Abstract
Serratia marcescens is an opportunistic pathogen and a major cause of ocular infections. In previous studies of S. marcescens MG1, we showed that biofilm maturation and sloughing were regulated by N-acyl homoserine lactone (AHL)-based quorum sensing (QS). Because of the importance of adhesion in initiating biofilm formation and infection, the primary goal of this study was to determine whether QS is important in adhesion to both abiotic and biotic surfaces, as assessed by determining the degree of attachment to hydrophilic tissue culture plates and human corneal epithelial (HCE) cells. Our results demonstrate that while adhesion to the abiotic surface was AHL regulated, adhesion to the HCE cell biotic surface was not. Type I fimbriae were identified as the critical adhesin for non-QS-mediated attachment to the biotic HCE cell surface but played no role in adhesion to the abiotic surface. While we were not able to identify a single QS-regulated adhesin essential for attachment to the abiotic surface, four AHL-regulated genes involved in adhesion to the abiotic surface were identified. Interestingly, two of these genes, bsmA and bsmB, were also shown to be involved in adhesion to the biotic surface in a non-QS-controlled fashion. Therefore, the expression of these two genes appears to be cocontrolled by regulators other than the QS system for mediation of attachment to HCE cells. We also found that QS in S. marcescens regulates other potential cell surface adhesins, including exopolysaccharide and the outer membrane protein OmpX. We concluded that S. marcescens MG1 utilizes different regulatory systems and adhesins in attachment to biotic and abiotic surfaces and that QS is a main regulatory pathway in adhesion to an abiotic surface but not in adhesion to a biotic surface.
Serratia marcescens is an emerging pathogen that is predominantly involved in nosocomial infections (25). However, it has also been implicated in a range of ocular infections (5, 8, 11, 37). As many S. marcescens strains are resistant to multiple antibiotics, infections by this organism represent a growing public health risk (25). In addition to causing mammalian infections, S. marcescens is also commonly found in soils and can attach to plant tissues (9). Indeed, S. marcescens MG1 was initially isolated from a rotting cucumber (21). Undoubtedly, colonization of both abiotic and biotic surfaces by S. marcescens is an integral component of its success as a pathogen. For example, colonization of abiotic materials, such as contact lenses, provides S. marcescens with a medium for facilitating contact with target human tissue (i.e., corneal cells) (30). However, to be entirely successful, S. marcescens must also have the ability to adhere to its target tissue. Adhesion is considered to be the first step in the development of bacterial biofilms and a critical step in initiating infection. A number of different factors affect adhesion, including physicochemical interactions between the bacterium and the substratum, flagella, fimbriae, outer membrane proteins, and the presence of extracellular polymers (18, 32). Previous studies have shown that N-acyl homoserine lactone (AHL)-based quorum sensing (QS) regulates several aspects of surface colonization in S. marcescens, including swarming motility, biofilm maturation, and biofilm sloughing (15, 34, 51). However, the factors that mediate and control adhesion during the initial stages of colonization in S. marcescens are less well understood, and in particular, the role of QS in attachment has not been investigated.
The QS system of S. marcescens MG1 is governed by luxI and luxR homologues, swrI and swrR, respectively (15). SwrI predominantly synthesizes N-butanoyl-l-homoserine lactone (C4-HSL) and is believed to bind SwrR and affect the expression of at least 28 proteins, as demonstrated by two-dimensional polyacrylamide electrophoresis (22). Some of these QS-regulated proteins have been shown to be involved in swarming motility and biofilm maturation (34, 39). Since adhesion to both abiotic and biotic surfaces is important in S. marcescens pathogenicity, we examined whether C4-HSL regulation was involved in adhesion to an abiotic hydrophilic surface and a biotic surface, human corneal epithelial (HCE) cells. In this study, we determined that C4-HSL regulation is involved in adhesion to the abiotic surface but not in adhesion to the biotic HCE cell line. Adhesion to the HCE cells is largely mediated by type I fimbriae, but these fimbriae play no role in adhesion to the abiotic surface. Therefore S. marcescens utilizes different regulatory systems and adhesins for adhesion to surfaces with different properties. While we were unable to determine a single C4-HSL-regulated adhesin that is essential for attachment to the abiotic surface, we identified four C4-HSL-regulated genes that, when mutated, reduced the level of attachment to 50% of the wild-type level. Furthermore, exopolysaccharide (EPS) and OmpX were found to be QS regulated, and the latter was not involved in adhesion to either the abiotic or biotic surfaces assayed in this study. We propose a model for the colonization of abiotic and corneal surfaces by S. marcescens. Different regulatory systems drove adhesion to the different surfaces, and QS was a major regulatory pathway in adhesion to and biofilm formation on the abiotic surface. Since abiotic surfaces are an important source of infective cells, QS may be an important but indirect regulatory system driving eye infection.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. All primers used in this study are listed in Table 2. All strains were routinely grown at 30°C in Luria-Bertani broth (54) with 1% NaCl (LB10) or in minimal broth Davis (MBD) without dextrose (Difco Laboratories, Detroit, MI) supplemented with 0.2% glucose and 0.5% Casamino Acids. Antibiotics were added as required at the following final concentrations: kanamycin, 100 μg ml−1; and streptomycin, 200 μg ml−1. Gentamicin was used at concentrations of 60 and 25 μg ml−1 for S. marcescens and Escherichia coli strains, respectively. For complementation of S. marcescens strain MG44 and transposon mutants, C4-HSL was added to the medium at a final concentration of 250 nM.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Promega |
