Abstract
We describe here details of the method we used to identify and distinguish essential from nonessential genes on the bacterial Escherichia coli chromosome. Three key features characterize our method: high-efficiency recombination, precise replacement of just the open reading frame of a chromosomal gene, and the presence of naturally occurring duplications within the bacterial genome. We targeted genes encoding functions critical for processes of transcription and translation. Proteins from three complexes were evaluated to determine if they were essential to the cell by deleting their individual genes. The transcription elongation Nus proteins and termination factor Rho, which are involved in rRNA antitermination, the ribosomal proteins of the small 30S ribosome subunit, and minor ribosome-associated proteins were analyzed. It was concluded that four of the five bacterial transcription antitermination proteins are essential, while all four of the minor ribosome-associated proteins examined (RMF, SRA, YfiA, and YhbH), unlike most ribosomal proteins, are dispensable. Interestingly, although most 30S ribosomal proteins were essential, the knockouts of six ribosomal protein genes, rpsF (S6), rpsI (S9), rpsM (S13), rpsO (S15), rpsQ (S17), and rpsT (S20), were viable.
A gene may either be essential or nonessential for viability of a cell. An essential gene encodes a function which is required under all growth conditions, and so its elimination is lethal to the cell. Hence, this is generally the most interesting, yet difficult, type of genes to identify and characterize. In an era when many genomes have been sequenced and their coding regions identified, the ability to distinguish essential from nonessential genes still requires careful experimental assessment (23, 24, 29, 48). Here, we create gene replacements to distinguish these two classes of genes via direct selection in Escherichia coli using phage λ Red-mediated homologous recombination, termed recombineering (12, 20, 69).
Recombineering uses the Red recombination functions of phage λ to manipulate the DNA on chromosomes or plasmids of enteric bacteria (12, 18). In this work, recombineering was adapted to directly identify essential genes in E. coli. The procedure is not intended for a high-throughput identification of essential genes, although it could certainly be upgraded to a larger-scale analysis. It should be most useful for a targeted, knowledge-based analysis of cellular systems and their individual components, as we have demonstrated in this paper. Our method was developed using three features: (i) an extremely high efficiency of recombineering (69); (ii) the ability of recombineering to provide precise replacement of just the open reading frame (ORF) of a chromosomal gene with the orf of an antibiotic resistance cassette, taking care to design and express such a replacement orf from the regulatory regions of the replaced gene (69) so as to avoid polar effects of the insert on transcription and translation; and (iii) naturally occurring duplications of regions in the bacterial genome among cells existing in the culture (34).
We have examined the functions of two cellular systems that are very important for bacterial macromolecular syntheses. One is the Nus-dependent transcription antitermination system, which has components that interact with RNA polymerase during transcription elongation and termination to modify the polymerase, allowing read-through of transcription terminators (21, 50, 54, 55). The second system we examined is the ribosome, and in particular the small subunit (30S) ribosomal proteins (r-proteins) as well as the four known minor ribosome-associated proteins. The minor r-proteins, as their name implies, are not found in equimolar amounts to conventional 30S and 50S r-proteins, but their ratio does increase in stationary phase and under some other specific conditions of growth (1, 28, 35, 64). We have shown that in E. coli the transcription antitermination factors, except for NusB, which is conditionally lethal, are essential to the bacterial cell. In contrast, all minor ribosomal proteins are dispensable for cell viability in agreement with published results. Unexpectedly, we found that six r-proteins of the 30S subunit are not absolutely required for growth and, like NusB, exhibit conditional cell lethality.
(Research performed by Theresa Baker was in partial fulfillment of the requirements for a Master of Science in Biomedical Sciences at Hood College, Frederick, MD.)
MATERIALS AND METHODS
Media and bacteriological techniques.
Standard bacteriological media and techniques were used to cultivate cells (37). Cells were grown in LB broth or on LB or M63 agar plates. The M63 plates were supplemented with 0.2% glucose, vitamin B1, and biotin. The following final concentrations of antibiotics were used, unless otherwise stated: ampicillin (Ap), 25 μg/ml; tetracycline (Tc), 12.5 μg/ml; kanamycin (Km), 25 or 50 μg/ml, and chloramphenicol (Cm), 10 or 30 μg/ml, for recombinant strains or maintenance of plasmids, respectively.
Strains and plasmids.
Strains and plasmids used in this study are shown in Table 1.
TABLE 1.
Strains and plasmids used
| Strain | Description | Source |
|---|---|---|
| W3110 | Wild type | |
| DY330 | W3110 ΔlacU169 gal490 pglΔ8 λ [ρ]cI857 Δ(cro-bioA) | 69 |
| DY331 | DY330 recA | 69 |
| DY432 | DY330 ΔN-kil | C. McGill (NCI) |
| IQ527 | MC4100 ssyB63 (nusB::IS10) zba525::Tn10 | 59 |
| NB132 | DY330 rpsF<>kan | This work |
| NB149 | DY330 rpsJ(nusE)<>kan/rpsJ(nusE)+ partial diploid | This work |
| NB207 | DY432 nusB<>cat/nusB+ partial diploid | This work |
| NB208 | DY330 nusA<>cat/nusA+ partial diploid | This work |
| NB209 | DY330 nusA<>cat pAB116 | This work |
| NB210 | NB207 nusB<>cat/nusB+thrA::Tn10 thrA+ | This work |
| NB213 | DY330 rpsM<>kan | This work |
| NB216 | DY330 rho<>kan/rho+ | This work |
| NB217 | DY330 rhoL-rho<>kan/rho+ | This work |
| NB218 | DY330 rhoL-rho<>kanSD/rho+ | This work |
| NB345 | DY330 rpsT<>kan | This work |
| NB350 | DY330 rpsQ<>kan | This work |
| NB421 | W3110 nusB<>cat | This work |
| NB429 | DY330 rpsO<>kan | This work |
| NB438 | W3110 rpsV<>kan | This work |
| NB439 | W3110 yfiA<>kan | This work |
| NB440 | DY330 rpsV<>kan | This work |
| NB441 | DY330 yfiA<>kan | This work |
| NB469 | DY330 rpsJ(nusE)<>kan pAB37 | This work |
| NB470 | DY330 rpsI<>kan | This work |
| NB483 | DY330 yhbH<>cat | This work |
| NB484 | DY330 rpsV<>spc | This work |
| NB485 | DY330 rmf<>bla | This work |
| NB493 | W3110 yhbH<>cat | This work |
| NB494 | W3110 rmf<>bla | This work |
| NB495 | W3110 rpsV<>spc | This work |
| NB539 | W3110 rmf<>bla rpsV<>spc yfiA<>kan yhbH<>cat | This work |
| NB606 | DY432 nusB<>cat | This work |
| NB610 | DY330 nusG<>cat nusG + partial diploid | This work |
| NB611 | DY330 nusG<>cat pAB90 | This work |
| NB747 | IQ527 ssyB63<>nusB<>cat | This work |
| pCR-Script | oriColE1 Cmr | Stratagene |
| pPCR-Blunt | oriColE1 Kmr | Invitrogen |
| pAB37 | pCR CmrnusE(rpsJ) | This work |
| pAB90 | pPCR KmrnusG | 30 |
| pAB116 | pPCR KmrnusA | This work |
Gene disruption by recombineering.
