Abstract
Recent work with bacteria and eukaryotes has shown that GTPases play important roles in ribosome assembly. Here we show that the essential GTPase YqeH is required for proper 70S ribosome formation and 30S subunit assembly/stability in Bacillus subtilis.
Several GTPases contain a circular permutation of the GTP-binding domain (cpGTPases) in which the normal G1-G2-G3-G4-G5 motif orientation of the GTP-binding domain has been rearranged to G4-G5-G1-G2-G3 (10). Structural, bioinformatic, and biochemical studies of cpGTPases revealed that this G domain permutation occurs in proteins implicated in ribosome assembly or with domains that interact with RNA (1, 6, 8, 11, 15). Several cpGTPases, including the bacterial RbgA protein and Saccharomyces cerevisiae homologs Nog2p, Nug1p, and Lsg1p, have been demonstrated to participate in large ribosomal subunit assembly (2, 8, 11, 15, 16).
The GTPase YqeH is a cpGTPase that has an interesting phylogenetic distribution and is found in many bacterial and eukaryotic genomes (10). YqeH contains an N-terminal zinc ribbon motif that has been implicated in RNA binding and is found in several ribosomal proteins (9). YqeH is essential for growth in several bacteria, but the biological function of YqeH has remained elusive (12, 17). For Bacillus subtilis, cells depleted of YqeH show a loss of coordination of DNA replication, suggesting that YqeH is somehow involved in the negative regulation of DNA replication (12). Interestingly, the Arabidopsis thaliana AtNOS1 protein, a mitochondrial nitric oxide synthase that is required for the plant to resist reactive oxygen stress, is the eukaryotic homolog with the highest degree of similarity to YqeH (5, 7). In this study, we show that YqeH is required for the efficient assembly of 70S ribosomes and of the small ribosomal subunit. Cells depleted of YqeH are greatly reduced in functional 70S ribosomes and 30S subunits. YqeH is therefore a member of the growing number of cpGTPases that have functional roles in ribosome biogenesis.
Effects of depletion of YqeH on cell growth.
Previous work has shown that yqeH, encoding a GTPase of unknown function in B. subtilis, is essential for growth (12). In order to study the effects of depleting YqeH on cell growth, we constructed strains that place the expression of yqeH under the control of the isopropyl-beta-d-thiogalactopyranoside (IPTG)-inducible, LacI-repressible promoter Pspank. A 283-bp fragment containing the ribosome-binding site and the 5′ end of the yqeH gene was cloned into the Pspank vector pJL86, which cannot replicate in B. subtilis. The resulting plasmid (pLS20) was transformed and recombined into the chromosome of B. subtilis by a single crossover. This recombination results in the removal of the full-length yqeH gene from the control of its native promoter and places it under the control of the Pspank promoter (generating strain RB286). Primer sequences for all strain constructions are available upon request.
The location of the yqeH gene suggests that it is coexpressed with a number of additional genes (aroD-yqeIJKLM). Therefore, we were concerned that construction of the Pspank-yqeH strain would result in a decreased expression of genes downstream of yqeH, making it difficult to assign observed phenotypes to yqeH depletion and not to the downstream genes. To address this problem, we constructed an additional strain (RB288) that places aroD, the gene immediately downstream of yqeH, under the control of Pspank. This was achieved by cloning a 282-bp fragment containing the ribosome-binding site and the 5′ end of the aroD gene into the Pspank vector pJL86 (resulting in plasmid pLS21). The Pspank-yqeH and Pspank-aroD strains were grown on LB medium in the presence of 1 mM IPTG and in the absence of IPTG. As expected, both strains formed large colonies after overnight incubation at 37°C in the presence of 1 mM IPTG. However, only the Pspank-yqeH strain showed reduced growth in the absence of IPTG, with small colonies visible after 24 h. The growth rate of the Pspank-aroD (RB288) strain grown in the absence of IPTG was indistinguishable from that of the strain grown with 1 mM IPTG, demonstrating that depletion of YqeH is required for the growth defect. To confirm this result, we constructed a second strain (RB406) in which the full-length yqeH gene was placed at the amyE locus under the control of the PxylA promoter, using the plasmid pSWEET (generating plasmid pMK1), and the native yqeH gene was replaced by a spectinomycin cassette (3). All of the growth, rRNA, and ribosome defects associated with RB286 (Pspank-yqeH) were observed in the RB406 strain grown in the absence of xylose (see below and data not shown). Because the expression of yqeH from the PxylA promoter was sufficient to complement the defects associated with the disruption of yqeH, polar effects on genes downstream of yqeH were unlikely to be involved in the observed phenotypes.
