Abstract
Glucose-grown cells of Bacillus cereus respond to anaerobiosis and low extracellular oxidoreduction potentials (ORP), notably by enhancing enterotoxin production. This response involves the ResDE two-component system. We searched the B. cereus genome for other redox response regulators potentially involved in this adaptive process, and we identified one gene encoding a protein predicted to have an amino acid sequence 58% identical (80% similar) to that of the Bacillus subtilis Fnr redox regulator. The fnr gene of the food-borne pathogen B. cereus F4430/73 has been cloned and partially characterized. We showed that fnr was up-regulated during anaerobic fermentation, especially when fermentation occurred at low ORP (under highly reducing conditions). The expression of fnr was down-regulated in the presence of O2 and nitrate which, unlike fumarate, stimulated the respiratory pathways. The inactivation of B. cereus fnr abolished fermentative growth but only moderately affected aerobic and anaerobic nitrate respiratory growth. Analyses of glucose by-products and the transcription profiles of key catabolic genes confirmed the strong regulatory impact of Fnr on B. cereus fermentative pathways. More importantly, the fnr mutation strongly decreased the expression of PlcR-dependent hbl and nhe genes, leading to the absence of hemolysin BL (Hbl) and nonhemolytic enterotoxin (Nhe) secretion by the mutant. These data indicate that fnr is essential for both fermentation and toxinogenesis. The results also suggest that both Fnr and the ResDE two-component system belong to a redox regulatory pathway that functions at least partially independently of the pleiotropic virulence gene regulator PlcR to regulate enterotoxin gene expression.
Bacillus cereus is a facultatively anaerobic, spore-forming, opportunistic human pathogen (25, 56). The majority of reported illnesses involving B. cereus are food-borne intoxications, classified as emetic and diarrheal syndromes (28, 48). Diarrheal syndrome may be the consequence of the production in the human host's small intestine of various extracellular factors including hemolysin BL (Hbl), nonhemolytic enterotoxin (Nhe), and cytotoxin CytK (28, 40). The genes encoding these potential virulence factors belong to the PlcR regulon (1, 14, 38, 51).
B. cereus can grow well in amino acid-rich media supplemented with glucose or other carbohydrates as the major source of carbon and energy (11, 39, 45). Such carbohydrates are available in the human small intestine (6), where B. cereus encounters an oxygen-deprived and highly reducing environment (32). We recently showed that B. cereus is able to grow efficiently by anaerobic glucose fermentation under highly reducing conditions (60) and that the ResDE two-component system is one redox response regulator involved in the metabolic adaptation to these conditions. Moreover, ResDE was also demonstrated to play a role in the regulation of enterotoxin expression (12). This led to the conclusion that enterotoxin expression and fermentative metabolism may be controlled coordinately at the transcription level. However, the ResDE system, although important, is not essential for both fermentative metabolism and enterotoxin expression, suggesting that another redox regulator may act in synergy with ResDE to control the expression of fermentation and enterotoxin genes. One candidate redox regulator may be a member of the cyclic AMP receptor protein (CRP)-Fnr (fumarate and nitrate reduction regulator) family (27). Indeed, the CRP-Fnr regulators play an important role in modulating the expression of numerous metabolic genes in many facultatively and strictly anaerobic bacteria. Their functions also include the control of virulence factors (3, 4, 24, 46, 58). Furthermore, one-component CRP-Fnr regulators are known to act coordinately with two-component regulators homologous to ResDE in response to two environmental signals, namely, oxygen availability and the presence of alternative electron acceptors (e.g., nitrate). This coordinate action has been particularly well documented for B. subtilis (43). Like all the members of the CRP-Fnr family, the B. subtilis regulator (FnrBac) is characterized by a large nucleotide-binding domain that extends from the N terminus over roughly 170 residues to a C-terminally located helix-turn-helix structural motif. This DNA-binding domain is followed by a short C-terminal sequence with four cysteine residues. Three Cys residues from this C terminus together with one Cys residue from the central part of the protein bind a [4Fe-4S]2+ center that serves as a redox sensor (42). The ability of FnrBac to function as a transcription factor depends on the integrity of the [4Fe-4S]2+ cluster, which promotes a conformation that is necessary for site-directed DNA binding and transcriptional activation. Under anaerobic growth conditions, FnrBac is directly responsible for the induction of the narGHJI operon encoding the respiratory nitrate reductase; the narK-fnr operon encoding both a potential nitrite extrusion protein and FnrBac itself; and the arfM gene, which encodes an anaerobic respiration and fermentation regulator (31, 43). By activating nitrate reductase formation, FnrBac also mediates the repression of aerobic genes (such as cydABCD, encoding cytochrome bd quinol oxidase) and key fermentation genes (such as ldh and lct, encoding lactate dehydrogenases) via the redox regulator YdiH. For these reasons, FnrBac is considered to be a major regulatory switch in the adaptation to anaerobic growth conditions. B. cereus, like B. subtilis, is able to grow under nitrate respiratory conditions with either glucose or glycerol as a carbon source (45). Due to the energy conserved, this mode of anaerobic growth is preferred over fermentation in B. cereus (Fig. 1). However, the fermentative metabolism in B. cereus is more efficient than that in B. subtilis (35, 37, 60). Therefore, it is expected that the metabolism of B. cereus is controlled differently in response to anaerobiosis.
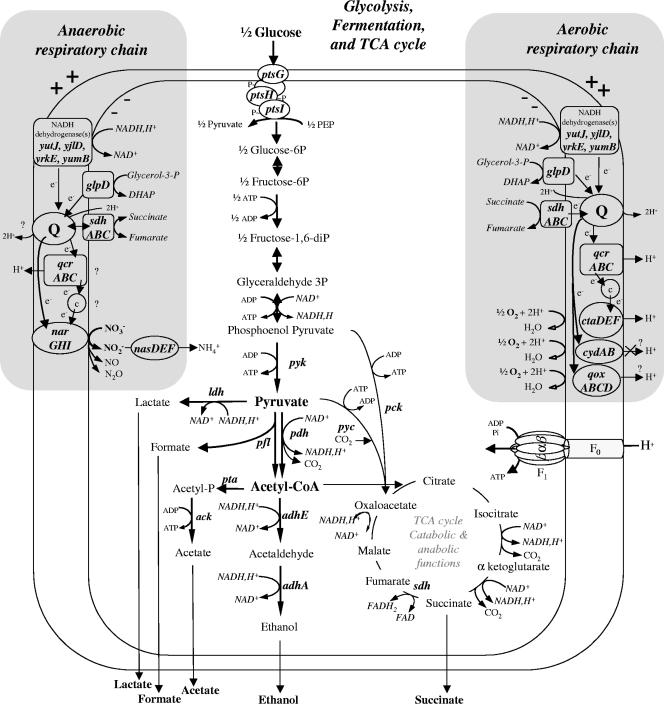
FIG. 1.
Architecture of fermentative and respiratory pathways in B. cereus. Genomic data (24) and information from references (12, 26, 45, 60) were used for this scheme. Most of the genes and functions are described in Table 4. TCA, tricarboxylic acid; Q, (mena)quinone; DHAP, dihydroxyacetone phosphate; e−, electron; c, cytochrome c; acetyl-CoA, acetyl coenzyme A; FAD, flavin adenine dinucleotide; FADH2, reduced flavin adenine dinucleotide. The glpD gene encoding sn-glycerol-3-phosphate dehydrogenase was repressed by glucose and induced during aerobic and anaerobic growth on glycerol (reference 60 and data not shown). Respiratory complex III encoded by the qcrABC operon was required for growth on glycerol, while it made only a minor contribution to growth on glucose (45).
In this study, we report the identification, cloning, and preliminary functional characterization of a homolog of B. subtilis fnr (42) in the food-borne pathogen B. cereus F4430/73 (52). We show that fnr expression is dispensable for nitrate respiration but essential for both fermentation and the production of the enterotoxins Hbl and Nhe.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strain TOP10 (Invitrogen) [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)-7697 galU galK rpsL (Strr) endA1 nupG] was used as the general cloning host, and E. coli strain ET12567 (F− dam-13::Tn9 dcm-6 hsdM hsdR recF143 zjj-202::Tn10 galK2 galT22 ara14 pacY1 xyl-5 leuB6 thi-1) was used to generate unmethylated plasmid DNA prior to B. cereus transformation. Both E. coli strains were routinely grown in Luria broth with vigorous agitation at 37°C. B. cereus F4430/73 (52) and derivatives thereof were grown in synthetic MOD medium (11, 60) supplemented with 30 mM glucose or 60 mM glycerol as the carbon source in the presence or absence of 20 or 60 mM nitrate or fumarate (potassium salts). Ampicillin (50 μg/ml for E. coli), kanamycin (50 μg/ml for E. coli and B. cereus), and erythromycin (10 μg/ml for B. cereus) were added to the media when needed.
pH-regulated batch cultures.