Genes were disrupted in the chromosome as described elsewhere (60) with some modifications. Briefly, DY330 cells were induced for λ Red functions at an A600 of 0.5 and prepared for electroporation with 300 ng of a linear DNA antibiotic resistance cassette. The cells were recovered in 0.9 ml LB, grown for 2 to 3 h at 32°C, spread as aliquots on LB agar plates with appropriate antibiotic, and incubated up to 7 days at 34°C. The 34°C temperature was chosen because the defective λ prophage contained in DY330 has a killing function that is expressed above 37°C. The antibiotic resistance cassettes for gene disruption were made by PCR amplification (Expand PCR kit; Roche Applied Science, Indianapolis, IN) of the antibiotic-resistant orfs (see reference 60 for the description of antibiotic resistance orfs) with hybrid primers (Table 2) that contained a 40- to 50-nucleotide (nt) sequence at their 5′ ends homologous to chromosomal regions surrounding the ORF of the gene targeted for disruption, followed by a 22- to 25-nt priming sequence complementary to the 5′ and 3′ ends of an antibiotic resistance gene ORF. Primers for inactivation of the 30S r-protein genes were designed in the same manner; their sequences are available upon request. The resulting PCR-generated cassettes have an antibiotic resistance orf flanked by the 40- to 50-nt regions of homology that are needed for recombination into the chromosomal gene, such that the chromosomal orf is precisely replaced with the antibiotic resistance orf.
TABLE 2.
List of primers
| Primera | Homology sequenceb |
|---|---|
| nusA-Cm(f) | CCCCACTTTTAATAGTCTGGATGAGGTGAAAAGCCCGCG-cat(f) |
| nusA-Cm(r) | CGTTACATCTGTCATGCTGTTCCTTCCTGCTACAGTTTA-cat(r) |
| nusA-pCR(f) | GCTGCCCGTAATATTTGCTGGTTCGGTGACGAAGCGTAA-pCR(f) |
| nusA-pCR(r) | ATTGGATACGGCTTCAACTACAGCCAAAATTTCTTTGTT-pCR(r) |
| nusA-check(f) | TAAAGATGAAGTGTTCGCGCTGAG |
| nusA-check(r) | GCTGTACCAGGCGTTCCACGGAGG |
| nusB-Cm(f) | TGAAAGCCATCAAGGCCTGAAATTAGTAAGGGGAAATCC-cat(f) |
| nusB-Cm(r) | CATGGAACGGTCTTCCGTGAATCTACCGGCCTGGATATCA-cat(r) |
| nusB-check(f) | AAGGTGCAGAAGCTGCACTGACCGCGC |
| nusB-check(r) | GGCAATCAGGGAGAACTCGCCACATGC |
| nusE-Km(f) | AATCATTTTCGTTTATAAAATAATTGGAGCTCTGGTCT-kan(f) |
| nusE-Km(r) | CAATCATTGTTTCAACCTCTCAATCGCTCAATGACCTGA-kan(r) |
| nusE-exp(f) | CAGAAGGAGATATTCATATGCAGAACCAAAGAATCCGTATC |
| nusE-exp(r) | TTGCATGCGCATCTAGATTAACCCAGGCTGATCTGCACGTC |
| nusE-check(f) | CTCCCATCAATCGTAATGGGTCTG |
| nusE-check(r) | ACGGGTCATACCCACTTTTTTACC |
| nusG-Cm(f) | GGTGAAAATGTTTGTAGAAAACTTCTGACAGGTTGGTTT-cat(f) |
| nusG-Cm(r) | TTGTGCAACGATTAAATCGCCGCTTTTTTGATCGCTGGG-cat(r) |
| nusG-check(f) | CGCAGTAATGTCACTGATCCTGTG |
| nusG-check(r) | GCGAAATTGTATTCCAATCTCACG |
| rho-Km(f) | ACATTAAGTTCGAGATTTACCCCAAGTTTAAGAACTCACACCACT-kan(f) |
| rhoL-Km(f) | TAAGTTTGAATCTTGTAATTTCCAACGCTTCCCGTTTTATCTTAA-kan(f) |
| rhoL-KmSD | rhoL-Km(f)-CAGGATGAGGATCGTTTCGCATG |
| rho-Km(r) | AGCAAAACGCCACGTAAACACGTGGCGTTTTTGGCATAAGACAAA-kan(r) |
| rho-check(f) | TGTTGACTTCGTATTAAACATACC |
| rho-check(r) | GATGAGATCAGTACTCACTGTCAG |
| rmf-Ap(f) | TTTCTTTTCCACCAGAAACCAGTATGAGGGAAACGAGGC-amp(f) |
| rmf-Ap(r) | TCCTCCGCAATGCGGAGGTTTCTTTTTAAAGAGACAGAA-amp(r) |
| rmf-check(f) | TGACGGCAGTTATGATTCGCGGTA |
| rmf-check(r) | AGGATACGTCTGCCTTCTGATTAT |
| rpsV-Km(f) | TGTTGTCCTTAAAACTAGCTACAGGATTGAGGAGTTAAA-kan(f) |
| rpsV-Km(r) | AATGGTGTTTAATCGTCATTGAGGACTGATGGTTATGAA-kan(r) |
| rpsV-check(f) | TTATGTGGTCAGTGGCCAGCACC |
| rpsV-check(r) | TTGGTTCCATGTCACTCACTCTT |
| yfiA-Km(f) | GCTGAATTCACCAAGACGGGAAGACAAGAGGTAAAATTT-kan(f) |
| yfiA-Km(r) | CGCCCGAAGGCGCGTTGGCGATACACTCAATATAAAGGA-kan(r) |
| yfiA-check(f) | CACATTTTGACATCAGGAACGG |
| yfiA-check(r) | GTACTGTTTTCACGCTGTCAAC |
| yhbH-Cm(f) | AACAACTCGTTTGACCCAACCGATAAGGAAGACACT-cat(f) |
| yhbH-Cm(r) | AACAGACCGCCATGCACATGCTAATTGCCCGGACAA-cat(r) |
| yhbH-check(f) | CACTGTTGCGAAGTACCGAGAGTCT |
| yhbH-check(r) | ATTAACCGTACAAATGGCCCGTTGT |
| amp(f) | ATGAGTATTCAACATTTCCGT |
| amp(r) | TTACCAATGCTTAATCAGTGA |
| cat(f) | ATGGAGAAAAAAATCACTGGATATAC |
| cat(r) | TTACGCCCCGCCCTGCCACTCATC |
| kan(f) | ATGATTGAACAAGATGGATTG |
| kan(r) | TCAGAAGAACTCGTCAAGAAG |
| pCR(f) | CCTGAATTCTGCAGATATCCATCAC |
| pCR(r) | CATAGCTGTTTCCTGTGTGAAATTG |
f and r indicate forward and reverse primers, respectively.
Sequences of primers used to generate gene cassettes for recombineering are shown in two parts. The first part is shown in normal font and represents sequences complementary to the flanking regions of the gene of interest. The second part is in bold and was used for amplification of the respective PCR fragments; the sequences for these primers are shown at the bottom of the table.
PCR analysis of gene replacements.
Recombinant colonies were purified on LB plates with the appropriate antibiotic at 34°C to avoid contamination with the background cells. Individual colonies were resuspended in 30 μl sterile H2O, and 1 μl was used as a template for PCR analysis using primers flanking the targeted gene's ORF (the set of checking primers is shown in Table 2) in a 50 μl-reaction mix. PCR products were visualized on a 1% agarose gel.
Construction of multiple knockouts for minor ribosome-associated proteins.
The individual gene knockouts were made by recombineering as described above using different antibiotic resistance gene cassettes. These knockouts were sequentially combined in W3110 by P1 transduction (37) to obtain a quadruple gene knockout, rmf<>bla, rpsV<>spc, yfiA<>kan, and yhbH<>cat. After each transduction, the transduced gene replacement was selected on LB plates with antibiotic and verified by PCR analysis. The final knockout was confirmed in the same manner for all four replaced genes.
Cloning of nus genes.