| SY327 λ pir | Δ(lac pro) argE(Am) rif nalA recA56 | 44 |
| SM10 λ pir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Tcr Kmr | 58 |
| Serratia marcescens strains | ||
| MG1 | Wild type, Apr Tcr | 21 |
| MG44 | MG1, swrI gene disrupted with a streptomycin cassette, Smr | 15 |
| PL10 | MG44, mini-Tn5 transposon insertion in swrA | 39 |
| MG3645 | MG44, mini-Tn5 transposon insertion in lipB | 52 |
| MG3646 | MG44, mini-Tn5 transposon insertion in bsmA | 34 |
| MG3651 | MG44, mini-Tn5 transposon insertion in bsmB | 34 |
| MG3633 to MG3651 | MG44, disruption in a C4-HSL responsive gene by a mini-Tn5 transposon carrying promoterless luxAB and a kanamycin marker, Kmr | 39 |
| M5 | MG1, deletion in ompX gene and disrupted with a gentamicin cassette, Gmr | This study |
| ML1 | MG1, fimA gene disrupted with a gentamicin cassette, Gmr | This study |
| Pseudomonas aeruginosa 6294 | Wild type | 17 |
| Plasmids | ||
| pBluescript SK II | Cloning vector, Apr | Stratagene |
| pGP704 | Apr | 43 |
| pUCGm | Gmr | 56 |
| pGEM-T Easy | Cloning vector, Apr | Promega |
| pBsmA | pBR322 bearing a 1.4-kb bsmA fragment and a gentamicin cassette, Gmr | 34 |
| pBsmB | pBR322 bearing a 1.2-kb bsmB fragment and a gentamicin cassette, Gmr | 34 |
| pWC202 | Plasmid bearing the rssA-rssB operon | 35 |
| pMWBCD10 | Plasmid carrying the lipBCD genes | 2 |
| pLOW2 | Cloning vector, Kmr | 24 |
| pOmpX | pBluescript SK II bearing a 1.9-kb ompX fragment, Apr | This study |
| pOmpXSphI | pOmpX with a 312-bp fragment deleted inside the ompX gene and SphI engineered in the remaining ompX gene, Apr | This study |
| pOmpXGm | pOmpXSphI with the 855-bp gentamicin cassette from pUCGM ligated into the SphI site, Apr Gmr | This study |
| pGP704OmpXGm | pGP704 bearing the disrupted ompX gene from pOmpXGm, Apr Gmr | This study |
| pFimA | 1.3-kb fimA fragment in pGEM-T Easy | This study |
| pLOW2fimA | 1.3-kb fimA fragment in pLOW2 | This study |
| pFimAGm | pFimA with the 0.9-kb gentamicin cassette from pUCGM ligated into the HindIII site, Apr Gmr | This study |
| pGPFimAGm | pGP704 bearing the disrupted fimA gene from pFimAGm, Apr Gmr | This study |
Tcr, tetracycline resistant; Apr, ampicillin resistant; Smr, streptomycin resistant; Kmr, kanamycin resistant; Gmr, gentamicin resistant.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| FompX | ATGTCTTTCCGCAGTAGC |
| RompX | TAGAAGCGGTAGCCTACG |
| ForO | GAGCGACAGGCCGTAAATC |
| RevO | GCGGAGCGTTACAACATGG |
| pOF | ACCCAATCGAAAAGCATGCCC |
| pOR | GCCTTGTTAGCATGCCCTTGG |
| F-FimA2 | GTIGTIGAYCARGAYCA |
| R-FimA2 | GGRTTRTCICCRTCIGC |
| FimAFor1 | TGTCGACAGACCATTTAA |
| FimARev1 | TTATCTTGCTTAGGCGTT |
| FimC-F | AGCGGGGATGATATTCTGC |
| FimC-R1 | AGCGAAAGYTCYTGRTAUCC |
| FimC-F2 | GCCTGCTGGTGAAATGGG |
| FimD-R | GGGGAGTTCATCATDATNGG |
| AraC-R2 | TGGATHGARAAYCATCTGGA |
The SphI sites are underlined.
Genetic methods.
Plasmid DNA preparations were obtained using the Wizard Plus Minipreps DNA purification system (Promega). Cloning, chemical transformation of E. coli, and electroporation of S. marcescens MG1 were performed using standard procedures (7). PCR was performed using either Taq DNA polymerase (Roche) or Pwo DNA polymerase (Boehringer Mannheim), and DNA fragments were purified using either a Prep-a-Gene DNA purification kit (Bio-Rad) or a QIAquick DNA purification kit (QIAGEN). Genomic DNA was extracted from 2 ml of overnight cultures of S. marcescens strains by using the XS buffer protocol (63). Sequencing of the area surrounding transposon insertion in S. marcescens MG3640 was carried out using panhandle PCR (57) and shotgun ligation and by screening a clone library and sequencing the insert. DNA sequences were compared to sequences in the GenBank database using the online BLAST search engine at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
Amplification and sequencing of the 16S rRNA gene from strain MG1 and phylogenetic analysis.
The 16S rRNA gene was amplified from genomic DNA by PCR performed with previously described standard primers corresponding to the E. coli 16S rRNA gene sequence (36). Single-band products were purified and sequenced, and the overlapping consensus sequence was compared with sequences in the GenBank database using BLAST sequence homology searches. Phylogenetic analysis was performed using the ARB software (www.arb-home.de) and 16S rRNA gene sequences of members of the genus Serratia and the family Enterobacteriaceae. Evolutionary distance and parsimonious analyses of 1,239 nucleotides were carried out with PAUP (version 4.08b), employing 1,000 bootstrap resamplings.
Abiotic surface adhesion assay.
S. marcescens strains were grown with gentle shaking to the early stationary phase (optical density at 610 nm [OD610], ∼3.0) in MBD supplemented with 0.2% glucose and 0.5% Casamino Acids. Cells were diluted to obtain an OD610 of 1.0, and 100-μl aliquots were added to the wells of a 96-well hydrophilic tissue culture microtiter plate (Co-Star 3595; Corning Incorporated, Corning, NY). The plates were statically incubated at 30°C for 2 h, and adhesion was quantified by staining with crystal violet as previously described (47).
In vitro assay for adhesion to human corneal epithelial cells.
An assay to determine adhesion to an immortalized HCE cell line (4) was carried out as previously described (61). Briefly, HCE cells were cultured in 24-well microtiter plates to obtain confluent monolayers and incubated with bacterial suspensions containing 106 CFU/ml. After incubation for 3 h, the total bacterial adhesion was measured by washing and lysing the HCE cells with Triton X-100 and determining the total number of bacterial cells by viable counting on nutrient agar.