B. cereus cells were grown in a 2-liter bioreactor (Discovery 100; Inceltech, Toulouse, France) as previously described (12, 60). Briefly, the temperature was maintained at 34°C and the pH was maintained at 7.2 by the automatic addition of 2N KOH. The bioreactor was equipped with a Mettler Toledo polarographic oxygen electrode coupled with feedback regulation to maintain the set-point dissolved-oxygen tension value (pO2) via air sparging and agitation speed (11). The bioreactor was sparged with air alone to set a pO2 value of 100%. The pO2 values of 21% and 1% were obtained by sparging the bioreactor with air balanced with pure nitrogen gas. In addition, the N2 flow was adjusted manually during growth to compensate for fluctuations 5% higher than the theoretical set value. A pO2 of 0% was obtained by continuously flushing the medium at 20 ml/h with either pure N2 gas or pure H2 gas previously passed through a Hungate column. The oxidoreduction potential (ORP) was measured using a redox-combined electrode (AgCl; Mettler Toledo), and the values were corrected according to the reference electrode value (+200 mV at 34°C). Each bioreactor was inoculated with a subculture grown overnight under aerobiosis or anaerobiosis on MOD medium supplemented with a carbon source and the required electron acceptors. A sample of the overnight culture was diluted in fresh medium to achieve an initial optical density of the culture at 560 nm of 0.02.
Analytical procedures and physiological parameters.
B. cereus growth was monitored spectrophotometrically at 560 nm and calibrated against cell (dry weight) measurements as previously described (11). The specific growth rate (μ) was determined using the modified Gompertz equation (15, 61, 62). For substrate, by-product, and enterotoxin determinations, a 4-ml sample was centrifuged twice at 10,000 × g for 3 min at 4°C and the supernatants were frozen at −80°C until analysis. Glucose, glycerol, lactate, ethanol, formate, acetate, succinate, and nitrate concentrations were determined using commercial enzyme kits (Diffchamb, Lyon, France; R-Biopharm, Saint-Didier-au-Mont-d'Or, France; and Roche Molecular Diagnostics). The nitrite concentration was determined using the Griess reagent (17).
Carbon and oxidation balances were calculated using the established values for glucose (6 mol of carbon/mol; oxidation state, 0), glycerol (3 mol of carbon/mol; oxidation state, −1), lactate (3 mol of carbon/mol; oxidation state, 0), acetate (2 mol of carbon/mol; oxidation state, 0), formate (1 mol of carbon/mol; oxidation state, +1), ethanol (2 mol of carbon/mol; oxidation state, −2), and CO2 (1 mol of carbon/mol; oxidation state, +2). Values for succinate were corrected for CO2 fixation, with 1 mol of CO2 being required for 1 mol of succinate (Fig. 1), giving 3 mol of carbon/mol of the product (instead of 4 mol) and −1 (instead of +1) for the oxidation state. The amount of CO2 produced via the Pdh-dependent oxidation of pyruvate (Fig. 1) was calculated using theoretical flux (CO2 = ethanol + acetate − formate). Biomass was also taken into account for calculations by factoring in a molecular mass of 100 g/mol (22), a 50% carbon content, and an oxidation state of −1.5 (44).
The production of enterotoxins Hbl and Nhe in cultures was quantified as previously described (60). The amounts of Hbl produced were determined from the titer, which was defined as the reciprocal of the highest dilution of crude supernatant that gave an Hbl-dependent agglutination signal. The amounts of Nhe were estimated by measuring the optical density at 420 nm (1 U of Nhe was defined as the equivalent of an optical density at 420 nm of 1.0). The specific level of enterotoxin production was defined as the amount (in units) produced per gram of cell (dry weight).
General molecular methods.
Restriction endonuclease and T4 DNA ligase were obtained from Promega and used in accordance with the manufacturer's instructions. Genomic DNA of B. cereus was purified as described by Guinebretiere and Nguyen-The (19). Plasmid DNA was purified by using anion exchange columns (Promega). PCR amplification of DNA was carried out with Taq polymerase by using the specifications of the manufacturer (Roche Molecular Biochemicals) for the reaction conditions. DNA was introduced into B. cereus by electroporation (30). Prior to sequencing or cloning, PCR fragments were purified with a High Pure PCR product purification kit (Roche Molecular Biochemicals). For reverse transcription (RT)-PCR assays, crude cell extracts were rapidly prepared from pellets obtained from 200-ml culture aliquots harvested during the mid-exponential growth phase (μ = maximum μ [μmax]) (12). Total RNA was isolated from these pellets using an RNeasy minikit (QIAGEN Inc., France) according to the manufacturer's instructions, with additional on-column DNase I digestion to eliminate genomic DNA contamination. The absence of contaminating DNA was then verified by real-time PCR with the fnr primer set (Table 1).
TABLE 1.
Oligonucleotide primers used for real-time reverse transcription-PCR
| Oligonucleotidea | Nucleotide sequence (5′ to 3′) |
|---|---|
| fnrF | GCAAACGAAGTTCCGAGATT |
| fnrR | GCAGAGCAATCTTCACAAGC |
| yrkEF | GCAGCAGCGTATGATCAAGA |
| yrkER | CTGCAAACTCTACGCCTTCC |
| yutJF | TGGCGTTGAAGTAGCAAGTG |
| yutJR | TAAGACAACCCGTCGCGTAC |
| yumBF | TGTTGGTGGTGCTGGATTTA |
| yumBR | ACACCTGCAGCCCAAACTAC |
| yjlDF | GCTGGCGGCACTACTTTATC |
| yjlDR | GGATTTTTGGACCTGCTTCA |
| narGF | TTATTCGTATGGCGTGCAAA |
| narGR | ATTCCAGTCAGAGCGTGCTT |
| nasDF | ACATCCAGCACGATGGTACA |
| nasDR | CAGAAGCGAGAACCAACACA |
F, forward; R, reverse.
Cloning of the B. cereus fnr locus.
The genome sequence database of the ATCC 14579 strain (http://www.integratedgenomics.com) was used to construct primers (5′-CGAACACTTCAGCAGGCATA-3′ and 5′-TCGTACAACAATTGGCCCTT-3′) for the amplification and cloning of the 1,405-bp fnr locus from the B. cereus F4430/73 strain into the TA cloning vector pCR-TOPO (Invitrogen).
Construction of fnr-disrupted mutant and complementation assays.
A DNA fragment of 288 bp corresponding to the internal region of the fnr open reading frame (codons 128 to 224) was first PCR amplified using the primer pairs 5′-GCAAACGAAGTTCCGAGATT-3′ and 5′-GCAGAGCAATCTTCACAAGC-3′ and then cloned into the TA cloning vector pCR-TOPO (Invitrogen). The PCR fragment was then cut with BamHI and NotI and ligated to similarly digested pMUTIN4 (54). The resulting recombinant plasmid was obtained in E. coli strain ET12567 and then used to transform B. cereus F4430/73. The integrants resulting from a single-crossover recombination event between the chromosomal locus and the cloned sequence were selected by using erythromycin resistance. Gene disruption was confirmed by sizing of the PCR fragments generated from this chromosomal region (data not shown).
To complement the fnr gene in trans, the 1,405-bp fnr locus (see above) was cloned into the plasmid pHT304aph3, which is a Kmr derivative (J. Brillard, unpublished data) of the shuttle vector pHT304 (2). Briefly, the 1,405-bp fnr locus was isolated from pCR-TOPO by digestion with EcoRV and BamHI and inserted into pHT304aph3 digested with the same restriction enzymes. The integrity of the fnr locus of the recombinant vector (pHT304aph3-fnr) was verified by sequencing, and the vector was then used to transform the wild-type F4430/73 and its derivative fnr mutant strain. Attempts to restore the wild-type growth phenotype were carried out in the presence of 50 μg of kanamycin added to the MOD medium. Uncontrolled anaerobic and aerobic cultures were performed as described previously (45). The amounts of enterotoxin produced were determined as described by Guinebretiere et al. (18).
Mapping of transcriptional start site.
The 5′ end of the fnr mRNA was mapped from a 5′ rapid amplification of cDNA ends (RACE) PCR product obtained with the 3′/5′ RACE kit (Roche Molecular Biochemicals). For this purpose, we used total RNA extracted from B. cereus F4430/73 cells harvested at μmax, i.e., the maximal expression of the fnr gene. Briefly, the first-strand cDNA was synthesized from total RNA with fnr-specific primer SP1 (5′GGGTAGAATACAGGGCACCTT 3′) (Fig. 1B), avian myeloblastosis virus reverse transcriptase, and the deoxynucleotide mixture from the 3′/5′ RACE kit as recommended by the manufacturer. After purification and dA tailing of the cDNA, a PCR with the dT anchor oligonucleotide primer and the specific fnr SP2 primer (5′-GTGTTAGTTCTTGTCCGTCAGCAC-3′) followed by a nested PCR with the SP3 primer (5′-ATAGAGTTCGGTTGCCTCCA-3′) (Fig. 2) led to a PCR product of 151 bp, as revealed by 2% agarose gel electrophoresis. This PCR product was purified and sequenced.
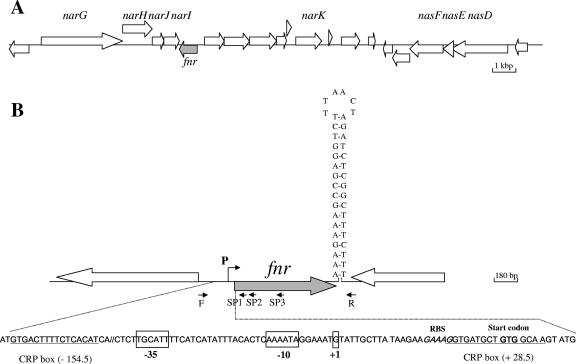
FIG. 2.