Cloning of the nusA and nusG open reading frames was done by retrieval (gap repair) recombineering (60) into a linear pPCR-Blunt vector (Invitrogen, Inc., Carlsbad, CA) exactly as described for nusG (30). The ORF of nusE, also known as rpsJ, was amplified with its own translation initiation region by PCR from the chromosome, using the Expand PCR kit (Roche Applied Science, Indianapolis, IN), and the PCR fragment was cloned into pCR-Cam (Stratagene, La Jolla, CA). Plasmids with the correct orientation of the insert relative to Plac were screened by restriction analysis and verified by DNA sequencing. See Table 1 for plasmids with cloned genes and Table 2 for primers used to clone the genes.
Disruption of chromosomal nusA and nusG by recombineering in the presence of plasmid-expressed genes.
The DY330 cells were transformed by electroporation with pAB90 or pAB116 expressing nusG or nusA, respectively. The freshly transformed cells were used for recombineering to replace chromosomal nusA and nusG genes with a cat cassette, as described above. Recombinants were selected at 34°C on LB-Cm plates and analyzed by PCR as above. Constitutive expression from Plac allowed sufficient expression of NusA or NusG proteins from the plasmid in the DY330 cells. Note that homologies used to replace the chromosomal nus genes by recombineering are not present on the plasmid clones.
Transfer of nusE disruption to the chromosome by P1 transduction in the presence of plasmid-expressed nusE.
The nusE<>kan allele was crossed into the recipient DY330 bearing a nusE-expressing plasmid, pAB37, by P1 transduction with a lysate grown on NB149, nusE<>kan/nusE+ partial diploid. DY330 recipient cells were used as a negative control for transduction of nusE<>kan. Transductants were spread on LB-Km plates and incubated at 34°C for several days. They were purified on LB-Km plates and verified by PCR for configuration of the nusE gene.
Analysis of size and stability of chromosomal duplications.
The extent of nusG and nusB chromosomal duplications was analyzed with a set of mini-Tn10-generated auxotrophic markers originally designed for fine gene mapping by P1 transduction (40, 52). Each of these markers was P1 transduced into the nusG and nusB duplication-containing strains, and transductants were selected on LB-Tc plates and then scored for prototrophy on M63 minimal medium. Growth of the Tcr cells on minimal medium indicated a duplication.
We examined the stability of duplications in two strains. Cultures of the nusG or nusB partial diploids were grown overnight in LB-Cm or LB-Cm, Tc broth. The overnight cell cultures were washed of antibiotic, and the rate of duplication segregation was determined by serial 300-fold dilution into fresh LB medium every 12 h without antibiotic for the next 8 days of incubation. Every 24 h, the number and frequency of Cmr cells in the total cell population were determined by counting colonies on LB-Cm versus LB plates. The experiments were repeated twice.
Phenotypic analysis of knockout mutants.
The nusB, rmf, rpsV, yfiA, and yhbH knockout mutants were P1 transduced into W3110, selecting for their respective antibiotic resistance marker. They were analyzed by phenotypic microarrays (7) for major metabolic functions, as recommended by the manufacturer (Biolog, Inc.). Cell growth was analyzed on LB and M63 agar plates without antibiotic at 30, 37, and 42°C.
To study the viability of knockouts for minor r-proteins in stationary phase, 3 ml LB was inoculated with either wild-type W3110 or a mutant knockout and grown at 37°C for 24 h. Equal volumes of each of the mutant and W3110 cell cultures were mixed together and continued to incubate under the same conditions for another 10 days. Controls included the initial mutant and W3110 monocultures. Samples were taken every 2 days, diluted, and plated on LB agar, either with or without its appropriate antibiotic, to determine the total number of cells and the number of mutant cells which survived in the mixed cultures over the incubation time.
Bioinformatic analysis.
The older version of COGnitor (58) available from the NCBI website was adapted for this study. The basic procedure included the generation of a phylogenetic pattern for the protein of interest and then screening it for protein conservation in minimal eubacterial genomes of less than 1.2 Mb in size. Since these genomes are severely limited in the number of their genes (38), they should contain a higher percentage of essential genes. The orthologous proteins encoded in these genomes were additionally analyzed by tree analysis of protein similarity followed by sequence analysis of the protein family, using ClustalW 1.74 with manual adjustment of aligned sequences. As a result of these manipulations, only the truly orthologous proteins were selected. The results of the COG analysis were verified by the more expansive Blastn and Blastp analyses of the whole bacterial genomes available to date, using the E. coli ortholog as bait. It was found that the basic procedure of screening for proteins that are conserved over 90% of the time in minimal genomes was sufficient to reliably predict proteins that are essential as determined by our gene replacement technology.
RESULTS
Rationale for use of recombineering to analyze essential genes.
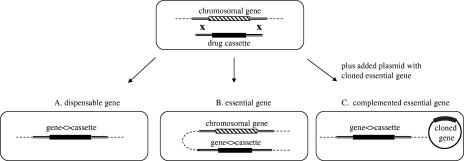
An essential gene encodes a function which is required under all growth conditions, and so its elimination is lethal to the cell. However, under our experimental conditions we define essential genes as those whose functions are required for colony formation on LB agar at 34°C. Genes have been inactivated in a manner so as not to affect the expression of adjacent genes. In general, the gene coding region from the start to stop codon was replaced by a precise in-frame substitution of an antibiotic resistance cassette orf (69), allowing direct selection for the replacement. Nonessential genes can be routinely replaced by this substitution technique (Fig. 1A) at frequencies that usually generate as many as 104 recombinants per 108 electroporated cells (69). Essential genes, on the other hand, cannot be replaced by recombination without a second wild-type copy of the gene being present in the cell (Fig. 1B and C).
FIG. 1.
Gene disruption recombineering in E. coli. Replacement of an entire chromosomal gene ORF with an antibiotic resistance cassette orf by recombineering is shown in the upper row. If the gene is dispensable (A), the antibiotic resistance orf simply replaces the gene ORF without affecting cell survival. In case of essential gene replacement, the cells with a replaced gene can only survive if there is an additional wild-type copy of the gene, a chromosomal duplication (B) or a complementing plasmid expressing this gene in trans (C).
Within a population of cells growing in culture, approximately 10−3 cells undergo a duplication of a large genomic segment, creating a partial diploid region of the chromosome (34). Independent duplications of most, if not all, regions of the genome occur and exist as significant subpopulations in any bacterial culture. When essential genes are targeted for gene replacement by recombineering, these partial diploid subpopulations are present and available for recombination. Recombinants can only survive if the essential gene to be deleted resides in a region of the genome that is diploid. Such recombinants are found at a very low frequency of ∼5 to 50 per 108 cells as antibiotic-resistant colonies, in which the duplicated state is stabilized by selection for the antibiotic resistance marker and for expression of the essential gene (Fig. 1B). If a gene cannot be replaced efficiently by recombineering and rare recombinants appear as partial diploids, then we assume that gene may be essential. Alternatively, it is possible that the gene is not essential, and the particular replacement caused some unexpected defect in an adjacent essential gene. To test this possibility, the open reading frame of each candidate essential gene is individually cloned onto a plasmid by recombineering. The knockout in the chromosome can then be recreated in the presence of this plasmid clone (Fig. 1C). These procedures were used for analysis of gene essentiality, as outlined in Fig. 2.
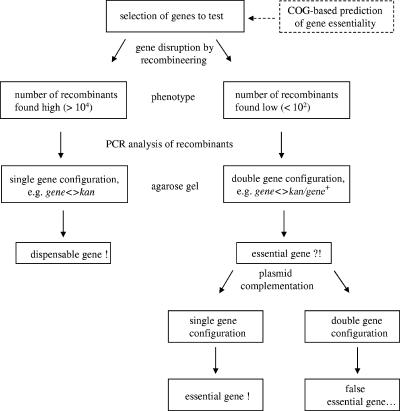
FIG. 2.