A decrease in the growth rate directly correlated with the amount of IPTG present in RB286 (Pspank-yqeH) cultures. Strain RB286 was grown in LB medium at 37°C in the presence of decreasing amounts of IPTG. RB286 cells grown in the presence of 1 mM IPTG grew at a rate indistinguishable from that of wild-type cells (doubling time, 28 min). The growth rate decreased until a doubling time of 100 min was achieved in the absence of IPTG. Cells grown without IPTG still grew exponentially, likely due to leaky expression from the Pspank promoter. Intermediate levels of growth (45-min doubling time with 10 μM IPTG and 70-min doubling time with 5 μM IPTG) were observed when the IPTG concentration was varied, showing that the growth rate correlated with the YqeH level. A dependence of growth rate on the level of a protein involved in translation is expected and has been demonstrated for the essential GTPase RbgA and for initiation factor 2 in Escherichia coli (4, 15).
Decreased levels of 16S rRNA are associated with YqeH depletion.
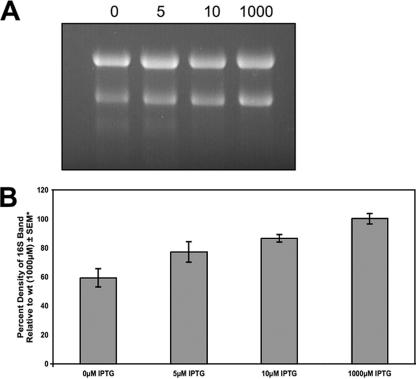
When isolating RNAs from YqeH-depleted cells, we frequently observed that the 16S rRNA band was much fainter than the 16S rRNA band for RNAs isolated from nondepleted cells. To quantify this reduction in 16S rRNA, RB286 (Pspank-yqeH) cells were grown in 1 mM IPTG, 10 μM IPTG, 5 μM IPTG, or no IPTG to identical optical densities at 600 nm, and cells were harvested for RNA isolation. RNAs were analyzed by formaldehyde-agarose gel electrophoresis, and band intensities were measured by densitometry with an Alpha Imager 2200 (Alpha Innotech) gel documentation system. rRNAs isolated from RB286 cells grown in 1 mM IPTG had a 23S/16S rRNA ratio of approximately 1.5, similar to what we observed for rRNAs isolated from wild-type B. subtilis cells (data not shown). rRNAs isolated from cells depleted of YqeH showed an increased ratio of 23S/16S rRNA (Fig. 1). The increasing ratio correlated inversely with the amount of IPTG added to the culture. Decreasing the amount of IPTG, and thus the YqeH level, resulted in higher 23S/16S rRNA ratios. For cultures grown in the absence of IPTG (100-min doubling time), the 23S/16S ratio was ∼2.6. Making the assumption that the level of 23S rRNA was not altered in these cells, the 16S rRNA was ∼40% less abundant in cells depleted of YqeH. These results were confirmed using strain RB406 by growing the strain in various levels of xylose (data not shown). Two conclusions about these experiments are worth noting. First, the decrease in 16S rRNA correlates well with the decrease in YqeH level and growth rate: as the YqeH level decreases, the amount of 16S rRNA decreases. Second, a 40% decrease in 16S rRNA is dramatic considering that the ribosome is one of the most abundant molecules in the cell. The addition of an RNase inhibitor to cells prior to RNA isolation had no effect on the stability of 16S rRNA, suggesting that 16S rRNA was not being degraded during RNA isolation. These results strongly suggest a role for YqeH in translation, possibly in 16S rRNA stability or 30S subunit assembly.
FIG. 1.
The ratio of 23S/16S rRNA is altered in RB286 (Pspank-yqeH) cells. Total RNA was run in a 1% formaldehyde-agarose gel. RNAs from induced (left) and fully YqeH-depleted cells are shown. The ratio of 23S/16S rRNA levels was quantitated. Results for three independent experiments are shown. Error bars depict the standard errors of the means (SEM).
Ribosome assembly defects in cells depleted of YqeH.