Genetic organization of the fnr chromosomal region of B. cereus F4430/73 and ATCC 14579. (A) Large arrows represent the open reading frames identified in strain ATCC 14579 (ERGO; Integrated Genomics, Inc.), and their orientation shows the transcriptional direction. Only the names of open reading frames encoding proteins with predicted metabolic functions are indicated. (B) The nucleotide sequences of the promoter region and the putative terminator of F4430/73 fnr are shown. Primers F and R were used for the amplification of the overall DNA of the fnr locus, and primers SP1, SP2, and SP3 were used to map the transcriptional start site. The transcriptional start site (+1) and the putative −35 and −10 motifs are boxed. Putative CRP boxes are underlined. RBS, ribosome-binding site.
Relative quantification of gene expression using SYBR green real-time PCR.
Real-time RT-PCR was performed using SYBR green technology on a LightCycler instrument (Roche Diagnostics) as previously described (11, 12). Most of the oligonucleotide sequences used for these experiments have been reported previously (12). Sequences specifically used in the present study are given in Table 1.
DNA sequencing and sequence analysis.
Sequencing was performed by MWG-Biotech (Roissy, France). DNA and amino acid sequences were analyzed using the Infobiogen and ExPASy servers (http://www.infobiogen.fr and http://au.expasy.org/). DNA and protein homology searches were carried out by performing BLAST analyses (http://www.ncbi.nlm.nih.gov). Conserved and functional domains were identified with rpsblast (http://www.ncbi.nlm.nih.gov). The search for a potential binding site in the promoter region was performed using the PRODORIC database (http://www.prodoric.de).
Nucleotide sequence accession number.
The DNA sequence corresponding to the fnr locus amplified from B. cereus F4430/73 was deposited in GenBank under accession no. DQ681074.
RESULTS
Fnr-like genes in B. cereus ATCC 14579 and F4430/73.
In silico analysis of the B. cereus ATCC14579 genome identified one open reading frame (RZC00220) homologous to the B. subtilis fnr gene (ERGO; Integrated Genomics Inc.). As shown in Fig. 2A, the fnr-like gene is located immediately downstream from and in the orientation opposite that of the putative narGHJI operon, which encodes the three-subunit nitrate reductase complex (NarGHI) and a fourth polypeptide required for the assembly of the complex (NarJ). This genomic organization differs from that found in B. subtilis, in which fnr is located far in the 5′ region of the narGHJI operon (9) and is surrounded upstream by the narK gene, encoding an integral membrane protein involved in nitrite extrusion, and downstream by the arfM gene, encoding a modulator of anaerobic respiration and fermentation (31). In B. cereus, the narK gene is located 5,169-bp downstream of fnr (Fig. 2A) and no arfM-like gene was found in the ATCC 14579 genome.
Given that the B. cereus F4430/73 strain belongs to the same genetic group as the ATCC 14579 strain (23), we designed primers corresponding to the ATCC 14579 fnr-like locus to clone and sequence its homolog from strain F4430/73. The DNA sequence of the 1,405-bp cloned fragment differed by only one base from that from the ATCC 14579 strain. This base change was located in the 3′ untranslated region (data not shown). The start codon (GTG) of the fnr-like gene was identified according to protein sequence alignments and the position of a potential ribosome-binding site. A transcriptional start site (G) located 30 bp from the translational start site (GTG) was determined by 5′ RACE PCR (Fig. 2B). Upstream of this transcriptional start site, we identified a potential housekeeping σA type −10 sequence, CAAAAT (underlining indicates deviations from the −10 σA consensus sequence of B. subtilis). This sequence is preceded by a poor −35 signal, TTGCAT, suggesting the involvement of a transcription activator. Interestingly, two putative CRP-Fnr-binding sites were mapped to the positions centered at −154.5 upstream and +28.5 downstream of the transcriptional start site (Fig. 2B) (27). Furthermore, the fnr locus appeared to be followed by an inverted repeat (ΔGo = −22.3 kcal/mol) that may be a transcriptional terminator, indicating that the fnr gene of B. cereus may be transcribed as a single unit.
Properties of B. cereus fnr.
The B. subtilis fnr-like gene in B. cereus encodes a protein of 230 amino acid residues with 58% sequence identity and 80% sequence similarity to FnrBac (42). While the Fnr-like protein and FnrBac do not share the same hydropathy profiles, they include the same two conserved domains that characterize the CRP-Fnr superfamily. In addition, the C-terminal DNA-binding regions of both proteins are followed by similar four-cysteine clusters (Cys-X3-Cys-X2-Cys-X4-Cys). Due to these protein similarities, the gene and protein from B. cereus were designated fnr and Fnr, respectively. Together with the four cysteines of the C-terminal cluster, B. cereus Fnr contains three other cysteine residues. Two of them (Cys61 and Cys174) are in conserved positions in relation to those of FnrBac, while the third (Cys85) is replaced by a valine in FnrBac (42). The latter difference might be a signature of B. cereus-type CRP-Fnr proteins, since it has been found in other sequenced members of the B. cereus group (21, 42).
Expression of fnr in aerobic and anaerobic B. cereus F4430/73 cells.
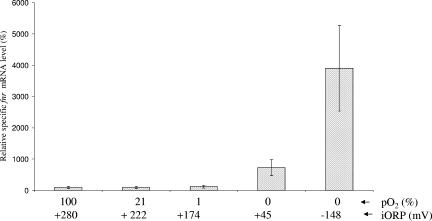
Using real-time RT-PCR, we investigated the expression of B. cereus fnr in aerobic (pO2 = 100% and 21%), microaerobic (pO2 = 1%), and anaerobic F4430/73 cells grown in pH-regulated batch cultures on glucose-containing MOD medium. Our focus was on a single physiological state, i.e., the mid-exponential growth phase (μ = μmax), which is the phase showing the highest level of transcription of fnr (data not shown). We used the transcription of the housekeeping ssu gene (11, 12, 60) as a normalization control. As shown in Fig. 3, levels of fnr transcripts remained unaffected by changes in O2 tension between 100 and 1%. In contrast, fnr mRNA levels were increased during anaerobic fermentative growth, especially when it took place at low ORPs (initial ORP = −148 mV). These results indicate that fnr expression in B. cereus is activated by anaerobiosis and highly reducing conditions for fermentative cells.
FIG. 3.
Effect of growth conditions on specific levels of fnr transcripts. Wild-type F4430/73 cells were grown in regulated batch cultures (pH 7.2) under aerobic (pO2 = 100% and 21%), microaerobic (pO2 = 1%), and anaerobic (pO2 = 0%) conditions at different initial ORPs (iORP) as described in Materials and Methods. Relative specific levels can be compared to the 100% value that was arbitrarily fixed for the fully aerobic condition (pO2 = 100%). For each experiment, two measures from two independent RNA samples from the same culture taken during the mid-exponential growth phase (μ = μmax) were analyzed in parallel. Each data point represents an average of the results for the combined experiments. RNA sampling and statistical methods were identical to those described in Table 4 and in Materials and Methods.
Impact of fnr mutation on the aerobic and anaerobic growth behavior of B. cereus F4430/73.
To investigate the role of B. cereus Fnr, an fnr-disrupted mutant was created by the integration of the suicide vector pMUTIN4 (54). No transcription of the disrupted gene was detected by RT-PCR analyses (data not shown), proving that the genomic disruption of the gene generated a null allele.
Growth features of the fnr mutant and the parental F4430/73 strain under aerobiosis (pO2 = 100%) and anaerobiosis (pO2 = 0%; generated under an N2 atmosphere) were compared. Both strains were also compared under conditions favoring anaerobic respiration by using 20 mM nitrate or fumarate as a potential terminal electron acceptor. As shown in Table 2, fnr disruption only moderately disturbed aerobic respiration but abolished anaerobic fermentative growth. This defect was restored by the addition of nitrate, which allowed the cells to harbor a mixed respirofermentative metabolism (45). Fumarate did not restore the growth of the fnr mutant and did not modify the fermentative growth parameters of the wild-type (Table 2). Thus, B. cereus, like B. subtilis (7, 10, 33, 59), was probably unable to perform anaerobic fumarate respiration. This conclusion was backed up by the absence of anaerobic growth of B. cereus F4430/73 on MOD medium with 60 mM glycerol (as a respiratory substrate) and 20 mM fumarate (data not shown). Taken together, these data indicate that the fnr mutant is unable to grow under glucose-based fermentative conditions and that this growth defect is repaired by respiratory functionality. Under anaerobic nitrate conditions, there was no difference in the μmax between the fnr mutant strain and the wild type but the percentage of glucose consumed by the mutant decreased by 88% compared to that consumed by the wild type and the final biomass was twofold lower. The major defect in glucose consumption was partially compensated for by a higher biomass yield (and ATP-generating acetate overproduction). From these results, we conclude that glucose catabolism is strongly affected in the fnr mutant. In order to determine whether fnr disruption also disturbed anaerobic respiration, we compared the growth of the fnr mutant with that of its parental F4430/73 strain on MOD medium supplemented with glycerol as a carbon source (Table 3). The data showed that the fnr mutant required a high level of nitrate in order to grow similarly to the wild type. This confirms that fnr plays a role in anaerobic nitrate respiration. However, this role is moderate compared to the role played in fermentation. Finally, we concluded that fnr is generally important for the anaerobic growth of B. cereus.