Construction of the ompX and fimA mutants and plasmid pLOW2fimA.
Fragments of the ompX and fimA genes were initially amplified from the MG1 chromosome using primers FompX and RompX and primers F-FimA2 and R-FimA2, respectively. The resulting PCR fragments were sequenced. The entire DNA coding sequence and surrounding sequence of each gene were obtained by panhandle PCR performed as described by Siebert et al. (57), with modifications (62). A 1.9-kb fragment containing the ompX gene and the surrounding sequence was amplified using primers ForO and RevO and ligated into the SmaI site of pBluescript SK II to obtain pOmpX. Primers pOF and pOR were designed to contain SphI sites and were used to amplify pOmpX. The resulting PCR fragment was digested with SphI, and the digested fragment was circularized by ligation to obtain pOmpXSphI, which resulted in a 312-bp deletion in the ompX gene. A 0.9-kb SphI fragment from pUCGm containing a gentamicin resistance cassette was ligated into the SphI site of pOmpXSphI, disrupting the ompX gene, to create pOmpXGm. The gentamicin-disrupted ompX gene and the surrounding sequence were then transferred to the suicide vector pGP704 to obtain plasmid pGP704OmpXGm. A 1.3-kb fragment bearing the fimA gene was amplified from chromosomal DNA using primers FimAFor1 and FimARev2. The fimA fragment was ligated into pGEM-T Easy (Promega) to obtain plasmid pFimA. Plasmid pFimAGm was constructed by ligating a 0.9-kb HindIII gentamicin cassette (from pUCGm) into the unique HindIII site identified in the fimA insert. The disrupted insert from pFimAGm was cut out using EcoRI and ligated into pGP704 to obtain pGPFimAGm. Plasmids pGPFimAGm and pGP704OmpXGm were electroporated into E. coli S17-1 and conjugated with S. marcescens MG1. Recombinants were selected for on MBD supplemented with 0.2% glucose and 60 μg/ml gentamicin, and double-crossover mutants were screened by PCR. To create pLOW2fimA, a 1.3-kb fragment bearing the fimA gene was cut out of pFimA using EcoRI and ligated into the EcoRI site of pLOW2.
Measurement of promoter induction by C4-HSL.
Induction of the transposon-disrupted genes by C4-HSL was determined by measuring luminescence (encoded by the promoterless luxAB genes on the transposon) before and after addition of C4-HSL. The assay was performed as previously described (34).
Calcofluor assay.
S. marcescens strains were streaked onto Kings agar (1% [wt/vol] peptone, 1% [wt/vol] NaCl, 1% [vol/vol] glycerol, 1.5% [wt/vol] agar) containing 0.002% calcofluor (fluorescent brightener 28; Sigma F-3397). Kings agar has previously been reported to enhance exopolysaccharide production in S. marcescens strains (6). To determine if a strain produced exopolysaccharide, agar plates were exposed to long-wavelength UV light, and the bacterial streaks on the agar plates were monitored for bright blue fluorescence. When necessary, C4-HSL was added to the agar medium to a final concentration of 250 nM.
Outer membrane protein analysis.
Outer membranes were prepared from mid-log-phase cells (OD610, ∼1.2) and early-stationary-phase cells (OD610, ∼3.6) as previously described (28). The outer membranes were resuspended in sterile Milli-Q water, and the total protein concentration of each preparation was determined using the bicinchoninic acid method (59). For each preparation, 15 μg of protein was electrophoresed on a 15% sodium dodecyl sulfate (SDS)-polyacrylamide gel containing 4 M urea in the separating gel. The gels were stained with Coomassie brilliant blue, and proteins of interest were excised from the gels and subjected to terminal sequencing for identification at the Australian National University, Canberra.
Extracellular protein and surface protein extraction.
Cells used for surface protein extraction were scraped from MBD agar plates supplemented with 0.2% glucose and 0.5% Casamino Acids and resuspended in 1.5 ml of phosphate-buffered saline (pH 7.4) to an OD610 of 5.0. Surface proteins were sheared off the bacterial surface by passing the suspension through a 26.5-gauge needle three times. Cells were removed by centrifugation (4,620 × g), and the supernatant was filter sterilized (0.2-μm Supor membrane filter; PALL Corporation). The proteins in the supernatant were precipitated by adding trichloroacetic acid to a final concentration of 20% (vol/vol) and were incubated on ice for 30 min. The surface proteins were collected by centrifugation (25,200 × g) for 10 min at 4°C. The pellet was washed twice with phosphate-buffered saline (pH 7.4) and resuspended in 2% SDS. Samples were loaded onto 12% SDS-acrylamide gels, and the gels were stained with Coomassie brilliant blue. Proteins of interest were cut out for identification and subjected to in-gel digestion with trypsin. The peptides were then analyzed using matrix-assisted laser desorption ionization (MALDI), which provided highly accurate masses for the peptides in the mixture and a fragment ion spectrum for the five highest-concentration peptides. The fragment ion spectrum was then run through the Mascot search engine (http://www.matrixscience.com/) database, which led to identification of proteins 1, 2, and 3 and the flagellin protein. Protein 4 was identified by analysis of the peptides using liquid chromatography-tandem mass spectrometry (LC-MS/MS). To do this, peptide sequences were obtained and run through BLASTP to determine possible homology to proteins in the database. The relative amounts of the FimA protein in the samples were determined by measuring the total pixels of the proteins in the gel picture using the NIH Image program and normalizing the results to the total pixels for flagellin protein.
Electron microscopy.
Cells were grown at 30°C with gentle agitation (50 rpm) in MBD supplemented with 0.2% glucose and 0.5% Casamino Acids. Cells were added to a carbon-coated copper grid and stained with 2% uranyl acetate. Transmission electron microscopy of the stained cells was carried out using a Hitachi H7000 electron microscope.