Analysis of ribosomes and ribosomal subunits from YqeH-depleted cells indicated that 30S ribosome assembly or stability is defective. Figure 2 shows the results of the ribosome profile analysis for RB286 cells grown in the presence (1 mM) or absence of IPTG. Cell lysates were prepared and ultracentrifuged over a 10 to 25% sucrose gradient, as previously described (14, 15). When RB286 was grown in LB medium at 37°C in the presence of 1 mM IPTG (Fig. 2A), the cells showed a distribution of 70S ribosomes and individual 50S and 30S subunits that was similar to that in wild-type cells. When cells were grown in the absence of IPTG (Fig. 2B), there was a decrease in the 30S subunit peak and a reduction of functional 70S ribosomes. The 50S subunits appeared to be increased, presumably due to a lack of 30S subunits to partner with to form a 70S ribosome. (The RB288 strain that depletes the expression of genes downstream of yqeH had a ribosome profile indistinguishable from a wild-type profile when grown without IPTG [data not shown].) This profile correlates well with our data showing that 16S rRNA is specifically depleted in YqeH-depleted cells (Fig. 1) and indicates that the biogenesis of the 30S subunit is defective. Notably, this ribosome profile is very different from that for cells depleted of known translation initiation and elongation factors, demonstrating that the YqeH-depleted profile is not simply an indirect effect of a defect in translation (15). With translation factor-depleted cells, we did not observe a loss of 30S subunits relative to 50S ribosomes. These ribosome assembly defects were also observed in the RB406 strain grown in the absence of xylose (data not shown).
FIG. 2.
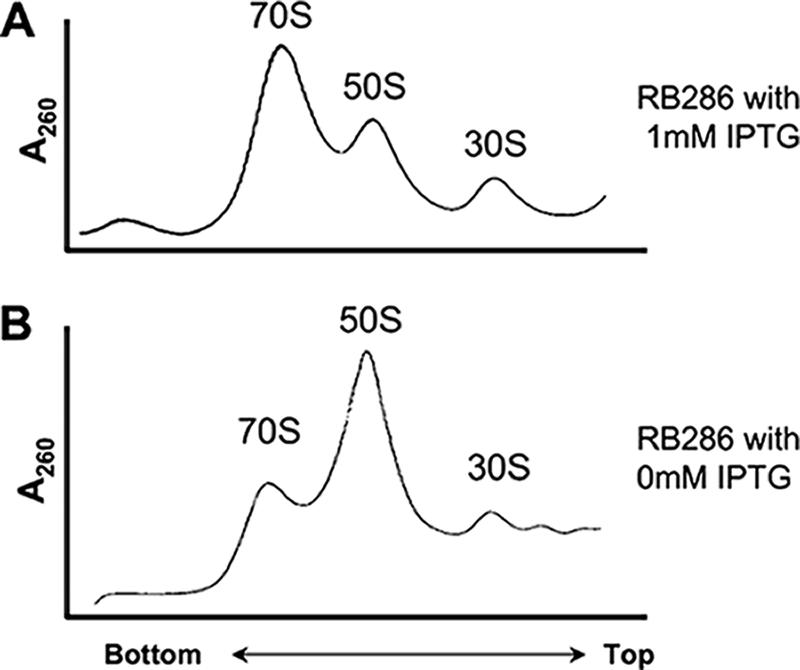
Ribosome profiling of YqeH-depleted cells. (A) Ribosome profile of RB286 grown in the presence of 1 mM IPTG (no depletion of YqeH). (B) Ribosome profile of RB286 grown in the absence of IPTG (YqeH-depleted cells). The direction of the 10 to 25% sucrose gradient is indicated on the x axis, and the A260 reading is indicated on the y axis.
The zinc ribbon motif and GTP-binding domain are required for YqeH function in vivo.