TABLE 2.
Results for pH-regulated batch cultures of the fnr mutant and its parent strain, B. cereus F4430/73a
| Parameter or yield | Value under indicated growth conditions
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Aerobic (respiration)
|
Anaerobic
|
|||||||
| Fermentation
|
Fumarateb respiration
|
Nitrateb respiration
|
||||||
| WT | fnr | WT | fnr | WT | fnr | WT | fnr | |
| Initial ORP (mV) | +280 | +280 | +47 | NG | +77 | NG | +180 | +180 |
| Final ORP (mV) | +160 | +160 | −57 | NG | −14 | NG | +5 | +73 |
| Growth parameters | ||||||||
| μmax (h−1) | 1.4 | 1.1 | 0.9 | NG | 0.9 | NG | 1.0 | 1.0 |
| Final biomass (g/liter) | 2.5 | 2.8 | 0.8 | NG | 0.8 | NG | 1.0 | 0.5 |
| Glucose consumed (%) | 100 | 100 | 100 | NG | 100 | NG | 100 | 12 |
| Nitrate consumed (%) | ND | ND | ND | NG | ND | NG | 100 | 99 |
| Residual nitrite (mM) | ND | ND | ND | NG | ND | ND | 0.1 | 0.7 |
| Yields of end products (mol/mol of glucose) | ||||||||
| Biomassc | 0.83 | 0.93 | 0.27 | NG | 0.25 | NG | 0.34 | 1.40 |
| Lactate | NZ | NZ | 1.40 | NG | 1.24 | NG | 0.90 | 0.5 |
| Acetate | 0.96 | 0.56 | 0.29 | NG | 0.22 | NG | 0.73 | 1.11 |
| Formate | NZ | NZ | 0.34 | NG | 0.52 | NG | 0.56 | 0.43 |
| Ethanol | NZ | NZ | 0.08 | NG | 0.18 | NG | NZ | NZ |
| Succinate | NZ | 0.94 | 0.04 | NG | 0.05 | NG | 0.10 | 0.21 |
| Carbon recovered (%)d | 39 | 73 | 92 | NG | 89 | NG | 86 | 91 |
| Oxidation state balancee | −1.24 | −2.33 | −0.21 | NG | −0.26 | NG | +0.29 | −0.50 |
Cultures were grown on MOD medium containing 30 mM glucose with or without an alternative electron acceptor. Values are means of results for triplicate cultures. For clarity, standard deviations (below 10%) are not shown. WT, wild type (B. cereus F4430/73); fnr, fnr mutant; NG, no growth; ND, not determined; NZ, yield was near zero or below 0.01.
Potassium salts of this acid were added to the medium at a final concentration of 20 mM.
The biomass yield was calculated (22) with the following approximate molecular formula: molecular mass of C4H7O2N ≈ 100 g/mol.
The values were corrected for CO2 fixation related to the formation of succinate (see Fig. 1). The biomass was taken into account as a carbon content of 50%.
The oxidation state balance was calculated as the sum of products with positive oxidation states and products with negative oxidation states per mole of consumed glucose. The balance was corrected for CO2 fixation. For more details, see Materials and Methods.
TABLE 3.
Results for pH-regulated batch cultures of the fnr mutant and its parent strain, B. cereus F4430/73a
| Parameter or yield | Value for anaerobic growth with nitrate
|
|||
|---|---|---|---|---|
| 20 mM
|
60 mM
|
|||
| WT | fnr | WT | fnr | |
| Initial ORP (mV) | +150 | +150 | +170 | +170 |
| Final ORP (mV) | +20 | +60 | +30 | +70 |
| Growth parameters | ||||
| μmax (h−1) | 0.8 | 0.4 | 0.9 | 0.8 |
| Final biomass (g/liter) | 0.5 | 0.2 | 0.6 | 0.6 |
| Glycerol consumed (%) | 75 | 75 | 91 | 91 |
| Nitrate consumed (%) | 100 | 100 | 99 | 99 |
| Residual nitrite (mM) | NZ | NZ | 0.04 | 0.05 |
| Yields of end products (mol/mol of glycerol) | ||||
| Biomass | 0.11 | 0.05 | 0.11 | 0.11 |
| Lactate | NZ | NZ | NZ | NZ |
| Acetate | 0.32 | 0.11 | 0.35 | 0.15 |
| Formate | 0.06 | 0.04 | 0.02 | 0.03 |
| Ethanol | NZ | NZ | NZ | NZ |
| Succinate | NZ | NZ | 0.03 | 0.10 |
| Carbon recovered (%) | 34 | 12 | 37 | 27 |
| Oxidation state balance | +0.41 | +0.14 | +0.56 | −0.11 |
Cultures were grown under anaerobic conditions on MOD medium with 60 mM glycerol as the carbon source and nitrate as the electron acceptor. Values are means of results for triplicate cultures. For clarity, standard deviations (below 10%) are not shown. Data are presented as described in the footnotes to Table 2. WT, wild type (B. cereus F4430/73); fnr, fnr mutant; NG, no growth; NZ, yield near zero or below 0.01.
Analysis of by-product profiles of the fnr mutant.
The amounts of by-products excreted by wild-type B. cereus and the fnr mutant under aerobic and anaerobic nitrate conditions were compared (Tables 2 and 3). During anaerobic respirofermentative growth on glucose plus nitrate, the amounts of lactate and formate in the fnr mutant fell by approximately 45% and 25%, respectively. In contrast, acetate and succinate were overproduced. As a consequence, the oxidation state balance was significantly modified (Table 2). These results suggested that fnr had an impact on carbon partitioning at the three putative branch points of the fermentative pathways, i.e., the phosphoenolpyruvate (if succinate is produced through phosphoenolpyruvate carboxykinase), pyruvate, and acetyl coenzyme A nodes (Fig. 1). Acetate overproduction and the concomitant inhibition of formate production might be the result of pyruvate dehydrogenase complex activation at the expense of pyruvate formate lyase (Pfl). In contrast, during anaerobic respiratory growth on glycerol, the fnr mutant excreted two- to threefold-lower levels of acetate (the usual, most persistent by-product) than the wild-type strain (Table 3). This decrease was not compensated for by an increase in formate levels, notably because Pfl is poorly functional under respiratory conditions. An additional finding was the effect of fnr on succinate production under almost all the growth conditions tested (Table 2 and 3). This impact was especially visible during aerobic glucose respiration. Therefore, fnr plays a key role in carbon metabolism and the production of acidic by-products under both anaerobic and aerobic growth conditions.
Expression profiles of genes involved in glucose catabolism and nitrate respiration under anaerobiosis.
Using real-time RT-PCR, we investigated the effect of the presence of nitrate on the transcription of both glucose catabolism genes and nitrate respiration genes under anaerobiosis (Table 4). We observed a strong change in the mRNA profiles of two (ldhA and ldhC) of the three (ldhA, ldhB, and ldhC) genes predicted to encode l-lactate dehydrogenase and also a significant decrease in the levels of transcripts of adhE (encoding acetaldehyde dehydrogenase) and adhA (encoding alcohol dehydrogenase) in the presence versus in the absence of nitrate. These variations in transcription levels were consistent with the lower production of lactate and ethanol (Table 2). The transcription of pdhA remained unchanged in the presence of nitrate, indicating that pyruvate dehydrogenase (Pdh) is essential for both the fermentative and respirofermentative anaerobic growth of B. cereus. In contrast, the transcription of pfl was strongly reduced when nitrate was added to the growth medium. This finding was in agreement with a diminished role of Pfl whenever respiration is present but did not match with formate production data (Table 2). Thus, it is still unclear how formate production is regulated in B. cereus. The level of sdhA mRNA was increased twofold in the presence of nitrate. Succinate levels exhibited a similar trend (Table 2). These data suggested that the anaerobic fumarate reductase activity involved in succinate production was due to the respiratory succinate dehydrogenase (SdhABC). As expected, the levels of transcripts of the genes encoding nitrate and nitrite reductases (narG and nasD) were much higher in the presence of nitrate. Nitrate also modified the expression profiles of genes encoding respiratory dehydrogenases (yutJ, yjlD, yrkE, and yumB). Interestingly, there was no difference in the transcription of resD and fnr under anaerobic nitrate conditions and aerobic conditions while fermentative conditions enhanced the transcription of these genes (Table 4). This suggests that nitrate respiration, unlike glucose fermentation, may not be governed by the overexpression of these anaerobic regulators.
TABLE 4.