Flow chart of the procedure for analysis of gene essentiality. A gene is disrupted with recombineering by exactly replacing the gene ORF with an antibiotic resistance cassette orf. A group of genes can be analyzed by choosing different antibiotic resistance cassettes for disruption (see “Construction of multiple knockouts for minor ribosome-associated proteins” in the text). A COG-based prediction procedure may aid in selecting the genes for analysis. Essential versus dispensable genes are determined by their different recombination frequencies and detection of a partial duplication in the chromosomal region for a targeted gene if the gene is essential. If needed, the gene essentiality can be further tested by gene disruption in the presence of a complementing plasmid carrying the wild-type allele of this gene.
Use of precise gene replacement by a selectable antibiotic resistance cassette might not always yield recombinants, even if a gene is not essential. Whenever the ORF of the target gene in question is replaced precisely by the ORF of the cassette, the promoter and translation signals of the target gene are used to express the cassette. We expect that for some genes, expression will be insufficient to allow antibiotic resistance. In cases where no recombinant colonies are found, alternative cassette structures must be used which contain the promoter and ribosome-binding site of the cassette, with or without its transcription terminator (see examples in reference 69). When a transcription terminator is used, the terminator may block expression of downstream genes in an operon. However, in the absence of a terminator, the cassette's promoter may be too strong and cause toxic overexpression of distal genes. For all genes tested below, the native expression system was sufficient to express the antibiotic resistance cassette, and insertion of additional regulatory elements was not required. If recombinants are not found even in the presence of a cloned copy, it is possible that a region of the chromosome may be refractory to recombination. Although a systematic search has not been completed, of several hundred gene replacements attempted in this laboratory, no example of such a refractory gene or region has yet been found.
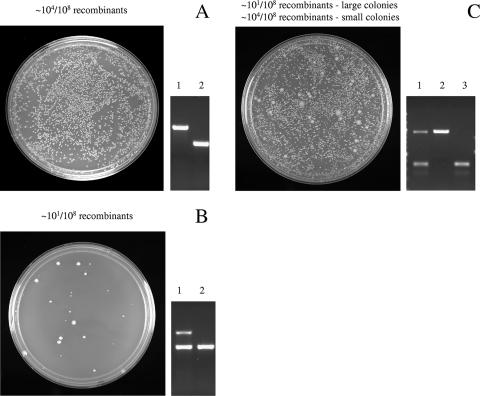
Three patterns observed for orf replacements.
Three typical patterns that can be ascribed to essential, growth-impaired, and nonessential gene knockouts are described in detail below (Fig. 3). The pattern observed for inactivation of an essential gene (Fig. 3B) is a resultant low number of normally growing recombinant colonies (<102/108 viable cells), all of which bear a duplication of the replaced gene carrying the wild-type allele (gene<>antibiotic resistance cassette/gene+). Another pattern is characterized by impaired growth caused by a gene replacement (Fig. 3C). In this case, after recombination a few normally growing recombinant colonies are observed among thousands of slow-growing ones. The large normal colonies contain a duplication in the region of the replaced gene, whereas the small slow-growing colonies contain only the disrupted gene. The nonessential gene pattern (Fig. 3A) is characterized by a high recombination frequency (103 to 104 recombinants/108 viable cells) of normally growing recombinant colonies bearing a single replaced chromosomal gene with an antibiotic resistance cassette (gene<>antibiotic resistance cassette).
FIG. 3.
Three patterns of gene knockouts observed during analysis of gene essentiality by recombineering. A. Nonessential gene pattern characterized by a standard high recombination frequency on LB plates (left panel) and single configuration of a replaced gene as analyzed by agarose gel electrophoresis (right panel). The agarose gel shows a gene replaced with an antibiotic resistance cassette (lane 1) versus the original wild-type gene (lane 2). B. Essential gene pattern characterized by a low recombination frequency of colonies and duplicated configuration of the disrupted gene (lane 1) versus its wild-type allele (lane 2) by gel analysis. The duplication includes the gene replaced with an antibiotic resistance cassette (lane 1, upper band) and the wild-type gene (lane 1, lower band). C. Growth-impaired pattern characterized by a mixed pattern of two colony sizes with different recombination frequencies. The large colonies appear first with a low recombination frequency and a duplicated configuration (lane 1) of the essential gene. The small colonies have a high recombination frequency and a single configuration of the replaced gene (lane 2) versus its wild-type allele (lane 3).
Essential nus genes.
Replacement of nusA, nusD/rho, nusE/rpsJ, and nusG by a cat or kan cassette resulted in a very low frequency of antibiotic-resistant recombinants (Table 3). PCR analysis showed (Fig. 3B, lane 1) that the antibiotic-resistant recombinants of nusA, nusE, nusG, and rho contained both a wild-type and an antibiotic resistance-substituted copy of the respective targeted gene, indicating that these genes are essential by our definition. Our results with rho disagree with those found by Baba et al. (5), who reported that rho is nonessential. We made three different knockouts for rho. The first removed just the rho gene, and the second removed the rho gene and the rho leader region including the small “gene” rhoL. For the third construct, since rhoL does not have a distinctive Shine-Dalgarno region, a rhoL-rho knockout was also made with the same kan cassette but containing its own Shine-Dalgarno region. Recombinants for all three constructs were made, and all showed the pattern of essential gene knockouts and carried the gene duplication.
TABLE 3.
Results of gene disruption recombineering applied to antitermination and ribosome-associated protein genes
| Gene | Recombination efficiencya | Gene configurationb | Gene essentiality |
|---|---|---|---|
| nusA | 1.8 × 101 | nusA<>cat/nusA+ | Essential |
| nusBc | 0.9 × 101 | nusB<>cat/nusB+ | NAe |
| 6.5 × 103 | nusB<>cat | Growth impaired | |
| nusD/rho | 3.7 × 102 | nusD<>kan/nusD+ | Essential |
| rhoL-nusD/rho | 2.7 × 102 | nusD<>kan/nusD+ | Essential |
| rhoL-nusD/rhod | 4.2 × 102 | nusD<>kanSD/nusD+ | Essential |
| nusE(rpsJ) | 0.7 × 101 | nusE<>kan/nusE+ | Essential |
| nusG | 1.2 × 101 | nusG<>cat/nusG+ | Essential |
| yfiA | 7.3 × 104 | yfiA<>kan | Nonessential |
| yhbH | 6.6 × 104 | yhbH<>cat | Nonessential |
| rmf | 3.2 × 104 | rmf<>amp | Nonessential |
| rpsV | 8.5 × 104 | rpsV<>kan | Nonessential |
Number of antibiotic-resistant DY330 recombinants per 5 × 108 viable cells.
Determined by PCR analysis of the chromosomal region with the replaced gene, as shown in Fig. 3. Partial diploids are shown as gene<>antibiotic-resistance cassette/gene+.
Two gene knockout patterns were observed for nusB knockouts. Cells with a diploid gene configuration appeared with a low frequency on the first day. After 2 days, they were followed by numerous nusB<>cat recombinants.
rhoL-rho was replaced with kan containing its own Shine-Dalgarno sequence, kanSD.
NA, not applicable.
Initially we had thought that the duplicated regions might have been formed by unequal crossing over between small homologous regions on the chromosome mediated by the host recombination system. However, when we repeated the rho gene replacement exactly as reported above but with a recA mutant derivative, we had the same result. Red-mediated replacements were found, but rarely, and only in cells with duplications.
The growth-impaired nusB gene.
In the case of nusB replacements (Table 3), a few normal-sized colonies appeared on the first day of incubation, mimicking the character of the other nus genes. However, unlike the other nus gene replacements, after 2 days a large number of much-smaller-sized colonies appeared (∼104 colonies/108 viable cells). These two types of isolates from the nusB gene replacement showed two distinct PCR patterns. The larger colonies contained both the wild-type nusB and the cat-replaced gene, whereas the small nusB<>cat colonies had only the replaced gene copy (Fig. 3C, lanes 1 and 2).
Replacement of 30S ribosomal protein genes.