YqeH contains a highly conserved zinc ribbon motif (CXXCN26CXXC), which has been found in ribosomal proteins and may participate in protein-RNA interactions (9). This motif is conserved in all bacterial and eukaryotic yqeH homologs sequenced to date. To test if the zinc ribbon motif was required for function, we created a site-directed mutation in this motif to determine if the mutant YqeH protein could support growth. Because yqeH is essential, we created a strain in which the wild-type copy of yqeH was under the control of the IPTG-inducible Pspank promoter (RB286 strain) and mutant copies of yqeH were under the control of the xylose-inducible promoter PxylA. Site-directed mutations were made in the yqeH gene of plasmid pMK1 by using a QuikChange II kit (Stratagene), generating a C36A/C39A double mutation (plasmid pWU1). These mutations alter the first two cysteines of the zinc ribbon motif. In addition, extra copies of lacI were provided to the RB286 strain by the addition of the pMAP65 plasmid (13) to reduce leaky transcription from Pspank, generating strain RB567. Plasmids pMK1 and pWU1 were transformed into the RB567 background, generating strains RB641 and RB643, respectively. The resulting strains produced wild-type YqeH when grown in the presence of IPTG but produced wild-type or mutated YqeH when grown in the presence of xylose without IPTG. Strain RB641 had wild-type versions of yqeH at both loci, whereas RB643 had a mutant version of yqeH (C36A/C39A) under the control of the xylose promoter. RB641 was able to form wild-type colonies in the presence of either 1 mM IPTG or 2% xylose but formed only small colonies in the absence of any inducer (likely due to leaky expression of yqeH from the Pspank promoter). RB643 formed wild-type colonies in the presence of IPTG but formed small colonies in the presence of only 2% xylose (Fig. 3). These colonies were indistinguishable from those of RB643 cells grown in the absence of either inducer, indicating that the C36A/C39A mutation in yqeH resulted in a severe, possibly null defect. These results demonstrate that the putative zinc ribbon motif is important for yqeH function or for the stability or folding of YqeH.
FIG. 3.
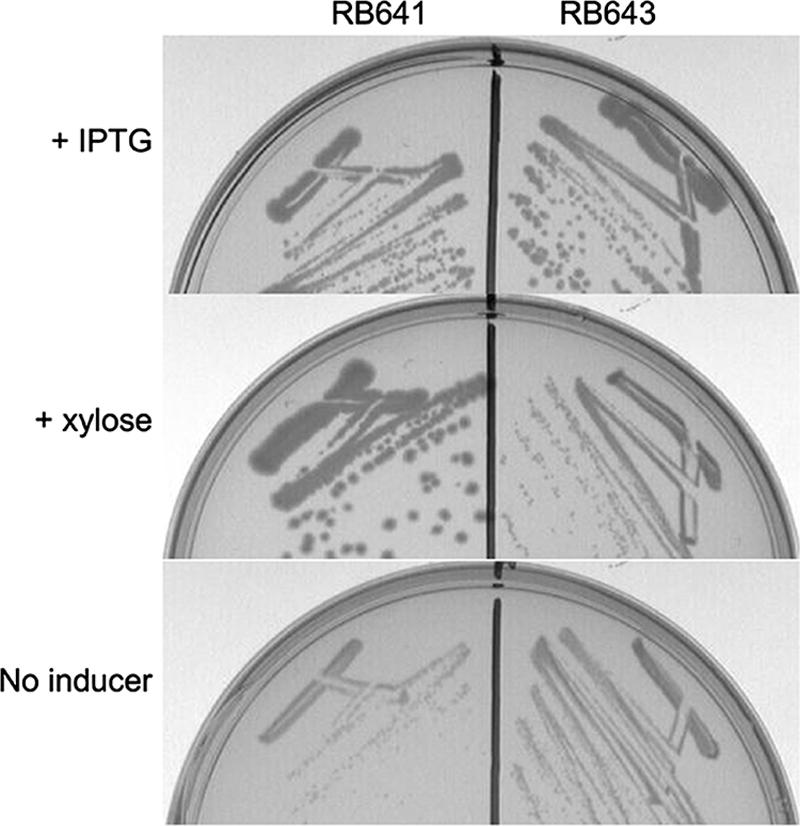
Mutation of the putative zinc ribbon motif in YqeH does not support wild-type growth. (Top) Strains RB641 and RB643 grown on LB plates in the presence of 1 mM IPTG. (Middle) Strains RB641 and RB643 grown on LB plates in the presence of 2% xylose. (Bottom) Strains RB641 and RB643 grown on LB plates without an inducer.
The same experimental approach was also used to determine if the GTP-binding domain is also required for yqeH function. A mutation in lysine 104 of the G1 domain (K104A), which participates in the binding of GTP, was cloned into the pSWEET vector (resulting in plasmid pWU16). This vector was then transformed into the RB567 background. The resulting strain (RB647) containing the K104A mutation behaved similarly to strain RB643 and was unable to support wild-type growth on LB medium supplemented with 2% xylose (data not shown). Thus, not surprisingly, the GTP-binding domain is also required for YqeH function.