Expression of genes involved in glucose metabolism, nitrate respiration, and enterotoxin production in B. cereus F4430/73 grown under anaerobic conditionsa
| Gene | Description of product | Induction or repression (n-fold) under indicated conditionsb
|
|
|---|---|---|---|
| Nitrate respiration | Fermentation | ||
| Genes for glucose catabolism | |||
| ptsG | Phosphotransferase system; glucose-specific ABC II component | +1.5 | +2.1 |
| ptsH | Phosphotransferase system; phosphocarrier protein HPr | +1.5 | 1.0 |
| pykA | Pyruvate kinase I | +1.1 | +1.1 |
| pykF | Pyruvate kinase II | −4.4 | +2.1 |
| pckA | Phosphoenolpyruvate carboxylase | +1.3 | −1.6 |
| pycA | Pyruvate carboxylase | +3.2 | +1.4 |
| ldhA | Lactate dehydrogenase I | −34.0 | +2083 |
| ldhB | Lactate dehydrogenase II | +5.5 | +8.4 |
| ldhC | Lactate dehydrogenase III | −16.5 | +12.3 |
| pfl | Pyruvate formate lyase | +3.7 | +86.8 |
| pdhA | Pyruvate dehydrogenase E1 alpha subunit | +6.0 | +6.9 |
| pta | Phosphate acetyltransferase | +17.5 | +11.6 |
| ackA | Acetate kinase | +4.2 | +11.2 |
| adhE | Acetaldehyde dehydrogenase | +6.7 | +40.1 |
| adhA | Alcohol dehydrogenase | −1.1 | +17.8 |
| sdhA | Succinate dehydrogenase flavoprotein subunit | +29.0 | +15.2 |
| Genes for nitrate respiration | |||
| yutJ | Respiratory dehydrogenase I | +7.9 | +3.8 |
| yjlD | Respiratory dehydrogenase II | +2.3 | +2.9 |
| yrkE | Respiratory dehydrogenase III | +10.5 | +31.2 |
| yumB | Respiratory dehydrogenase IV | −2.1 | +2.7 |
| narG | Nitrate reductase alpha subunit | +174.8 | +19.8 |
| nasD | NADH-dependent nitrite reductase | +807.1 | +116.3 |
| Genes for enterotoxin production | |||
| hblA | Hemolysin BL component L2 | −5.5 | +10.2 |
| nheC | Nonhemolytic enterotoxin component A | −11.0 | +6.8 |
| Regulators | |||
| resD | Two-component response regulator | +1.7 | +4.6 |
| fnr | One-component regulator | −1.6 | +6.9 |
| plcR | Phospholipase C regulator | +2.5 | +17.0 |
F4430/73 anaerobic cells were grown in pH-regulated batch cultures on MOD medium with 30 mM glucose as the carbon source and 20 mM nitrate when required.
Each value represents the mean specific level of the mRNA in the anaerobic sample in relation to that in the aerobic sample. For each experiment, two measures from two independent RNA samples from the same culture taken during the mid-exponential growth phase (μ = μmax) were analyzed in parallel. Each value is an average of the results from the combined experiments. Only ratios of ≤0.5 and ≥2 were considered to be significant (i.e., P of ≤0.05) according to the precision of the method. + and − indicate up- and down-regulation of genes, respectively.
Involvement of fnr as a metabolic transcriptional regulator.
Since the fnr mutant cannot grow under fermentative conditions, mRNA levels from both wild-type and mutant strains under aerobic and anaerobic nitrate conditions were compared (Table 5). During aerobic glucose respiration, most of the genes tested were deregulated in the fnr mutant. Among the genes tested, fermentative genes such as ldhA, adhE, and adhA were highly up-regulated while ldhB, pfl, and pdhA showed more moderate deregulation. These fermentative genes are poorly expressed under aerobiosis compared to anaerobiosis (Table 4) according to the mode of energy generation used (Fig. 1), which means that changes in their expression levels had no impact on bacterial growth (Table 2). Under nitrate respirofermentative conditions, the fnr mutation caused no significant deregulation of genes required for glucose catabolism and nitrate respiration. This finding confirms the moderate contribution of fnr to nitrate respiration. However, this result partially conflicts with the disrupted glucose catabolism (Table 2) and suggests that Fnr may modulate glucose catabolism at a mainly posttranscriptional level in the presence of nitrate. Finally, these data suggest that fermentative gene expression in the absence of nitrate depends on fnr expression. As a consequence, the lack of high fnr expression under anaerobiosis could not allow B. cereus to complete fermentation, resulting in the absence of growth. Interestingly, fnr disruption had no impact on resD expression (Table 5), indicating that resD expression was not dependent on fnr expression.
TABLE 5.
Glucose metabolism genes of B. cereus F4430/73 differentially expressed in the fnr mutant strain grown under aerobic and anaerobic nitrate conditionsa
| Gene | Difference in mRNA level (n-fold) under the indicated conditionsb
|
|
|---|---|---|
| Without O2, with nitrate | With O2 | |
| Genes for glucose catabolism | ||
| ptsG | +1.0 | −2.5 |
| ptsH | −1.2 | −4.1 |
| pykA | −2.4 | +8.1 |
| pykF | −1.7 | +8.6 |
| pckA | +1.4 | −13.3 |
| pycA | −1.1 | −2.8 |
| ldhA | +1.0 | +24.1 |
| ldhB | +1.3 | +3.2 |
| ldhC | +1.1 | −1.1 |
| pfl | −1.3 | −3.5 |
| pdhA | +1.3 | +2.8 |
| pta | +1.1 | +1.5 |
| ackA | +1.2 | +1.3 |
| adhE | −1.3 | +26.6 |
| adhA | −1.4 | +10.7 |
| sdhA | +1.2 | +34.0 |
| Genes for nitrate respiration | ||
| yutJ | −1.6 | −1.8 |
| yjld | +1.6 | −2.5 |
| yrkE | −1.3 | +2.3 |
| yumB | +1.5 | −2.5 |
| narG | −1.2 | +2.2 |
| nasD | −1.4 | +2.8 |
| Regulator | ||
| resD | −1.2 | +1.5 |
Cells were grown in pH-regulated batch cultures on MOD medium with 30 mM glucose as the carbon source, and 20 mM nitrate was used if needed.
Each value represents the mean specific level of the mRNA in the mutant sample in relation to that in the wild-type sample. Data are presented as described in Table 4 footnote b.
Analysis of enterotoxin production by the fnr mutant.
To determine the effect of fnr expression on the production of enterotoxins, the specific levels of Hbl and Nhe were quantified at the end points for pH-regulated batch cultures of mutant and wild-type B. cereus strains (Table 6). In the parental F4430/73 strain, the highest production of Hbl and Nhe was observed under anaerobiosis when cells were grown fermentatively, i.e., on glucose-MOD medium in the presence or absence of fumarate. This finding indicates that enterotoxin production is not enhanced by anaerobic conditions per se (60) but rather by the mode of anaerobic growth. Interestingly, the fnr mutation inhibited Hbl and Nhe production to almost undetectable levels under both aerobic and anaerobic conditions (more than 99% reduction compared to that in the parental strain). This result indicates that fnr expression is essential for Hbl and Nhe production whatever the mode of growth.
TABLE 6.
Comparison of specific levels of enterotoxin production between the fnr mutant and its parent strain, B. cereus F4430/73a
| Growth conditions | Amt of enterotoxin (U g−1 [dry weight] of cells)
|
|||
|---|---|---|---|---|
| Hbl
|
Nhe
|
|||
| WT | fnr | WT | fnr | |
| Aerobic | ||||
| Glucose | 1.0 ± 0.2 | NZ | 1.5 ± 0.3 | NZ |
| Anaerobic | ||||
| Glucose | 12.0 ± 2.0 | NG | 8.0 ± 1.0 | NG |
| Glucose and 20 mM fumarate | 10.0 ± 2.0 | NG | 5.5 ± 0.5 | NG |
| Glucose and 20 mM nitrate | 1.0 ± 0.2 | NZ | 2.1 ± 0.2 | NZ |
| Glycerol and 20 mM nitrate | 0.16 ± 0.03 | NZ | 0.37 ± 0.03 | NZ |
| Glycerol and 60 mM nitrate | NZ | NZ | NZ | NZ |
Hbl and Nhe levels were measured in culture supernatants at the end points for pH-regulated batch cultures. Cells were grown under aerobiosis or anaerobiosis on MOD medium supplemented with glucose (30 mM) or glycerol (60 mM) as a carbon source and with or without an external electron acceptor (nitrate or fumarate). Values are means ± standard deviations of three independent measurements. WT, wild type (B. cereus F4430/73); fnr, fnr mutant; NG, no growth; NZ, yield near zero or below 0.01.
The fnr gene is involved in enterotoxin gene activation.
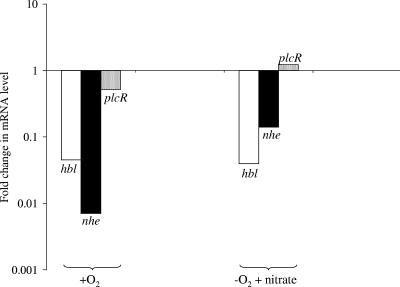
The fnr-dependent transcription of hbl and nhe was investigated using real-time RT-PCR with total RNA obtained from fnr mutant cells cultivated on glucose under aerobic (respiratory) and anaerobic nitrate (respirofermentative) conditions. As shown in Fig. 4, transcription of hbl and nhe was dramatically (>90%) down-regulated. This explains why enterotoxins were almost undetectable in the culture supernatants of the fnr mutant under all the growth conditions tested. Therefore, these results identify fnr as a crucial component of the regulatory system of enterotoxin expression in B. cereus. Furthermore, the observed variations in levels of PlcR-regulated hbl and nhe transcripts were not correlated with the variations in plcR mRNA levels (Fig. 4). This indicates that the fnr-dependent regulation of enterotoxin expression could not be mediated directly by PlcR.