Each of the 21 30S ribosomal protein genes was evaluated for essential function by replacement of the orf with a kan cassette. Fifteen of these gene replacements displayed the pattern of essential gene knockout: a low recombinant frequency and a duplication as confirmed by PCR analysis (Fig. 3). Six knockouts (rpsF, rpsI, rpsM, rpsO, rpsQ, and rpsT) formed as well numerous, small, slow-growing colonies, containing only the disrupted gene, as described for the growth-impaired nusB knockout. From these data we conclude that the majority of the 30S protein genes are essential, except rpsF, rpsI, rpsM, rpsO, rpsQ, and rpsT.
Nonessential ribosome-associated protein genes.
Replacement of the minor ribosome-associated protein genes by kan yielded a high frequency (>103 per 108 viable cells) of Kmr recombinants (Table 3). All isolates contained only the inactivated form of the targeted gene and formed normal-sized colonies (Fig. 3A). A fully viable quadruple knockout of all four minor protein genes (rmf<>amp, rpsV<>spc, yfiA<>kan, and yhbH<>cat) was generated by serial P1 transduction from the individual knockouts. Thus, all of the minor ribosome-associated proteins appear to be dispensable.
Inactivation of nus genes in the presence of complementing plasmids.
To inactivate essential chromosomal nus genes, two methods were used that required the presence of a complementing plasmid bearing the cloned nus genes. In the first method, the nusA or nusG genes were replaced with a cat cassette by recombineering. In the second method, the nusE or nusG genes were replaced by transduction with P1 grown on the previously made partial diploid nusE<>kan/nusE+ or nusG<>cat/nusG+. Both methods in the presence of the complementing plasmid yielded the single chromosome configuration of the replaced nus genes in all colonies tested (Table 3). In contrast, in the absence of the complementing plasmid, recombineering generated the gene replacement in the duplicated configuration; however, P1 transduction did not generate any recombinants because of its low frequency relative to recombineering. Note that each nus gene on the plasmid is not detected by the PCR analysis because the primers used are in the chromosomal flanking regions outside of the nus gene tested.
Characteristics of gene duplication.
We attempted to P1 transduce a number of essential gene replacements, including those of the nus genes, to another strain. In no case could nusE or nusG replacements be transduced unless the recipient strain contained plasmids expressing the respective nus gene. Since P1 is known to transduce nearly a 100-kb segment of chromosomal DNA, this suggests the duplication is at least 50 kbp. On the other hand, the nusB<>cat replacement could be transduced to other strains, but these transductants were always small colony formers lacking an intact nusB gene.
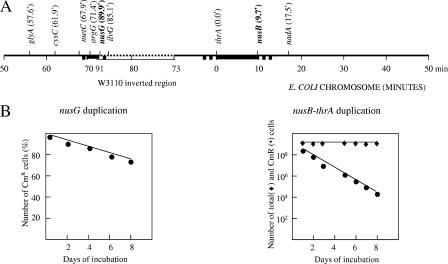
To determine how extensive the duplications might be, the duplicated nusG and nusB mutants were subjected to further tests. Several auxotrophic strains have been made by Tn10 transposon insertion (40, 52). We have transduced the tet markers from these auxotrophs into the duplicated nusG<>cat or nusB<>cat strains, selecting for tetracycline resistance (Tcr), respectively, on LB-Tc agar and scoring for prototrophy on M63 minimal agar. Transductants that were both Tcr and prototrophic were presumed to be diploid for that region. The results indicated that the duplicated regions extend far from the nus genes tested into the segments comprising at least 10 and 4 min of the chromosome for nusB and nusG duplications, respectively (Fig. 4A). The two duplications, however, exhibited different stability over time (Fig. 4B). The nusG duplication was quite stably inherited such that it persisted in more than 70% of the cells in the population after 9 days of growth. The nusB duplication, on the contrary, was quite unstable, with only 50% of cells bearing the duplication after the first 12 h of growth (see Materials and Methods).
FIG. 4.
Characterization of gene duplications with essential (nusG) and impaired (nusB) gene knockout patterns. A. Genetic mapping of nusG and nusB duplications with a set of Tn10 (Tcr) auxotrophic markers was done by plating the cells on LB-Tc and M63 minimal agar to select for Tcr and prototrophy, respectively. The gene markers and their position (min) in the E. coli chromosome are indicated. Positions of nusG and nusB are shown in bold. The dotted line defines the inverted chromosomal region found in W3110 (25). The duplicated regions are shown in bold. The bold dotted line indicates that duplication was not precisely mapped and may extend to the flanking area. B. Stability of nusG and nusB diploids. Every 12 h the diploid cultures were passed through LB medium. Every 24 h the number of diploids in cultures was estimated as a ratio of Cmr cells to the total number of cells in the cultures by plating them on LB-Cm and on plain LB plates, respectively. Note that because of the very different stabilities of diploids, the days of incubation of nusG and nusB diploids are plotted against the percentage or the actual number of Cmr cells in the cell cultures and are shown as a linear or a log plot, respectively.
Functional studies of replacement mutants.
An advantage of single-copy, nonpolar, gene replacements is that they can be transferred to a different genetic background and directly used for functional studies. In this work, we transferred the disrupted alleles of nusB and nonessential ribosomal protein genes to the wild-type W3110 background to further investigate their properties.
We found that the growth of the single-copy nusB<>cat is impaired under normal growth conditions and greatly impaired at temperatures between 23° and 30°C, indicating that under these circumstances NusB is very important for cell growth, although not absolutely essential. Initially, we found that nusB<>cat was less cold sensitive than ssyB63, another loss-of-function nusB::IS10 insertion mutant in strain MC4100 (59). However, the difference in growth was caused by the strain background, as we demonstrated by directly replacing ssyB63 in MC4100 with nusB<>cat using P1 transduction. In MC4100, nusB<>cat grows like the ssyB63 mutant (data not shown).
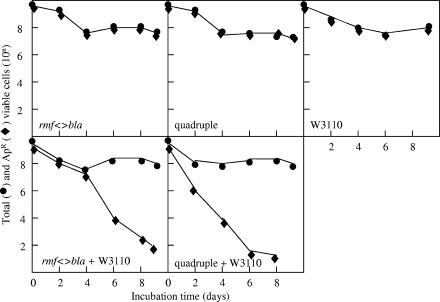
The minor ribosome-associated protein gene replacements were fully functional at all temperatures either in rich or in minimal medium and did not show gross impairment in major biochemical pathways as judged by phenotypic microarrays (data not shown). Although these proteins have increased levels at stationary phase, their gene knockouts grew normally in stationary phase for a long period of time. Even a quadruple knockout mutant defective for all four minor r-proteins was fully viable as well as normal in the phenotypic microarray analysis (data not shown), demonstrating that the whole module of minor r-proteins is dispensable for most cell functions. These data are consistent with the published data on the function of the rpsV product, SRA (28), and with the recently published data on the characterization of yfiA and yhbH knockouts (62), but not for the function of the rmf product. The rmf::kan cells have been shown to gradually lose viability in stationary phase (68). Our rmf knockout was viable but could be competed out in a mixed culture with wild-type W3110 cells after 4 to 8 days of incubation (Fig. 5). Other r-protein knockouts grew normally in the mixed cultures over the same period of time (data not shown). In our rmf<>bla knockout, the rmf orf is precisely replaced by bla, whereas Yamaguchi et al. (68) placed a promoter-containing kan cassette in the middle of the rmf gene, potentially causing polar effects on the expression of genes surrounding rmf. The competitive growth disadvantage of the rmf knockout was more evident in the quadruple gene knockout, being the only defect found for the quadruple knockout under our experimental conditions (Fig. 5). Properties of the six conditionally essential ribosomal protein gene knockouts, rpsF, rpsI, rpsM, rpsO, rpsQ, and rpsT, are reported elsewhere (10).
FIG. 5.