Nucleotide sequence accession numbers.
The nucleotide sequences of ompX and the sequence surrounding the transposon insertion in MG3640 have been deposited in the GenBank database under accession numbers AY498858 and AY498857, respectively. The nucleotide sequence of a contig containing fimA, fimB, and part of fimC has been deposited in the GenBank database under accession number AY730610. Partial sequences of the PCR products amplified from the S. marcescens MG1 fimbrial operon have been deposited in the GenBank database under accession numbers EF193059 and EF193060. The S. marcescens MG1 16S rRNA gene sequence has been deposited in the GenBank database under accession number AY498856.
RESULTS
Reclassification of Serratia liquefaciens MG1 as S. marcescens MG1 based on 16S rRNA gene phylogeny.
S. marcescens MG1 was previously classified as Serratia liquefaciens MG1; however, phylogenetic analysis of the 16S rRNA gene sequence of strain MG1 revealed that this strain is closely related to the type strain of S. marcescens subsp. marcescens (99% identity) and forms part of a monophyletic S. marcescens cluster with a high level of statistical support. As a result, S. liquefaciens MG1 was reclassified as S. marcescens MG1 and is thus a nonpigmented S. marcescens strain, which is typical of pathogenic strains (10, 25).
S. marcescens utilizes different regulatory pathways for adhesion to abiotic and biotic surfaces.
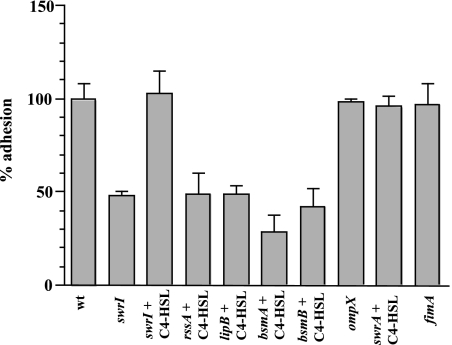
The importance of C4-HSL regulation in surface motility, biofilm formation, and detachment of S. marcescens prompted us to test whether C4-HSL-mediated gene regulation also played a role in adhesion. Since adhesion to both abiotic and biotic surfaces is integral for mediating infection, the role of C4-HSL regulation was examined by determining adhesion to a hydrophilic plastic surface and to HCE cells. In the adhesion assay with the abiotic surface, the swrI mutant S. marcescens strain MG44 adhered at a level that was 50% of the wild-type level (Fig. 1). The adhesion deficiency of the swrI mutant could be restored by exogenously providing C4-HSL at a final concentration of 250 nM, confirming that C4-HSL plays a significant role in regulating adhesion (Fig. 1). In contrast, the swrI mutant adhered to the HCE cells at the same level as the wild type (Fig. 2). Therefore, it appeared that C4-HSL signaling played no role in adhesion to HCE cells. Importantly, the levels of adhesion of S. marcescens MG1 to HCE cells were higher than those of the clinical corneal isolate Pseudomonas aeruginosa 6294, suggesting that the levels of S. marcescens MG1 adhesion to the HCE cell line might be clinically relevant (Fig. 2).
FIG. 1.
Adhesion of S. marcescens MG1 (wt), Tn5 mutants, and the ompX and fimA mutants to a hydrophilic surface. Cells were grown with gentle shaking to the early stationary phase, diluted to obtain an OD610 of 1.0, and allowed to adhere for 2 h. The levels of adhesion are expressed as percentages of the wild-type (wt) adhesion level. The error bars indicate standard deviations.
FIG. 2.
Adhesion to the human corneal epithelial cell line by S. marcescens MG1 (wt), the swrI mutant, Tn5 mutant strains, the ompX mutant, and the fimA mutant. The assay to determine adhesion to the HCE cell line was carried out for 3 h in a 24-well microtiter plate containing confluent HCE cells. The levels of adhesion are expressed as percentages of the wild-type adhesion level. The error bars indicate standard deviations.
Type I fimbriae are crucial for non-QS-mediated adhesion to the HCE biotic surface but have no role in adhesion to the abiotic surface.
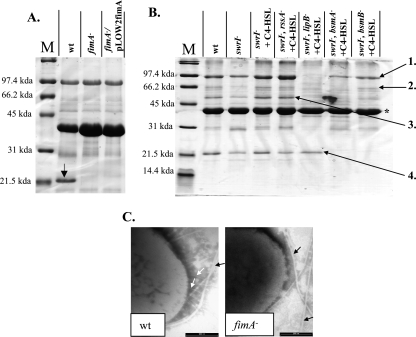
S. marcescens is an important eye pathogen; however, little is known about the adhesins utilized by this organism during attachment to corneal tissue. Since fimbriae are some of the most important adhesins involved in attachment to epithelial cells (32), fimbriae were considered to be the most likely adhesin responsible for attachment to the HCE cells. Surface proteins of S. marcescens MG1 were extracted and visualized on an acrylamide gel to determine the main fimbrial type expressed on the surface. A predominant protein band was observed at approximately 21 kDa, a size which is consistent with the sizes of major fimbrial subunits (Fig. 3A) (53). This protein band was excised and identified by LC-MS/MS as FimA, the major fimbrial subunit of type I fimbriae (46). The peptide sequences obtained by LC-MS/MS (FTGELVQS, VVDQDQNQEVYLGK, TTAFPTAGS, EAGNYTLR, VTVAADGDNP, and NPGAATFNLR) were aligned with the S. marcescens FimA protein sequence in the GenBank database (accession number YQSE1) and used to design degenerate primers F-FimA2 and R-FimA2 (Table 2) for amplification of fimA from S. marcescens MG1 chromosomal DNA. The fimA open reading frame (ORF) of S. marcescens MG1 encodes a 175-amino-acid protein exhibiting 66% identity and 75% similarity to the FimA protein of S. marcescens (GenBank accession number YQSE1). S. marcescens ML1, a fimA disruption mutant, was constructed and was confirmed not to produce fimbriae by gel electrophoresis of surface protein extracts and by electron microscopy (Fig. 3A and C). No other fimbrial structures were observed by electron microscopy, indicating that type I fimbriae were the only fimbriae present under the conditions tested. The S. marcescens ML1 fimA mutant adhered at a level that was only 6% of the level of the wild type, thereby demonstrating that type I fimbriae are the main adhesin in attachment to HCE cells (Fig. 2). Complementation of the HCE adhesion phenotype by supplementing fimA in trans was unsuccessful (Fig. 3A), presumably due to the polar effect on other downstream type I fimbrial genes in this operon. The known fimbrial nucleotide sequence of S. marcescens MG1 was found to be highly related to the