FIG. 4.
Changes in specific levels of transcripts of the PlcR-regulated genes hbl, nhe, and plcR induced by fnr mutation under aerobic (respiratory) and anaerobic nitrate (respirofermentative) growth conditions. The change in the mRNA level for each gene in the fnr mutant was calculated in relation to the mRNA level for the wild-type. Data presentation is as described in the legend to Fig. 3.
Complementation of the fnr mutant.
To confirm that the defect in fermentative growth and enterotoxin production was specifically ascribable to the disruption of fnr, a wild-type copy of the fnr locus cloned into the pHT304aph3 vector (2) and the vector was used to complement the fnr mutant strain. The trans complementation with fnr restored the fermentative growth of the fnr mutant (Table 7) and permitted the mutant to produce enterotoxins at levels comparable to those obtained with the wild-type strain (data not shown).
TABLE 7.
Results for uncontrolled batch cultures of the wild-type F4430/73 and the derivative fnr mutant transformed with the pHT304aph3-fnr recombinant vectora
| Strain | Final OD560 under indicated growth conditions
|
|
|---|---|---|
| Aerobic | Anaerobic | |
| WT | 0.60 ± 0.02 | 0.32 ± 0.01 |
| WT(pHT304aph3) | 0.57 ± 0.02 | 0.30 ± 0.02 |
| WT(pHT304aph3-fnr) | 0.60 ± 0.02 | 0.36 ± 0.01 |
| fnr | 0.55 ± 0.03 | NG |
| fnr(pHT304aph3) | 0.45 ± 0.01 | NG |
| fnr(pHT304aph3-fnr) | 0.45 ± 0.01 | 0.28 ± 0.02 |
Cells were grown as described in Material and Methods. Values are the means ± standard deviations of results for triplicate cultures. WT, wild type; fnr, fnr mutant; OD560, optical density at 560 nm; NG, no growth.
DISCUSSION
This research provides additional data on the anaerobic metabolic features of B. cereus. As has been well demonstrated with other facultative anaerobes (49), our data notably showed that key anaerobic catabolic enzymes were transcriptionally regulated by the availabilities of O2 and nitrate (as terminal electron acceptors). Similar to B. subtilis, B. cereus was unable to grow anaerobically by fumarate respiration but likely uses the succinate dehydrogenase SdhABC (respiratory complex II) for the reduction of fumarate to succinate during anaerobic fermentation and respirofermentation (with nitrate). Both bacilli possess genes encoding the putative fumarate transporter (ERGO; Integrated Genomics, Inc.; RZC06566) but lack the frd gene encoding fumarate reductase as well as genes resembling nuoAN encoding the subunits of the proton-pumping NADH dehydrogenase I of E. coli. It is thus likely that, as that of B. subtilis, the lack of anaerobic respiratory growth of B. cereus on fumarate is due to the absence of Nuo-type NADH dehydrogenase, which was reported as the only site for H+ translocation in fumarate respiration (53). It is still unknown whether the fumarate reductase activity of B. cereus could generate an electrochemical membrane potential via a redox half-loop (47). The NADH-recycling succinate pathway (also known as the reducing branch of the tricarboxylic acid cycle) is not a major fermentation route in B. cereus. Consistent with this hypothesis, succinate is formed as a minor by-product (Table 2), genes encoding key fermentative enzymes (ldhA, pfl, and adhA) were more highly up-regulated than sdhA during glucose fermentation (Table 4), and conversely, succinate levels and sdhA transcription were increased under anaerobic nitrate (respirofermentative) conditions while the activities of true fermentative pathways were reduced (Tables 2 and 4).
Unlike B. subtilis (33, 59), B. cereus has a very efficient glucose-fermentative metabolism which allows it to grow under highly reducing (low-ORP) conditions (60). In the fermentation pathways of B. cereus (Fig. 1), the conversion of pyruvate to acetyl coenzyme A is catalyzed by both pyruvate formate lyase (Pfl, which is absent in B. subtilis) and the pyruvate dehydrogenase complex (Pdh). Fermentation appeared to require high levels of Pfl compared to Pdh (Table 4), probably to avoid excessive NADH formation (Fig. 1). In contrast, Pdh may catabolize pyruvate preferentially under nitrate respiratory conditions, during which NADH reoxidation is less limiting. The main NAD+-regenerating fermentative pathway in B. cereus is the pyruvate-to-lactate pathway (Table 2). In B. cereus, this pathway involved three putative lactate dehydrogenases, instead of two as in B. subtilis. The anaerobic expression patterns of ldhA, ldhB, and ldhC clearly depend on the availability of the external electron acceptor (Table 4). In more specific terms, ldhA and ldhC were strongly down-regulated in the presence of nitrate, resulting in lower lactate production. The regulation of formate production was less clear, since pfl transcript levels and formate levels showed opposite trends under anaerobic nitrate conditions (Tables 2 and 4). The involvement (not yet excluded) of one or more putative anaerobic formate dehydrogenases (ERGO; Integrated Genomics Inc.) may complicate the analysis of formate levels. Our data also showed that the expression of the four genes encoding putative respiratory NADH dehydrogenases was also strongly influenced by nitrate and O2. This result will be further examined together with the levels of expression of the other aerobic or anaerobic potential dehydrogenases and respiratory components.
In B. subtilis, respiratory and fermentative enzymes are regulated by oxygen availability via a signal transduction system, which includes the one-component Fnr and the two-component ResDE systems (35, 36). A regulatory model was recently proposed (43) taking into account that B. subtilis fnr directly activates nitrate respiration via induction of the nitrate reductase operon (narGHJIK) and nitrate and nitrite transporter gene (narK) and indirectly controls the transcriptional activity of fermentative genes via the fermentation modulator ArfM (31) and the redox regulator YdiH (29). In this study, we identified in B. cereus a gene homologous to B. subtilis fnr located in a chromosomal region that, like the corresponding region in B. subtilis, comprises the genes encoding nitrate and nitrite reductase (9). However, these genes showed a genetic organization different from that observed in B. subtilis, and apart from fnr we found no other genes encoding proteins similar to regulatory proteins (such as arfM) in the vicinity of these nitrate respiratory genes. It should be emphasized that this organization appeared to be a characteristic of the sequenced members of the B. cereus group (references 21 and 41 and data not shown). The genetic organization of the fnr locus (Fig. 2) indicates that B. cereus fnr transcription is probably monocistronic, whereas B. subtilis fnr was expressed either as an fnr-specific transcript or as a bicistronic narK-fnr mRNA (9). Therefore, in B. cereus the absence of fnr up-regulation in the presence of nitrate (Table 4) could reflect the absence of the control via the narK promoter (which induced fnr transcription in B. subtilis). Putative Fnr-binding sites were found centered at positions −154.5 and +28.5 relative to the transcriptional start point of the fnr promoter of B. cereus. The positions of these Fnr boxes are different from those of B. subtilis Fnr-activated promoters (the palindromic sequence is centered on bp −40.5) (43) but consistent with those of CRP-activated promoters (27), suggesting that fnr autoregulation patterns may be different in B. cereus and B. subtilis. Analysis of the B. cereus fnr promoter region also revealed the presence of a putative binding sequence for the response regulator ResD (12). Furthermore, data presented in the study (Table 4) showed a correlation between the anaerobic changes in fnr expression and resD expression. This finding suggests that in B. cereus, as in B. subtilis (31), the anaerobic condition-dependent fnr signal transduction system is probably controlled by ResD. We have previously shown that like Fnr, the B. cereus ResDE system does not play a major role in aerobic respiration but unlike Fnr, it does not play a major role in fermentation under classical (N2 atmosphere) anaerobic conditions (12). In contrast, the ResDE-dependent regulation is essential under highly reducing (low-ORP) fermentative conditions generated under an H2 atmosphere. From additional experiments (data not shown), we observed that fnr expression was strongly reduced in a resD mutant when the mutant was grown under fermentative conditions (5- and 25-fold decreases under high- and low-ORP conditions, respectively) while no significant effect under respiratory conditions was observed. Therefore, the beginning of the regulatory pathway controlling the catabolic gene expression under anaerobiosis in B. cereus could involve, in hierarchical order, first the two-component ResDE system and second Fnr, as in B. subtilis (43). Subsequently, Fnr may act differently in B. cereus than in B. subtilis since Fnr is essential for glucose fermentation and dispensable for anaerobic nitrate respiration in B. cereus while it is essential for nitrate respiration and dispensable for glucose fermentation in B. subtilis. In B. cereus, Fnr may directly regulate certain key fermentative genes (such as ldh, pfl, and adh) via a cis-acting Fnr box in the corresponding promoter regions (such Fnr boxes were found using bioinformatics approaches; data not shown) and indirectly regulate the genes involved in nitrate formation. The link between nitrate respiration and fermentation may involve a redox regulator such as YdiH (29), since a gene encoding this B. subtilis homolog has been found in the B. cereus genome (ERGO; Integrated Genomics, Inc.; RZC01387). Nevertheless, this putative regulatory model requires experimental validation.