Survival of rmf<>bla and quadruple ribosome-associated protein mutants in stationary phase. Stationary-phase cultures of rmf and quadruple knockouts, as well as of W3110, were grown at 37°C for 24 h. The mixed cultures were obtained by combining 1 ml of each of the 24-h culture knockouts together with 1 ml of W3110 and grown at 37°C for 10 days (bottom panel) along with an equal volume (2 ml) of the original monoculture (top panel). Samples were taken as indicated, and the number of Apr survivors in the total cell population was counted by plating them on LB-Ap and plain LB plates, respectively.
DISCUSSION
An understanding of a complex biological process can be greatly facilitated by identification of its essential components. We developed a procedure to identify the key components of bacterial systems, based on their essentiality for growth and viability of cells. Central to this procedure (Fig. 1) is the technique of gene replacement in the chromosome using recombineering (12) with antibiotic resistance orfs. The high efficiency of recombineering and lack of polar effects of the chromosomal orf gene replacements allow positive selection for even an inactivated essential gene. Normally, for an essential gene, the recombination event replaces the gene in the cell and that cell is lost because it can never form a colony. However, in rare cases, the recombination event occurs within a cell with a naturally occurring duplication, replacing one copy of the essential gene with an antibiotic resistance cassette and leaving the second copy intact (Fig. 2). This gene duplication is stably inherited and may comprise a large segment of the chromosome.
This approach was utilized to evaluate components of two macromolecular cellular systems, the Nus transcription antitermination functions (21, 50, 55) and ribosomal proteins (9, 16, 47), including a set of minor ribosome-associated proteins (64). The Nus antitermination functions were originally identified because they play a critical role in the life cycle of phage λ by temporal activation of genes distal to terminators (21). In E. coli, these factors modulate the synthesis of rRNA (44, 54, 70). Although rRNA synthesis is vitally important for bacterial cells, E. coli can tolerate a substantial variability in expression of rRNA genes, being able to survive the deletion of all but one copy of the seven rRNA genes (4). The NusA and NusD/Rho functions were previously suspected to be essential for bacterial growth, since conditional lethal mutants of these genes had been isolated (13, 17, 39, 56). A nusG knockout was shown to be nonviable (72), while a nusA knockout could be obtained only in the presence of attenuating rho mutations (71). At the same time, rho (45) and nusG (27) knockouts in Bacillus subtilis, as well as a Staphylococcus aureus rho knockout (66), had been reported as viable. The NusB function has not been shown to be essential (44, 46, 59).
In this study, we demonstrate that nusA, nusD/rho, nusE/rpsJ, and nusG are indeed essential for cell growth, whereas nusB is required only at temperatures less than 30°C. Since NusB is required for rRNA antitermination both in vitro (54) and in vivo (61), but not for cell growth, the Nus-dependent rRNA type of antitermination may not be an essential cellular process. Residual transcription read-through may be sufficient in the absence of antitermination at least at higher temperatures. NusB binds to NusE and to an RNA sequence, boxA, in the rRNA leader (41). The boxA site is important for antitermination (6, 22, 63). NusA and NusG are known to interact with RNA polymerase and are involved not only in antitermination but often in elongation and termination steps of transcription (11, 32, 56, 63). Rho is a transcription termination factor that binds nascent RNA and moves along the RNA to interact with RNA polymerase, causing termination at “Rho sites” (49). It is tempting to hypothesize that NusB may be the only specific antitermination factor, whereas the other Nus factors may function more generally in modulating basic RNA polymerase activities in transcription elongation. Perhaps these other functions of NusA, NusG, and Rho proteins determine their essential nature. NusE is also essential but probably because it is a structural component of ribosomes.
Ribosomal proteins have long been thought of as being essential (16, 31, 67). We examined the essentiality of the 30S r-proteins of E. coli. Six of the 21 ribosomal protein gene knockouts, namely rpsF (S6), rpsI (S9), rpsM (S13), rpsO (S15), rpsQ (S17), and rpsT (S20), were viable as defined by our conditions of colony growth on LB medium at 34°C. However, these knockouts dramatically reduced the cell growth at 37°C in W3110 and did not grow at some other temperatures (data not shown). The other 15 r-proteins (S1 to S5, S7, S8, S10 to S12, S14, S16, S18, S19, and S21) were essential. The essential properties of S1 (53), S2 (33), and S16 (43), as well as the viability of the S9 (26), S13 (14), S15 (36), and S20 (51) gene knockouts, have been demonstrated previously. Spontaneous mutants lacking either S6, S9, S13, S17, or S20 protein have been obtained by Dabbs (16).
Analysis of the six viable ribosomal protein gene knockouts provides interesting implications for understanding the mechanisms of ribosome biogenesis and functioning and is detailed elsewhere (10). Initial analysis of essentiality of the 30S ribosomal proteins has revealed a number of intriguing observations. All six nonessential proteins are located along the rim of the 30S subunit from the platform side (47), away from the important functional centers of the ribosome. Three out of six viable knockouts affected the primary assembly proteins S15, S17, and S20, which are supposed to be crucial for initiating the subsequent cooperative binding of secondary and late assembly proteins. The secondary and late assembly proteins are essential, with the exception of the secondary protein S6. So it seems that ribosomes might assemble in vivo without primary assembly proteins, as has been directly demonstrated for ribosomes lacking S15 (10) or S20 (51). The principles of the cooperative and ordered assembly of proteins into ribosomes that had been deduced from in vitro experimentation (15, 42) may not quite mimic the ribosome biogenesis in vivo. Alternatively, the order of protein assembly into ribosomes may be somewhat different in vivo.
Among the set of four minor ribosome-associated proteins, RMF (65, 68) and YfiA (1, 2) were proposed to have important functions, directly inhibiting protein synthesis and reducing translation errors in vitro. However, as we have shown in this study, none of the minor proteins is required in vivo, even in stationary phase. They are weakly conserved among other bacteria, and all four genes can be inactivated together without affecting the growth or physiology of the cells. Within the group, RMF may be needed for cell survival in stationary phase, albeit only under competitive growth conditions, and in this respect distinguishes itself from the other three proteins. At the same time, the other three proteins, SRA, YfiaA, and YhbH, may support RMF function in the cells, as the rmf knockout by itself survives the stationary phase somewhat better than the quadruple knockout for all minor r-proteins.
While this paper was in preparation, a global analysis of individual gene knockouts of all E. coli genes was reported (5). Those authors employed an approach of orf gene replacement in the chromosome with a kan cassette carrying its own promoter and used a plasmid-based Red recombination system, which has at least a 10-fold lower recombination efficiency than the prophage-based system (18) used in this work. Although the method lacks a means of direct analysis of essential genes (see the introduction for features of our gene replacement assay), a list of essential genes was compiled there by default from genes for which no knockout could be obtained. Their analysis of nonessential and essential genes does not entirely agree with our analysis for the genes tested in this work, i.e., for nus and the ribosomal protein genes. They identified rpsI (S9), rpsM (S13), and rpsQ (S17) as essential, although all three of these genes were dispensable in our assay as well as in work by other researchers (14, 26). Another gene in their list of essential genes for which a viable E. coli knockout has been reported is the RNase III gene, rnc, for which a gene replacement was made and tested in the same way as described here (69). Baba et al. (5) also found rho and rpsU to be dispensable, although both genes are essential by our analyses.
We suggest a number of reasons for the source of disagreement in assessing gene essentiality by the method of Baba et al. (5). The rnc gene is known to be translationally coupled to the essential era gene downstream (8, 57). The rpsM and rpsQ genes are parts of large gene operons and may be translationally coupled to their essential neighboring genes, as well. The fusions generated by Baba et al. (5) do not ensure translational coupling between the downstream gene and the intermediate antibiotic resistance cassette replacement. The rpsI, rpsM, and rpsQ knockouts might also have problems due to the conditions of knockout selection. All of these knockouts exhibit poor growth in our hands and are quite sensitive to the concentration of Km in the selection medium.