nucleotide sequence of a fimbrial operon found in Serratia proteamaculans 568 (draft genome at http://genome.jgi-psf.org/draft_microbes/serpr/serpr.home.html). In S. proteamaculans 568, the type I fimbrial operon consists of fimA, fimB, fimC, and fimD. Immediately downstream of fimD is a gene coding in the reverse orientation for an AraC-like transcriptional regulator. To determine if the genetic context was identical in S. marcescens MG1, PCR products were amplified from MG1 using specific primers for the known MG1 sequence and degenerate primers designed using the S. proteamaculans 568 sequence. An initial 2.4-kb sequence encompassing the fimC gene was amplified using primers FimC-F and FimC-R1, and the 3′ end of the amplicon was sequenced. A second product and a third product were amplified using a specific primer designed using the 3′ sequence of the initial product (FimC-F2) and degenerate primers FimD-R and AraC-R2 encompassing the fimD gene (1.1 kb) and fimD/araC-like gene (1.7 kb), respectively. Sequences of the 3′ ends of all the amplicons verified their identities and confirmed that the genetic organization of the fimbrial operon and gene immediately downstream in S. marcescens MG1 was identical to the genetic organization in S. proteamaculans 568. Therefore, a polar effect on downstream genes would not affect any phenotype other than type I fimbrial assembly. This strongly supports the hypothesis that type I fimbriae were the adhesin driving adhesion to HCE cells. Type I fimbriae were not involved in adhesion to the abiotic surface since the levels of adhesion of the fimA mutant were identical to those of the wild type on this surface (Fig. 1).
FIG. 3.
(A and B) Surface and extracellular protein analysis of S. marcescens MG1 (wt), the fimA mutant, and the complemented fimA mutant (A) and of the swrI mutant and adhesion-deficient Tn5 mutants (B). Proteins 1 to 4 were identified by MALDI or LC-MS/MS analysis as S-layer, serralysin protease, triacylglycerol lipase, and FimA, respectively. The position of flagellin is indicated by an asterisk. (C) Electron micrographs of uranyl acetate-stained wild-type and fimA mutant S. marcescens, demonstrating the presence and absence of fimbriae, respectively. The white arrows indicate fimbrial structures, and the black arrows indicate flagella.
Identification of C4-HSL-regulated genes involved in QS-mediated abiotic adhesion.
In an effort to identify the specific C4-HSL-regulated genes required for adhesion by S. marcescens MG1, 19 previously isolated C4-HSL-responsive reporter transposon S. marcescens MG44 mutants (39) were screened for defective surface attachment in the presence of C4-HSL. From this screen four adhesion-deficient mutants were isolated: MG3640, MG3645, MG3646, and MG3651 (Fig. 1). Mutants MG3646 and MG3651 carry Tn5 insertions in the bsmA and bsmB genes, respectively, which have previously been reported to be important for biofilm maturation in this organism (34). Mutant MG3645 has previously been found to have an insertion in the lipB gene, which is part of a type I secretion system (52). Sequence data for areas from around the site of the transposon insertion in MG3640 indicated that the transposon disrupted a 1,307-bp ORF 98 nucleotides downstream of the putative translational start site. The 1,307-bp ORF encodes a 435-amino-acid protein having 96% identity and 98% similarity (over 395 amino acid residues) to the sensor kinase RssA (GenBank accession number AAN28325) from S. marcescens. Upstream, in the same putative operon, is a 519-bp ORF that encodes a 172-amino-acid protein that has 91% identity and 94% similarity (over 157 amino acid residues) to the response regulator protein RssB (GenBank accession number AAN28326) from S. marcescens. The rssA-rssB two-component system has been reported to affect the cellular fatty acid composition and cell surface topography of S. marcescens, both of which could affect adhesion (35). Measurement of promoter activity of the transposon mutants by detection of luminescence after the addition of C4-HSL revealed that rssA was induced approximately 2-fold and lipB was induced approximately 3.5-fold. Interestingly, rssA was maximally expressed in the mid-logarithmic growth phase and not in the transition to the stationary phase (data not shown), where AHL-regulated genes are often reported to be maximally activated. The bsmA and bsmB genes have previously been reported to be induced 2.8- and 9.8-fold, respectively, by C4-HSL in the early stationary phase (34).
The adhesion phenotypes of the rssA, bsmA, and bsmB transposon mutants could be partially restored through complementation of the mutated gene in trans. The rssA, bsmA, and bsmB complemented strains exhibited 85, 23, and 7% improvement in adhesion, respectively, compared to the uncomplemented strains. The partial restoration was most likely due to the presence of multiple copies of the complementation plasmids, and since the genes affect multiple phenotypes, the overexpression of the genes may have diluted the adhesion phenotype. Nevertheless, the results of the partial restoration analysis support the hypothesis that the rssA, bsmA, and bsmB genes are involved in adhesion. The complemented lipB mutant did not show an increase in adhesion compared to the uncomplemented strain. The complemented strain also did not exhibit other lip secretion functions, such as protease production. The reason for this is unknown; however, any polar effect exerted by the transposon mutation would have affected only genes in the lip operon. The lipB mutant has previously been well characterized, and the transposition did not appear to have any unusual effects (52); our analysis of the lipB mutant (Fig. 3B) supports these findings. Therefore, we propose that the effects on adhesion seen here were due to the lack of a lip secretion system.