In B. subtilis, the ability of Fnr to function as a transcription factor depends on its level of synthesis and on the integrity of its [4Fe-4S]2+ cluster, which promotes a conformation that is necessary for site-directed DNA binding and transcriptional activation. From a structural point of view, the predicted Fnr of B. cereus resembles the B. subtilis Fnr (42). It is thus tempting to speculate that the [4Fe-4S]2+ cluster could also be a key component required for the DNA-binding activity of B. cereus Fnr under anaerobiosis. However, the data presented here indicated that B. cereus Fnr modulates the transcription of some catabolic genes under both anaerobic and aerobic conditions (Table 5). For instance, the fnr mutation led to an abnormal overproduction of succinate during aerobic glucose respiration. This finding suggests that the B. cereus Fnr conserved some site-specific DNA-binding properties under aerobiosis, thus supporting the notion that the stability of the [4Fe-4S] cluster could be conserved, at least partially, in aerobically grown B. cereus cells, as reported for E. coli (5, 16). No such result has been reported for B. subtilis Fnr. This led us to consider that the B. subtilis and B. cereus Fnr proteins could not show the same sensitivities to oxygen due to a difference in their cluster stabilities. The findings that there is a significant variation in the amino acid residues around the cysteine ligands in the two proteins (42) and that the stability of the Fe-S cluster is dependent on this composition (55) further support this hypothesis. Clearly, pioneering experimental approaches are required to address this issue. In any case, the most convincing evidence that B. cereus Fnr can exist in an active state under aerobiosis was provided by the studies of enterotoxin expression (Table 6; Fig. 4). Indeed, B. cereus Fnr is required for the expression of PlcR-regulated genes hbl and nhe with or without the presence of oxygen. Surprisingly, enterotoxin gene expression was lower under anaerobic nitrate conditions than under aerobiosis while levels of fnr expression were not significantly different (Table 4). Given that B. cereus cells grown anaerobically with nitrate as the terminal electron acceptor generate significant NO (26), we propose that the [4Fe-4S] cluster of Fnr could be NO labile, as reported for E. coli (8). Assuming that Fnr binds at an Fnr box within the hbl and nhe promoter (data not shown), NO-inactivated Fnr may have a lower affinity for DNA than O2-inactivated Fnr and consequently could have a lower inducible capacity. However, while the expression of enterotoxin genes was lower under anaerobic nitrate conditions, specific levels of production of Hbl and Nhe remained similar, suggesting posttranscriptional regulation of enterotoxin expression. Under anaerobiosis, the expression of enterotoxin genes is thus controlled by at least two separate pathways, i.e., the PlcR-dependent pathway and the Fnr (ResDE)-dependent pathway. The signals triggering or blocking the Fnr (ResDE) pathway may be O2 and NO, which may interact not only with Fnr but also ResE (34) and other small redox effector molecules produced in response to anaerobic and highly reducing conditions.
In conclusion, this study showed that Fnr plays a more essential role than the ResDE two-component system in B. cereus fermentative growth. In addition, Fnr, like ResE, is essential for the production of the major virulence factors, i.e., enterotoxins Hbl and Nhe (13, 57). Therefore, the interplay between fermentative growth and virulence in a pathogen such as B. cereus F4430/73 undoubtedly implicates Fnr as a major regulatory protein. In the human intestine, B. cereus encounters anaerobic, highly reducing fermentative conditions (6, 20, 32, 50). The diarrheal syndrome may thus result from the capacity of a B. cereus strain to efficiently ferment carbohydrate in the small intestine environment.
Acknowledgments
A.Z. received a fellowship from the Franco-Algerian intergovernmental program.
Footnotes
Published ahead of print on 26 January 2007.
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Okstad, A. B. Kolsto, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. [DOI] [PubMed] [Google Scholar]
- 3.Baltes, N., M. N′Diaye, I. D. Jacobsen, A. Maas, F. F. Buettner, and G. F. Gerlach. 2005. Deletion of the anaerobic regulator HlyX causes reduced colonization and persistence of Actinobacillus pleuropneumoniae in the porcine respiratory tract. Infect. Immun. 73:4614-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartolini, E., E. Frigimelica, S. Giovinazzi, G. Galli, Y. Shaik, C. Genco, J. A. Welsch, D. M. Granoff, G. Grandi, and R. Grifantini. 2006. Role of FNR and FNR-regulated, sugar fermentation genes in Neisseria meningitidis infection. Mol. Microbiol. 60:963-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates, D. M., C. V. Popescu, N. Khoroshilova, K. Vogt, H. Beinert, E. Munck, and P. J. Kiley. 2000. Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S](2+) cluster to oxygen. J. Biol. Chem. 275:6234-6240. [DOI] [PubMed] [Google Scholar]
- 6.Bernier, J. J., J. Adrian, and N. Vidon. 1988. Localisation de la digestion des glucides dans le tube digestif, p. 211-215. In Doin (ed.), Les aliments dans le tube digestif. Collection Biosciences et Techniques, Paris, France.
- 7.Clements, L. D., U. N. Streips, and B. S. Miller. 2002. Differential proteomic analysis of Bacillus subtilis nitrate respiration and fermentation in defined medium. Proteomics 2:1724-1734. [DOI] [PubMed] [Google Scholar]
- 8.Cruz-Ramos, H., J. Crack, G. Wu, M. N. Hughes, C. Scott, A. J. Thomson, J. Green, and R. K. Poole. 2002. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 21:3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz Ramos, H., L. Boursier, I. Moszer, F. Kunst, A. Danchin, and P. Glaser. 1995. Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 14:5984-5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz Ramos, H., T. Hoffmann, M. Marino, H. Nedjari, E. Presecan-Siedel, O. Dreesen, P. Glaser, and D. Jahn. 2000. Fermentative metabolism of Bacillus subtilis: physiology and regulation of gene expression. J. Bacteriol. 182:3072-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duport, C., S. Thomassin, G. Bourel, and P. Schmitt. 2004. Anaerobiosis and low specific growth rates enhance hemolysin BL production by Bacillus cereus F4430/73. Arch. Microbiol. 182:90-95. [DOI] [PubMed] [Google Scholar]
- 12.Duport, C., A. Zigha, E. Rosenfeld, and P. Schmitt. 2006. Control of enterotoxin gene expression in Bacillus cereus F4430/73 involves the redox-sensitive ResDE signal transduction system. J. Bacteriol. 188:6640-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gohar, M., N. Gilois, R. Graveline, C. Garreau, V. Sanchis, and D. Lereclus. 2005. A comparative study of Bacillus cereus, Bacillus thuringiensis and Bacillus anthracis extracellular proteomes. Proteomics 5:3696-3711. [DOI] [PubMed] [Google Scholar]
- 14.Gohar, M., O. A. Okstad, N. Gilois, V. Sanchis, A. B. Kolsto, and D. Lereclus. 2002. Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics 2:784-791. [DOI] [PubMed] [Google Scholar]
- 15.Gompertz, B. 1925. On the nature of the function expressive of the law of human mortality, and on a new mode of determining the value of life contingencies. Philos. Trans. R. Soc. 115:513-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green, J., and M. S. Paget. 2004. Bacterial redox sensors. Nat. Rev. Microbiol. 2:954-966. [DOI] [PubMed] [Google Scholar]
- 17.Green, L. C., D. A. Wagner, J. Glogowski, P. L. Skipper, J. S. Wishnok, and S. R. Tannenbaum. 1982. Analysis of nitrate, nitrite, and [15N] nitrate in biological fluids. Anal. Biochem. 126:131-138. [DOI] [PubMed] [Google Scholar]
- 18.Guinebretiere, M. H., V. Broussolle, and C. Nguyen-The. 2002. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 40:3053-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guinebretiere, M. H., and C. Nguyen-The. 2003. Sources of Bacillus cereus contamination in a pasteurized zucchini purée processing line, differentiated by two PCR-based methods. FEMS Microbiol. Ecol. 43:207-215. [DOI] [PubMed] [Google Scholar]
- 20.Guyton, A. C. 1977. Normal function and mechanisms of diseases, p. 408-423. W. B. Saunders, Philadelphia, PA.
