ala mutations selectively alter helix-helix packing in the transmembrane segment of phage M13 coat protein. Proc. Natl. Acad. Sci. USA 90:11648-11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fürste, J. P., W. Pansegrau, R. Frank, H. Blöcker, P. Scholz, P. M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 4.Glucksman, M. J., S. Bhattacharjee, and L. Makowski. 1992. Three-dimensional structure of a cloning vector. X-ray diffraction studies of filamentous bacteriophage M13 at 7 A resolution. J. Mol. Biol. 226:455-470. [DOI] [PubMed] [Google Scholar]
- 5.Greenwood, J., G. J. Hunter, and R. N. Perham. 1991. Regulation of filamentous bacteriophage length by modification of electrostatic interactions between coat protein and DNA. J. Mol. Biol. 217:223-227. [DOI] [PubMed] [Google Scholar]
- 6.Haigh, N. G., and R. E. Webster. 1998. The major coat protein of filamentous bacteriophage f1 specifically pairs in the bacterial cytoplasmic membrane. J. Mol. Biol. 279:19-29. [DOI] [PubMed] [Google Scholar]
- 7.Hunter, G. J., D. H. Rowitch, and R. N. Perham. 1987. Interactions between DNA and coat protein in the structure and assembly of filamentous bacteriophage fd. Nature 327:252-254. [DOI] [PubMed] [Google Scholar]
- 8.Ito, K., T. Date, and W. Wickner. 1980. Synthesis, assembly into the cytoplasmic membrane, and proteolytic processing of the precursor of coliphage M13 coat protein. J. Biol. Chem. 255:2123-2130. [PubMed] [Google Scholar]
- 9.Kaufmann, A., E. H. Manting, A. K. J. Veenendaal, A. Driessen, and C. van der Does. 1999. Cysteine-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighboring SecE. Biochemistry 38:9115-9125. [DOI] [PubMed] [Google Scholar]
- 10.Kiefer, D., and A. Kuhn. 1999. Hydrophobic forces drive the spontaneous membrane insertion of the bacteriophage Pf3 coat protein without topological control. EMBO J. 18:6299-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn, A. 1995. Bacteriophage Pf3 and M13 as model systems for Sec-independent protein transport. FEMS Microbiol. Rev. 17:185-190. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn, A., and W. Wickner. 1985. Isolation of mutants in M13 coat protein that affect its synthesis, processing and assembly into phage. J. Biol. Chem. 260:15907-15913. [PubMed] [Google Scholar]
- 13.Kuhn, A., and W. Wickner. 1985. Conserved residues of the leader peptide are essential for cleavage by leader peptidase. J. Biol. Chem. 260:15914-15918. [PubMed] [Google Scholar]
- 14.Lee, G. F., G. C. Burrows, M. R. Lebert, D. P. Dutton, and G. L. Hazelbauer. 1994. Deducing the organization of a transmembrane domain by disulfide cross-linking. J. Biol. Chem. 269:29920-29927. [PubMed] [Google Scholar]
- 15.Lee, J., A. Kuhn, and R. E. Dalbey. 1992. Distinct domains of an oligotopic membrane protein are Sec-dependent and Sec-independent for membrane insertion. J. Biol. Chem. 267:938-943. [PubMed] [Google Scholar]
- 16.Linderoth, N. A., M. N. Simon, and M. Russel. 1997. The filamentous phage pIV multimer visualized by scanning transmission electron microscopy. Science 278:1635-1638. [DOI] [PubMed] [Google Scholar]
- 17.Lynch, B. A., and D. E. Koshland. 1991. Disulfide cross-linking studies of the transmembrane regions of the aspartate sensory receptor of Escherichia coli. Proc. Natl. Acad. Sci. USA 88:10402-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyons, L. B., and N. Zinder. 1972. The genetic map of the filamentous bacteriophage f1. Virology 49:45-60. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis, T., E. F. Fritsch, and J. Sambrock. Molecular. cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 20.Marassi, F. M., and S. J. Opella. 2003. Simultaneous assignment and structure determination of a membrane protein from NMR orientational restraints. Protein Sci. 12:403-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marvin, D. A. 1998. Filamentous phage structure, infection and assembly. Curr. Opin. Struct. Biol. 8:150-158. [DOI] [PubMed] [Google Scholar]
- 22.Marvin, D. A., R. D. Hale, C. Nave, and M. Helmer-Citterich. 1994. Molecular models and structural comparisons of native and mutant class I filamentous bacteriophages Ff (fd, f1, M13), If1 and IKe. J. Mol. Biol. 235:260-286. [DOI] [PubMed] [Google Scholar]
- 23.Marzec, C. J., and L. A. Day. 1983. DNA and protein lattice-lattice interactions in the filamentous bacteriophages. Biophys. J. 42:171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melnyk, R. A., S. Kim, A. R. Curran, D. M. Engelman, J. U. Bowie, and C. M. Deber. 2004. The affinity of GXXXG motifs in transmembrane helix-helix interactions is modulated by long-range communication. J. Biol. Chem. 279:16591-16597. [DOI] [PubMed] [Google Scholar]
- 25.Mori, H., N. Shimokawa, Y. Satoh, and K. Ito. 2004. Mutational analysis of transmembrane regions 3 and 4 of SecY, a central component of protein translocase. J. Bacteriol. 186:3960-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overman, S. A., M. Tsuboi, and G. J. Thomas. 1996. Subunit orientation in the filamentous virus Ff (fd, f1, M13). J. Mol. Biol. 259:331-336. [DOI] [PubMed] [Google Scholar]
- 27.Paetzel, M., A. Karla, N. C. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem. Rev. 102:4549-4580. [DOI] [PubMed] [Google Scholar]
- 28.Pakula, A. A., and M. I. Simon. 1992. Determination of transmembrane protein structure by disulfide cross-linking: the Escherichia coli Tar receptor. Proc. Natl. Acad. Sci. USA 89:4144-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohrer, J., and A. Kuhn. 1990. The function of a leader peptide in translocating charged amino acyl residues across a membrane. Science 250:1418-1421. [DOI] [PubMed] [Google Scholar]
- 30.Russ, W. P., and D. M. Engelman. 2000. The GXXXG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296:911-919. [DOI] [PubMed] [Google Scholar]
- 31.Serek, J., G. Bauer-Manz, G. Struhalla, L. van den Berg, D. Kiefer, R. Dalbey, and A. Kuhn. 2004. Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J. 23:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studier, F. W., and B. A. Moffat. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 33.Veenendaal, A. K., C. van der Does, and A. J. Driessen. 2001. Mapping the sites of interaction between SecY and SecE by cysteine scanning mutagenesis. J. Biol. Chem. 276:32559-32566. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Q., and H. R. Kaback. 1999. Helix packing in the lactose permease of Escherichia coli determined by site-directed thiol cross-linking: helix I is close to helices V and XI. Biochemistry 38:3120-3126. [DOI] [PubMed] [Google Scholar]
- 35.Whitley, P., L. Nilsson, and G. von Heijne. 1993. Three-dimensional model for the membrane domain of Escherichia coli leader peptidase based on disulfide mapping. Biochemistry 32:8534-8539. [DOI] [PubMed] [Google Scholar]