C4-HSL regulation of surface properties in S. marcescens MG1.
Surface properties, such as EPS, outer membrane proteins, and surface appendages, including fimbriae and flagella, are commonly involved in adhesion and biofilm formation. Since adhesion and biofilm development are regulated by C4-HSL in S. marcescens MG1, suggesting that one or more surface properties are regulated by this signal, the surface properties of the adhesion mutants were characterized.
(i) Exopolysaccharide is regulated by C4-HSL in S. marcescens MG1.
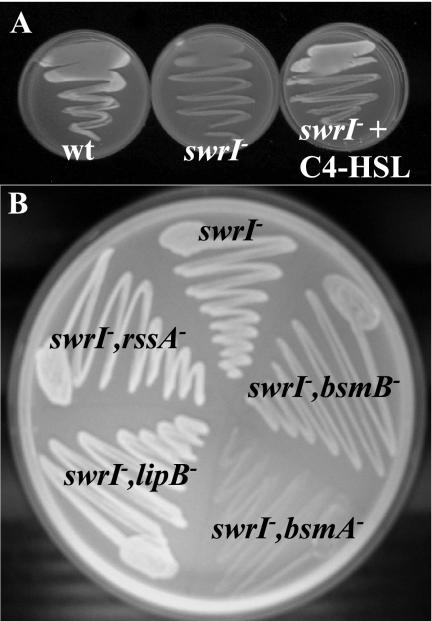
We noticed that on solid LB10, colonies of the wild-type strain appeared to be more opaque than colonies of the swrI mutant (data not shown). This was more evident on Kings agar, which has previously been reported to enhance EPS production in Serratia sp. (6), suggesting that EPS production was regulated by C4-HSL. Calcofluor binds polysaccharide, and colonies producing polysaccharide emit bright blue fluorescence when they are exposed to long-wavelength UV light. On Kings agar, LB10 agar, and MBD agar (supplemented with 0.2% glucose and 0.5% Casamino Acids) containing 0.02% calcofluor, the wild type consistently emitted bright blue fluorescence when it was exposed to long-wavelength UV light, whereas the swrI mutant did not (Fig. 4A). The inability of the swrI mutant to produce exopolysaccharide could be restored by supplementing the agar with 250 nM C4-HSL (Fig. 4A). To determine whether any of the adhesion-deficient transposon mutants could produce EPS, the mutants were streaked on Kings agar supplemented with 250 nM C4-HSL and calcofluor. Mutants MG3640 (rssA), MG3645 (lipB), and MG3651 (bsmB) produced wild-type levels of fluorescence; however, mutant MG3646 (bsmA) was found to emit no fluorescence, indicating that bsmA has a role in exopolysaccharide production (Fig. 4B). The lack of EPS may explain the adhesion-deficient phenotype of the swrI mutant and MG3646 (bsmA); however, the defective adhesion phenotype of the other transposon adhesion-defective mutants may not have been due to the absence of EPS but rather may have been due to the lack of a different adhesin.
FIG. 4.
Exopolysaccharide production in S. marcescens MG1 (wt), the swrI mutant, and the swrI mutant with C4-HSL (A) and in adhesion-deficient Tn5 mutants (B). Strains were streaked on Kings agar supplemented with 0.02% calcoflour. The organisms were grown on plates and exposed to long-wavelength UV light, and bright blue fluorescence was observed.
(ii) Outer membrane protein analysis.
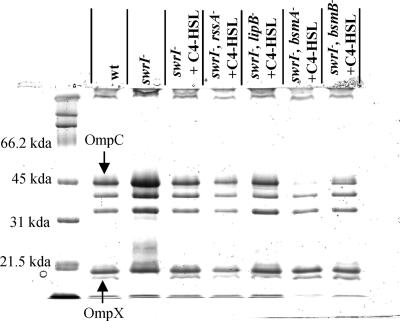
In the mid-logarithmic phase of growth, an 18-kDa protein was found to be dependent on C4-HSL (present in the wild-type and absent in the swrI mutant), and it was subsequently excised from the gel for N-terminal sequencing (Fig. 5). Twelve residues were identified as GQSTVSAGYAQG and were recognized as OmpX from S. marcescens N28b (23) based on a comparison with the BLASTP database. OmpX belongs to a family of outer membrane barrel proteins found in the family Enterobacteriaceae. The ompX gene of S. marcescens MG1 was sequenced and was found to contain a 519-bp ORF with 76% identity and 84% similarity to an ORF encoding the OmpX protein of S. marcescens (accession number CAA85513). Since OmpX proteins have previously been implicated in adhesion to epithelial cells (64), an ompX mutant of S. marcescens MG1 was constructed, but it was found to adhere normally to the abiotic and biotic surfaces (Fig. 1 and 2). No other outer membrane protein was identified as a QS-regulated protein or as a protein that was commonly absent (or present) in the adhesion mutants compared to the wild type, indicating that an outer membrane protein was not involved in C4-HSL-mediated adhesion (Fig. 5).
FIG. 5.
Outer membrane protein profiles of S. marcescens MG1 (wt) and adhesion-deficient Tn5 mutants. Labeled proteins were excised and identified by N-terminal sequencing.
We also observed that the MG3646 (bsmA) mutant had reduced levels of an outer membrane protein (Fig. 5) subsequently identified as OmpC by N-terminal sequencing (AEIYNKDGNKLD). OmpC is a major porin that is induced at high osmolarity and is important for maintaining proper osmotic pressure within the cell (45). Although bsmA is C4-HSL regulated, a similar effect was not observed with the swrI mutant, indicating that this effect was likely a result of the bsmA mutation. The effect of the bsmA mutation on OmpC levels was further confirmed by the poor growth of the MG3646 (bsmA) mutant observed at high salt concentrations (Luria-Bertani medium containing 46.75 to 58.44 g/liter NaCl) compared to the growth of the wild type and the swrI mutant (data not shown).
(iii) Surface and extracellular protein analysis.