- 21.Han, C. S., G. Xie, J. F. Challacombe, M. R. Altherr, S. S. Bhotika, D. Bruce, C. S. Campbell, M. L. Campbell, J. Chen, O. Chertkov, C. Cleland, M. Dimitrijevic, N. A. Doggett, J. J. Fawcett, T. Glavina, L. A. Goodwin, K. K. Hill, P. Hitchcock, P. J. Jackson, P. Keim, A. R. Kewalramani, J. Longmire, S. Lucas, S. Malfatti, K. McMurry, L. J. Meincke, M. Misra, B. L. Moseman, M. Mundt, A. C. Munk, R. T. Okinaka, B. Parson-Quintana, L. P. Reilly, P. Richardson, D. L. Robinson, E. Rubin, E. Saunders, R. Tapia, J. G. Tesmer, N. Thayer, L. S. Thompson, H. Tice, L. O. Ticknor, P. L. Wills, T. S. Brettin, and P. Gilna. 2006. Pathogenomic sequence analysis of Bacillus cereus and Bacillus thuringiensis isolates closely related to Bacillus anthracis. J. Bacteriol. 188:3382-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbert, D., P. J. Phipps, and R. E. Strange. 1971. Chemical analysis of microbial cells, p. 209-344. In J. R. Norris and D. W. Ribbons (ed.), Methods in microbiology, vol. 5B. Academic Press, London, England. [Google Scholar]
- 23.Hill, K. K., L. O. Ticknor, R. T. Okinaka, M. Asay, H. Blair, K. A. Bliss, M. Laker, P. E. Pardington, A. P. Richardson, M. Tonks, D. J. Beecher, J. D. Kemp, A. B. Kolsto, A. C. Wong, P. Keim, and P. J. Jackson. 2004. Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 70:1068-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Householder, T. C., W. A. Belli, S. Lissenden, J. A. Cole, and V. L. Clark. 1999. cis- and trans-acting elements involved in regulation of aniA, the gene encoding the major anaerobically induced outer membrane protein in Neisseria gonorrhoeae. J. Bacteriol. 181:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen, G. B., B. M. Hansen, J. Eilenberg, and J. Mahillon. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5:631-640. [DOI] [PubMed] [Google Scholar]
- 26.Kalkowski, I., and R. Conrad. 1991. Metabolism of nitric oxide in denitrifying Pseudomonas aeruginosa and nitrate-respiring Bacillus cereus. FEMS Microbiol. Lett. 66:107-111. [DOI] [PubMed] [Google Scholar]
- 27.Korner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27:559-592. [DOI] [PubMed] [Google Scholar]
- 28.Kotiranta, A., K. Lounatmaa, and M. Haapasalo. 2000. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2:189-198. [DOI] [PubMed] [Google Scholar]
- 29.Larsson, J. T., A. Rogstam, and C. von Wachenfeldt. 2005. Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis. Microbiology 151:3323-3335. [DOI] [PubMed] [Google Scholar]
- 30.Mahillon, J., and D. Lereclus. 2000. Electroporation of Bacillus thuringiensis and Bacillus cereus, p. 242-252. In N. Eynard and J. Teissie (ed.), Electroporation of bacteria. Springer-Verlag Berlin, Berlin, Germany.
- 31.Marino, M., H. C. Ramos, T. Hoffmann, P. Glaser, and D. Jahn. 2001. Modulation of anaerobic energy metabolism of Bacillus subtilis by arfM (ywiD). J. Bacteriol. 183:6815-6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriarty-Craige, S. E., and D. P. Jones. 2004. Extracellular thiols and thiol/disulfide redox in metabolism. Annu. Rev. Nutr. 24:481-509. [DOI] [PubMed] [Google Scholar]
- 33.Nakano, M. M., Y. P. Dailly, P. Zuber, and D. P. Clark. 1997. Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth. J. Bacteriol. 179:6749-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano, M. M., H. Geng, S. Nakano, and K. Kobayashi. 2006. The nitric oxide-responsive regulator NsrR controls ResDE-dependent gene expression. J. Bacteriol. 188:5878-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakano, M. M., and F. M. Hulett. 1997. Adaptation of Bacillus subtilis to oxygen limitation. FEMS Microbiol. Lett. 157:1-7. [DOI] [PubMed] [Google Scholar]
- 36.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52:165-190. [DOI] [PubMed] [Google Scholar]
- 37.Nakano, M. M., P. Zuber, and A. L. Sonenshein. 1998. Anaerobic regulation of Bacillus subtilis Krebs cycle genes. J. Bacteriol. 180:3304-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okstad, O. A., M. Gominet, B. Purnelle, M. Rose, D. Lereclus, and A. B. Kolsto. 1999. Sequence analysis of three Bacillus cereus loci carrying PlcR-regulated genes encoding degradative enzymes and enterotoxin. Microbiology 145:3129-3138. [DOI] [PubMed] [Google Scholar]
- 39.Ouhib, O., T. Clavel, and P. Schmitt. 2006. The production of Bacillus cereus enterotoxins is influenced by carbohydrate and growth rate. Curr. Microbiol. 53:222-226. [DOI] [PubMed] [Google Scholar]
- 40.Ramarao, N., and D. Lereclus. 2006. Adhesion and cytotoxicity of Bacillus cereus and Bacillus thuringiensis to epithelial cells are FlhA and PlcR dependent, respectively. Microbes Infect. 8:1483-1491. [DOI] [PubMed] [Google Scholar]
- 41.Rasko, D. A., M. R. Altherr, C. S. Han, and J. Ravel. 2005. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol. Rev. 29:303-329. [DOI] [PubMed] [Google Scholar]
- 42.Reents, H., I. Gruner, U. Harmening, L. H. Bottger, G. Layer, P. Heathcote, A. X. Trautwein, D. Jahn, and E. Hartig. 2006. Bacillus subtilis Fnr senses oxygen via a [4Fe-4S] cluster coordinated by three cysteine residues without change in the oligomeric state. Mol. Microbiol. 60:1432-1445. [DOI] [PubMed] [Google Scholar]
- 43.Reents, H., R. Munch, T. Dammeyer, D. Jahn, and E. Hartig. 2006. The Fnr regulon of Bacillus subtilis. J. Bacteriol. 188:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riondet, C., R. Cachon, Y. Wache, G. Alcaraz, and C. Divies. 2000. Extracellular oxidoreduction potential modifies carbon and electron flow in Escherichia coli. J. Bacteriol. 182:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenfeld, E., C. Duport, A. Zigha, and P. Schmitt. 2005. Characterisation of aerobic and anaerobic vegetative growth of the food-borne pathogen Bacillus cereus. Can. J. Microbiol. 51:149-158. [DOI] [PubMed] [Google Scholar]
- 46.Schmiel, D. H., G. M. Young, and V. L. Miller. 2000. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J. Bacteriol. 182:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnorpfeil, M., I. G. Janausch, S. Biel, A. Kroger, and G. Unden. 2001. Generation of a proton potential by succinate dehydrogenase of Bacillus subtilis functioning as a fumarate reductase. Eur. J. Biochem. 268:3069-3074. [DOI] [PubMed] [Google Scholar]
- 48.Schoeni, J. L., and A. C. Wong. 2005. Bacillus cereus food poisoning and its toxins. J. Food Prot. 68:636-648. [DOI] [PubMed] [Google Scholar]
- 49.Shalel-Levanon, S., K. Y. San, and G. N. Bennett. 2005. Effect of ArcA and FNR on the expression of genes related to the oxygen regulation and the glycolysis pathway in Escherichia coli under microaerobic growth conditions. Biotechnol. Bioeng. 92:147-159. [DOI] [PubMed] [Google Scholar]
- 50.Skinner, F. A., and J. G. Carr. 1995. The normal microbial flora of man. Academic Press, London, England.
- 51.Slamti, L., and D. Lereclus. 2002. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 21:4550-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spira, W. M., and J. M. Goepfert. 1975. Biological characteristics of an enterotoxin produced by Bacillus cereus. Can. J. Microbiol. 21:1236-1246. [DOI] [PubMed] [Google Scholar]
- 53.Unden, G. 1998. Transcriptional regulation and energetics of alternative respiratory pathways in facultatively anaerobic bacteria. Biochim. Biophys. Acta 1365:220-224. [Google Scholar]
- 54.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 55.Van Spanning, R. J., A. P. De Boer, W. N. Reijnders, H. V. Westerhoff, A. H. Stouthamer, and J. Van Der Oost. 1997. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcriptional activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Mol. Microbiol. 23:893-907. [DOI] [PubMed] [Google Scholar]
- 56.Vilain, S., Y. Luo, M. B. Hildreth, and V. S. Brozel. 2006. Analysis of the life cycle of the soil saprophyte Bacillus cereus in liquid soil extract and in soil. Appl. Environ. Microbiol. 72:4970-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wassenaar, T. M., and W. Gaastra. 2001. Bacterial virulence: can we draw the line? FEMS Microbiol. Lett. 201:1-7. [DOI] [PubMed] [Google Scholar]
- 58.Wood, G. E., N. Khelef, N. Guiso, and R. L. Friedman. 1998. Identification of Btr-regulated genes using a titration assay. Search for a role for this transcriptional regulator in the growth and virulence of Bordetella pertussis. Gene 209:51-58. [DOI] [PubMed] [Google Scholar]
- 59.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zigha, A., E. Rosenfeld, P. Schmitt, and C. Duport. 2006. Anaerobic cells of Bacillus cereus F4430/73 respond to low oxidoreduction potential by metabolic readjustments and activation of enterotoxin expression. Arch. Microbiol. 185:222-233. [DOI] [PubMed] [Google Scholar]
- 61.Zwietering, M., I. Jongenburger, F. Rombouts, and K. Van't Riet. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zwietering, M. H., F. M. Rombouts, and K. van 't Riet. 1992. Comparison of definitions of the lag phase and the exponential phase in bacterial growth. J. Appl. Bacteriol. 72:139-145. [DOI] [PubMed] [Google Scholar]