In the case of rho and rpsU, we found an unusually high frequency of antibiotic-resistant recombinants for these two genes compared to other essential gene knockouts tested (Table 3, rho data). This might be explained because these regions of the chromosome undergo recombination more efficiently, as has been shown for some segments of chromosomal DNA (20). Alternatively, the frequency of partial diploids for this region might be much higher than for other regions of the chromosome. Note that according to the assay of Baba et al. (5), dispensable gene knockouts are the ones that can be detected. Their definition does not take into account the possibility of partial diploids. Experimental approaches to analyze gene essentiality must be examined carefully, and an assay with direct selection and screening for essential and dispensable genes, like the one described in this study, is required. We also want to point out that analysis of a gene knockout must take into account problems of polarity both for transcription and translation, the expression level of the antibiotic resistance cassette used, the presence of duplications, and the specific growth conditions used.
This study has clarified our understanding of the importance of several cellular systems: the transcription antitermination system and conventional ribosome and minor ribosome-associated proteins. Many other cellular systems can be analyzed in this manner, e.g., the modules of the ribosome biogenesis network, in which only rRNAs (4, 47) and the rRNA maturation system (3, 19) are partially understood.
Acknowledgments
We thank Nina Costantino, Simanti Datta, Craig Laufer, James Sawitzke, Lynn Thomason, and Xiaomei Zhou for many discussions and help with the manuscript. We are especially thankful to Mathias Springer for his valuable comments on the ribosomal part of the manuscript. We also wish to thank Nina Bubunenko for her help in preparation of purified oligonucleotides and bioinformatics analysis of essential genes.
This research was supported in part by the Intramural Research Program of the NIH National Cancer Institute, Center for Cancer Research (N01-CO-12400). The research was supported by a Trans-NIH/FDA Intramural Biodefense Program grant from the National Institute of Allergy and Infectious Diseases (D.L.C.).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Agafonov, D. E., V. A. Kolb, and A. S. Spirin. 2001. A novel stress-response protein that binds at the ribosomal subunit interface and arrests translation. Cold Spring Harbor Symp. Quant. Biol. 66:509-514. [DOI] [PubMed] [Google Scholar]
- 2.Agafonov, D. E., and A. S. Spirin. 2004. The ribosome-associated inhibitor A reduces translational errors. Biochem. Biophys. Res. Commun. 320:354-358. [DOI] [PubMed] [Google Scholar]
- 3.Apirion, D., and A. Miczak. 1993. RNA processing in prokaryotic cells. Bioessays 15:113-120. [DOI] [PubMed] [Google Scholar]
- 4.Asai, T., D. Zaporojets, C. Squires, and C. L. Squires. 1999. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl. Acad. Sci. USA 96:1971-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg, K., C. Squires, and C. L. Squires. 1989. Ribosomal RNA operon antitermination. Function of leader and spacer region boxB-boxA sequences and their conservation in diverse microorganisms. J. Mol. Biol. 209:345-358. [DOI] [PubMed] [Google Scholar]
- 7.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high throughput phenotypic testing and assay of gene function. Genome Res. 11:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britton, R. A., B. S. Powell, S. Dasgupta, O. Sun, W. Margolin, J. R. Lupski, and D. L. Court. 1998. Cell cycle arrest in Era GTPase mutants: a potential growth rate-regulated checkpoint in Escherichia coli. Mol. Microbiol. 28:1391-1393. [DOI] [PubMed] [Google Scholar]
- 9.Brodersen, D. E., and P. Nissen. 2005. The social life of ribosomal proteins. FEBS J. 272:2098-2108. [DOI] [PubMed] [Google Scholar]
- 10.Bubunenko, M., A. Korepanov, D. L. Court, I. Jagannathan, D. Dickinson, B. R. Chaudhuri, M. B. Garber, and G. M. Culver. 2006. 30S ribosomal subunits can be assembled in vivo without a primary binding ribosomal protein S15. RNA 12:1229-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns, C. M., L. V. Richardson, and J. P. Richardson. 1998. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J. Mol. Biol. 278:307-316. [DOI] [PubMed] [Google Scholar]
- 12.Court, D. L., J. A. Sawitzke, and L. C. Thomason. 2002. Genetic engineering using homologous recombination. Annu. Rev. Genet. 36:361-388. [DOI] [PubMed] [Google Scholar]
- 13.Craven, M. G., and D. I. Friedman. 1991. Analysis of the Escherichia coli nusA10(Cs) allele: relating nucleotide changes to phenotypes. J. Bacteriol. 173:1485-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cukras, A. R., and R. Green. 2005. Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation. J. Mol. Biol. 349:47-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Culver, G. M. 2003. Assembly of the 30S ribosomal subunit. Biopolymers 68:234-249. [DOI] [PubMed] [Google Scholar]
- 16.Dabbs, E. R. 1991. Mutants lacking individual ribosomal proteins as a tool to investigate ribosomal properties. Biochimie 73:639-645. [DOI] [PubMed] [Google Scholar]
- 17.Das, A., D. Court, and S. Adhya. 1976. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc. Natl. Acad. Sci. USA 73:1959-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datta, S., N. Costantino, and D. L. Court. 2006. A set of recombineering plasmids for gram-negative bacteria. Gene 379:109-115. [DOI] [PubMed] [Google Scholar]
- 19.Deutscher, M. P., and Z. Li. 2001. Exoribonucleases and their multiple roles in RNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 66:67-105. [DOI] [PubMed] [Google Scholar]
- 20.Ellis, H. M., D. Yu, T. DiTizio, and D. L. Court. 2001. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA 98:6742-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman, D. I., and D. L. Court. 1995. Transcription antitermination: the lambda paradigm updated. Mol. Microbiol. 18:191-200. [DOI] [PubMed] [Google Scholar]
- 22.Friedman, D. I., and M. Gottesman. 1981. Lytic mode of lambda development, p. 21-51. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Gerdes, S. Y., M. D. Scholle, J. W. Campbell, G. Balazsi, E. Ravasz, M. D. Daugherty, A. L. Somera, N. C. Kyrpides, I. Anderson, M. S. Gelfand, A. Bhattacharya, V. Kapatral, M. D'Souza, M. V. Baev, Y. Grechkin, F. Mseeh, M. Y. Fonstein, R. Overbeek, A. L. Barabasi, Z. N. Oltvai, and A. L. Osterman. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glass, J. I., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchinson III, H. O. Smith, and J. C. Venter. 2006. Essential genes of minimal bacterium. Proc. Natl. Acad. Sci. USA 103:425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill, C. W., and B. W. Harnish. 1981. Inversions between ribosomal RNA genes in Escherichia coli. Proc. Natl. Acad. Sci. USA 78:7069-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoang, L., K. Fredrick, and H. F. Noller. 2004. Creating ribosomes with all-RNA 30S subunit P-site. Proc. Natl. Acad. Sci. USA 101:12439-12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingham, C. J., J. Dennis, and P. A. Furneaux. 1999. Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol. Microbiol. 31:651-663. [DOI] [PubMed] [Google Scholar]
- 28.Izutsu, K., C. Wada, Y. Komine, T. Sako, C. Ueguchi, S. Nakura, and A. Wada. 2001. Escherichia coli ribosome-associated protein SRA, whose copy number increases during stationary phase. J. Bacteriol. 183:2765-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Judson, N., and J. J. Mekalanos. 2000. Transposon-based approaches to identify essential bacterial genes. Trends Microbiol. 8:521-526. [DOI] [PubMed] [Google Scholar]