The levels of four proteins (proteins 1 to 4) were found to be reduced in the adhesion-deficient mutants, or they were absent (Fig. 3B). MALDI analysis of extracted proteins resulted in identification of proteins 1, 2, and 3 as an S-layer protein, serralysin protease, and triacylglycerol lipase, respectively, all of which have previously been reported to be secreted through the lip secretion system (1, 2, 53). This conclusion was supported by our finding that these three proteins were not present in the extracellular fraction of the MG3645 (lipB) mutant. The bsmA mutant lacked serralysin protease, while the MG3646 (bsmA) and MG3651 (bsmB) mutants did not produce triacylglycerol lipase and had reduced levels of the S-layer protein. Reduced protease activities in the supernatants of the MG3646 (bsmA) and MG3651 (bsmB) mutants were confirmed by measuring azocasein hydrolysis (data not shown). It also appeared that smaller amounts of the FimA protein were produced in the swrI, rssA, and lipB mutants and that this protein was completely absent in the bsmA and bsmB mutants compared to the wild-type strain and the C4-HSL-complemented swrI mutant. Interestingly, the bsmA and bsmB genes are necessary for FimA production, and both are regulated by C4-HSL, while FimA production is largely QS independent. This suggests that there is an additional level of complexity in the regulation of these genes.
bsmA and bsmB genes are important in non-QS-mediated adhesion to the HCE cells.
While we determined that C4-HSL does not regulate adhesion to HCE cells and that type I fimbriae are the major adhesin driving the process, we were interested in whether genes previously observed to be important for colonization were also involved in adhesion to HCE cells. As a result, the transposon adhesion mutants MG3640, MG3645, MG3646, and MG3651, the ompX mutant, and the swrA swarming mutant were tested in HCE cell adhesion assays. While mutants MG3640 (rssA) and MG3645 (lipB), the ompX mutant, and the swrA mutant were unchanged in adhesion to the HCE cells, mutants MG3646 (bsmA) and MG3651 (bsmB) consistently adhered at levels that were approximately 50% of the wild-type level (Fig. 2). C4-HSL regulation is unimportant for adhesion to HCE cells; however, the C4-HSL-regulated bsmA and bsmB genes play an important role. This suggests that other regulatory processes induce these genes during adhesion to HCE cells. It is likely that the lack of type I fimbriae in the bsmA and bsmB mutants (Fig. 3B) is the cause for their poor adhesion to the HCE cells.
DISCUSSION
One factor that may enable S. marcescens MG1 to thrive in a variety of different environments is its ability to colonize surfaces and form biofilms, which have generally been associated with increased survival in the face of a variety of stresses (e.g., oxidative stress, antibiotics, and protozoan grazing) (16, 29, 41, 49). Surface colonization is a process consisting of attachment, surface motility, maturation into a biofilm, and detachment of cells, resulting in the planktonic forms, completing the life cycle. It is now appreciated that one of the factors influencing surface colonization is the cell-cell signaling system known as AHL-mediated QS (12, 40). Indeed, for S. marcescens, multiple processes of surface colonization are QS regulated, including swarming motility, biofilm maturation, and detachment (15, 34, 51). S. marcescens is widespread in the environment, particularly in soils in association with plants, but it is also increasingly being detected in nosocomial and ocular infections of humans. Moreover, this organism is a good colonizer of living and inanimate surfaces. For example, S. marcescens strains can consistently be isolated from contact lenses and contact lens cases (19, 31), and S. marcescens MG1 has previously been shown to colonize tomato roots (60). In light of this general control, we studied QS regulation of attachment and found that QS mediates attachment to some surfaces, such as hydrophilic, polystyrene microtiter plates. Interestingly, QS was not required for attachment to tissue culture cells (HCE cells).
The regulation of adhesion by extracellular signaling appears to be unusual; it has been reported only here and by Koutsoudis et al. (33), who showed that attachment of Pantoea stewartii is repressed by QS. This phase of colonization has not been studied as intensively in relation to QS since signaling compounds are often thought to act as sensors of population density, such that certain genes are switched on only when a sufficient cell density is reached (48). While adhesion is not a population-dependent phenotype, providing the signal compound can be contained within an enclosed environment, in one or a few cells the genes could be upregulated by a threshold concentration of signal. Hence, it is conceivable that as a bacterium approaches a surface, the surface acts as a diffusion barrier, resulting in an increased concentration of AHLs and the activation of genes coding for adhesins (50, 55).
As noted above, S. marcescens uses QS to regulate multiple stages of colonization and biofilm formation, which raises the question of how these temporally different, and in the case of detachment seemingly antagonistic, processes can be controlled by the same regulatory mechanism. The most likely solution to this conundrum is that the QS system does not operate alone but rather operates in conjunction with other regulatory systems which combine to ensure that phenotypic genes are expressed at the correct times. This conclusion is supported by observations made by Whiteley et al. (66), who found that the QS-regulated genes of P. aeruginosa could be divided into genes that are expressed early and late. Moreover, there are several examples of interactions of other global regulatory systems with QS, such as cAMP and the catabolite repressor protein in Vibrio fischeri and the P. aeruginosa homologue Vfr (3, 13). Additionally, the QS system of P. aeruginosa has been shown to be hierarchical; the las system is dominant with respect to the rhl system, and the two systems are linked through a quinolone signaling system (42). In S. marcescens, swarming motility is QS regulated, but it is also dependent on correct expression of the flhDC master regulator (14). Thus, it is likely that at least in S. marcescens, the QS system interacts with accessory regulators to induce the expression of genes that are involved in attachment, swarming, biofilm maturation, and detachment.
This notion is further supported by the observations that QS is important for adhesion to abiotic surfaces but not for adhesion to biotic surfaces and that different adhesins are involved in attachment to these surfaces. Type I fimbriae were identified as the main adhesin for the HCE cells but had no role in adhesion to the abiotic surface, which is consistent with the finding that different regulatory systems drive adhesion to these different surfaces. A similar result has been obtained for Vibrio cholerae, which utilizes different fimbrial appendages in adhesion to chitin and to an abiotic borosilicate substratum (65). In S. marcescens, type I fimbriae have previously been observed to be important for adhesion to a bladder carcinoma cell line (27) and for adhesion to uroepithelial cells (38); however, this is the first report of type I fimbriae mediating adhesion to HCE cells.