Conclusions.
Our results indicate that the essential GTPase YqeH is involved in ribosome assembly. The specific loss of 30S ribosomes and degradation of 16S rRNA suggest that YqeH depletion results in a defect in the small ribosomal subunit. Future studies are planned to dissect the roles of YqeH in ribosome assembly and to determine how YqeH may be involved in coordinating ribosome biogenesis and the initiation of DNA replication.
Acknowledgments
This work was supported by an IRGP New Investigator award from Michigan State University and by startup funds from Michigan State University provided to R.A.B.
Footnotes
Published ahead of print on 19 January 2007.
REFERENCES
- 1.Anand, B., S. K. Verma, and B. Prakash. 2006. Structural stabilization of GTP-binding domains in circularly permuted GTPases: implications for RNA binding. Nucleic Acids Res. 34:2196-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. [DOI] [PubMed] [Google Scholar]
- 3.Bhavsar, A. P., X. Zhao, and E. D. Brown. 2001. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl. Environ. Microbiol. 67:403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole, J. R., C. L. Olsson, J. W. Hershey, M. Grunberg-Manago, and M. Nomura. 1987. Feedback regulation of rRNA synthesis in Escherichia coli. Requirement for initiation factor IF2. J. Mol. Biol. 198:383-392. [DOI] [PubMed] [Google Scholar]
- 5.Crawford, N. M. 2006. Mechanisms for nitric oxide synthesis in plants. J. Exp. Bot. 57:471-478. [DOI] [PubMed] [Google Scholar]
- 6.Du, X., M. R. Rao, X. Q. Chen, W. Wu, S. Mahalingam, and D. Balasundaram. 2006. The homologous putative GTPases Grn1p from fission yeast and the human GNL3L are required for growth and play a role in processing of nucleolar pre-rRNA. Mol. Biol. Cell 17:460-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo, F. Q., M. Okamoto, and N. M. Crawford. 2003. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302:100-103. [DOI] [PubMed] [Google Scholar]
- 8.Hedges, J., M. West, and A. W. Johnson. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24:567-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laity, J. H., B. M. Lee, and P. E. Wright. 2001. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11:39-46. [DOI] [PubMed] [Google Scholar]
- 10.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317:41-72. [DOI] [PubMed] [Google Scholar]
- 11.Matsuo, Y., T. Morimoto, M. Kuwano, P. C. Loh, T. Oshima, and N. Ogasawara. 2006. The GTP-binding protein YlqF participates in the late step of 50S ribosomal subunit assembly in Bacillus subtilis. J. Biol. Chem. 281:8110-8117. [DOI] [PubMed] [Google Scholar]
- 12.Morimoto, T., P. C. Loh, T. Hirai, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2002. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148:3539-3552. [DOI] [PubMed] [Google Scholar]
- 13.Petit, M. A., E. Dervyn, M. Rose, K. D. Entian, S. McGovern, S. D. Ehrlich, and C. Bruand. 1998. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol. 29:261-273. [DOI] [PubMed] [Google Scholar]
- 14.Schaefer, L., W. C. Uicker, C. Wicker-Planquart, A. E. Foucher, J. M. Jault, and R. A. Britton. 2006. Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis. J. Bacteriol. 188:8252-8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uicker, W. C., L. Schaefer, and R. A. Britton. 2006. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol. Microbiol. 59:528-540. [DOI] [PubMed] [Google Scholar]
- 16.West, M., J. B. Hedges, A. Chen, and A. W. Johnson. 2005. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol. Cell. Biol. 25:3802-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zalacain, M., S. Biswas, K. A. Ingraham, J. Ambrad, A. Bryant, A. F. Chalker, S. Iordanescu, J. Fan, F. Fan, R. D. Lunsford, K. O'Dwyer, L. M. Palmer, C. So, D. Sylvester, C. Volker, P. Warren, D. McDevitt, J. R. Brown, D. J. Holmes, and M. K. Burnham. 2003. A global approach to identify novel broad-spectrum antibacterial targets among proteins of unknown function. J. Mol. Microbiol. Biotechnol. 6:109-126. [DOI] [PubMed] [Google Scholar]