- 30.Knowlton, J. R., M. Bubunenko, M. Andrykovitch, W. Guo, K. M. Routzahn, D. S. Waugh, D. L. Court, and X. Ji. 2003. A spring-loaded state of NusG in its functional cycle is suggested by X-ray crystallography and supported by site-directed mutants. Biochemistry 42:2275-2281. [DOI] [PubMed] [Google Scholar]
- 31.Koonin, E. V. 2000. How many genes can make a cell: the minimal-gene set concept. Annu. Rev. Genomics Hum. Genet. 1:99-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, J., S. W. Mason, and J. Greenblatt. 1993. Elongation factor NusG interacts with termination factor Rho to regulate termination and antitermination of transcription. Genes Dev. 7:161-172. [DOI] [PubMed] [Google Scholar]
- 33.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lupski, J. R., J. R. Roth, and G. M. Weinstock. 1996. Chromosomal duplications in bacteria, fruit flies, and humans. Am. J. Hum. Genet. 58:21-27. [PMC free article] [PubMed] [Google Scholar]
- 35.Maki, Y., H. Yoshida, and A. Wada. 2000. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells 5:965-974. [DOI] [PubMed] [Google Scholar]
- 36.Mathy, N., O. Pellegrini, A. Serganov, D. J. Patel, C. Ehresmann, and C. Portier. 2004. Specific recognition of rpsO mRNA and 16S rRNA by Escherichia coli ribosomal protein S15 relies on both mimicry and site differentiation. Mol. Microbiol. 52:661-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Experiments in molecular genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Moran, N. A.. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583-586. [DOI] [PubMed]
- 39.Nakamura, Y., A. Mizusawa, A. Tsugawa, and M. Imai. 1986. Conditionally lethal nusAts mutation of Escherichia coli reduces transcription termination but does not affect antitermination of bacteriophage lambda. Mol. Gen. Genet. 204:24-28. [DOI] [PubMed] [Google Scholar]
- 40.Nichols, B. P., O. Shafiq, and V. Meiners. 1998. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J. Bacteriol. 180:6408-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nodwell, J. R., and J. Greenblatt. 1993. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell 72:261-268. [DOI] [PubMed] [Google Scholar]
- 42.Nomura, M. 1973. Assembly of bacterial ribosomes. Science 179:864-873. [DOI] [PubMed] [Google Scholar]
- 43.Persson, B. C., G. O. Bylund, D. E. Berg, and P. M. Wikstrom. 1995. Functional analysis of the ffh-trmD region of the Escherichia coli chromosome by using reverse genetics. J. Bacteriol. 177:5554-5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quan, S., N. Zhang, S. French, and C. L. Squires. 2005. Transcription polarity in rRNA operons of Escherichia coli nusA and nusB mutant strains. J. Bacteriol. 187:1632-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quirk, P. G., E. A. Dunkley, Jr., P. Lee, and T. A. Krulwich. 1993. Identification of a putative Bacillus subtilis rho gene. J. Bacteriol. 175:647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajapandi, T., and D. Oliver. 1994. ssaD1, a suppressor of secA51(Ts) that renders growth of Escherichia coli cold sensitive, is an early amber mutation in the transcription factor gene nusB. J. Bacteriol. 176:4444-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramakrishnan, V. 2002. Ribosome structure and the mechanism of translation. Cell 108:557-572. [DOI] [PubMed] [Google Scholar]
- 48.Reich, K. A. 2000. The search for essential genes. Res. Microbiol. 151:319-324. [DOI] [PubMed] [Google Scholar]
- 49.Richardson, J. P. 2002. Rho-dependent termination and ATPase in transcript termination. Biochim. Biophys. Acta 1577:251-260. [DOI] [PubMed] [Google Scholar]
- 50.Roberts, J. W. 1993. RNA and protein elements of E. coli and lambda transcription antitermination complexes. Cell 72:653-655. [DOI] [PubMed] [Google Scholar]
- 51.Ryden-Aulin, M., Z. Shaoping, P. Kylsten, and L. A. Isaksson. 1993. Ribosome activity and modification of 16S RNA are influenced by deletion of ribosomal protein S20. Mol. Microbiol. 7:983-992. [DOI] [PubMed] [Google Scholar]
- 52.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sorensen, M. A., J. Fricke, and S. Pedersen. 1998. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J. Mol. Biol. 280:561-569. [DOI] [PubMed] [Google Scholar]
- 54.Squires, C. L., J. Greenblatt, J. Li, C. Condon, and C. Squires. 1993. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl. Acad. Sci. USA 90:970-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Squires, C. L., and D. Zaporojets. 2000. Proteins shared by the transcription and translation machines. Annu. Rev. Microbiol. 54:775-798. [DOI] [PubMed] [Google Scholar]
- 56.Sullivan, S. L., and M. Gottesman. 1992. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell 68:989-994. [DOI] [PubMed] [Google Scholar]
- 57.Takiff, H. E., S. M. Chen, and D. L. Court. 1989. Genetic analysis of the rnc operon of Escherichia coli. J. Bacteriol. 171:2581-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tatusov, R. L., E. E. Koonin, and D. J. Lipman. 1997. A genome perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 59.Taura, T., C. Ueguchi, K. Shiba, and K. Ito. 1992. Insertional disruption of the nusB (ssyB) gene leads to cold-sensitive growth of Escherichia coli and suppression of the secY24 mutation. Mol. Gen. Genet. 234:429-432. [DOI] [PubMed] [Google Scholar]
- 60.Thomason, L. C., M. Bubunenko, N. Costantino, H. R. Wilson, A. Openheim, S. Datta, and D. L. Court. 2005. Recombineering: genetic engineering in bacteria using homologous recombination, p. 1.16.11-1.16.21. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Wiley, New York, N.Y. [DOI] [PubMed]
- 61.Torres, M., J.-M. Balada, M. Zellars, C. Squires, and C. L. Squires. 2004. In vivo effect of NusB and NusG on rRNA transcription termination. J. Bacteriol. 186:1304-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ueta, M., H. Yoshida, C. Wada, T. Baba, H. Mori, and A. Wada. 2005. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells 10:1103-1112. [DOI] [PubMed] [Google Scholar]
- 63.Vogel, U., and K. F. Jensen. 1997. NusA is required for ribosomal antitermination and for modulation of the transcription elongation rate of both antiterminated RNA and mRNA. J. Biol. Chem. 272:12265-12271. [DOI] [PubMed] [Google Scholar]
- 64.Wada, A. 1998. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells 3:203-208. [DOI] [PubMed] [Google Scholar]
- 65.Wada, A., K. Igarashi, S. Yoshimura, S. Aimoto, and A. Ishihama. 1995. Ribosome modulation factor: stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem. Biophys. Res. Commun. 214:410-417. [DOI] [PubMed] [Google Scholar]
- 66.Washburn, R. S., A. Marra, A. P. Bryant, M. Rosenberg, and D. R. Gentry. 2001. rho is not essential for viability or virulence in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1099-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilson, D. N., and K. H. Nierhaus. 2005. Ribosomal proteins in the spotlight. Crit. Rev. Biochem. Mol. Biol. 40:243-267. [DOI] [PubMed] [Google Scholar]
- 68.Yamaguchi, M., H. Matsushima, A. Wada, M. Sakagami, N. Fujita, and A. Ishihama. 1993. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control. EMBO J. 12:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zellars, M., and C. L. Squires. 1999. Antiterminaor-dependent modulation of transcription elongation rates by NusB and NusG. Mol. Microbiol. 32:1296-1304. [DOI] [PubMed] [Google Scholar]
- 71.Zheng, C., and D. I. Friedman. 1994. Reduced Rho-dependent transcription termination permits NusA-independent growth of Escherichia coli. Proc. Natl. Acad. Sci. USA 91:7543-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou, Y., J. J. Filter, D. L. Court, M. E. Gottesman, and D. L. Friedman. 2002. Requirement for NusG for transcription antitermination in vivo by the lambda N protein. J. Bacteriol. 184:3416-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]