Characterization of the surface properties of S. marcescens did not elucidate the specific adhesin driving C4-HSL-mediated adhesion. However, two different surface characteristics, exopolysaccharide and OmpX, were found to be C4-HSL regulated. While mutations in fimA and ompX did not affect adhesion to the abiotic surface, it is possible that exopolysaccharide could play a role. Of the four C4-HSL-regulated genes involved in abiotic adhesion, the bsmA and bsmB genes were of particular interest. The bsmA and bsmB genes are involved in abiotic adhesion (Fig. 1), in late-stage biofilm development (34), and in adhesion to HCE cells (Fig. 2). The exact mechanism by which these genes exert their effects is unclear; however, the genes appear to be important for full expression of multiple extracellular properties. Both the bsmA and bsmB mutants had reduced levels of S-layer protein, triacylglycerol lipase, and serralysin protease and undetectable levels of the type I fimbrial major protein subunit FimA. Furthermore, mutant MG3646 (bsmA) was also unable to produce exopolysaccharide and had reduced levels of OmpC and reduced hemolytic and HCE cell cytotoxicity activities (data not shown). Given the effects that these genes have on the presence of extracellular products, it is probable that they are involved in secretion. Since BsmB contains a putative signal peptide, it is possible that this protein acts in the periplasmic space by assisting delivery of secreted products.
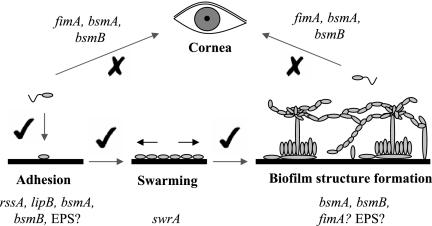
A model for abiotic and biotic surface colonization by S. marcescens MG1 is shown in Fig. 6. Colonization of an abiotic surface has been divided into three stages, adhesion, swarming, and biofilm structure formation. In all cases, C4-HSL signaling regulates these steps; however, colonization of the cornea does not require C4-HSL. The bsmA and bsmB genes are important for colonization of both surfaces and for multiple biofilm stages. This is due to the pleiotropic effects that mutations in these genes have on the bacterium's physiology. The absence of a specific bsmA- or bsmB-dependent adhesin is likely to be responsible for the defects seen in each stage. For example, bsmA/bsmB control of type I fimbriae (FimA) is important for colonization of HCE cells, whereas another unknown bsmA/bsmB-controlled adhesin is required for adhesion to the abiotic surface. A role for FimA in biofilm structure has not been ruled out. AHL regulation of the OmpX protein does not appear to be involved in any stage of colonization, and swrA is important only for swarming colonization (data not shown). However, this does not mean that SwrA and OmpX do not have a role in adhesion to other surfaces not tested here. However, AHL regulation of EPS may affect biofilm structure by providing scaffolding material. Assessment on how EPS affects biofilm structure and identification of the signaling pathway that controls bsmA and bsmB in S. marcescens MG1 are topics for further research.
FIG. 6.
Hypothetical model for colonization of abiotic and corneal surfaces by S. marcescens MG1. Biofilm formation on an abiotic surface is divided into three stages: adhesion, swarming, and biofilm structure formation. All three stages require C4-HSL signaling, which is indicated by a check mark. Cells adhering to corneal tissue are derived from the environment or from colonized abiotic surfaces. Adhesion to HCE cells is not regulated by C4-HSL, and the process is indicated by an X. The bsmA and bsmB genes are both regulated by C4-HSL and are important in adhesion to both the corneal and abiotic surfaces and for biofilm maturation. In addition to bsmA/bsmB, rssA and lipB drive adhesion to the abiotic surface, while type I fimbriae encoded by fimA drive adhesion to the corneal surface. The swrA gene is required only for the swarming stage, and the role of QS-regulated EPS in all stages is unknown.
The dependence on C4-HSL for colonization of abiotic surfaces in S. marcescens MG1 suggests that it is important to explore whether other S. marcescens strains (particularly clinical isolates) also utilize AHL regulation for surface colonization. If this is the case, it may be possible to use AHL inhibitors, such as furanones (20, 26), for prevention of adhesion and biofilm formation on medical equipment by this organism. Since S. marcescens strains are becoming increasingly resistant to antibiotics, new methods are necessary to control infections by this organism.
Acknowledgments
This work was funded by the Centre for Marine Biofouling and Bio-Innovation. Martin R. Larsen was supported by the Danish Natural Research Council.
We thank Greg Crocetti for assistance with the phylogenetic analysis.
Footnotes
Published ahead of print on 19 January 2007.
REFERENCES
- 1.Akatsuka, H., R. Binet, E. Kawai, C. Wandersman, and K. Omori. 1997. Lipase secretion by bacterial hybrid ATP-binding cassette exporters: molecular recognition of the LipBCD, PrtDEF, and HasDEF exporters. J. Bacteriol. 179:4754-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akatsuka, H., E. Kawai, K. Omori, and T. Shibatani. 1995. The three genes lipB, lipC and lipD involved in the extracellular secretion of the Serratia marcescens lipase which lacks an N-terminal signal peptide. J. Bacteriol. 177:6381-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. H. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araki-Sasaki, K., Y. Ohashi, T. Sasabe, K. Hayashi, H. Watanabe, Y. Tano, and H. Handa. 1995. An SV40-immortalized human corneal epithelial cell line and its characterization. Investig. Ophthalmol. Vis. Sci. 36:614-621. [PubMed] [Google Scholar]
- 5.Atlee, W. E., R. P. Burns, and M. Oden. 1970. Serratia marcescens keratoconjunctivitis. Am. J. Ophthalmol. 70:31-33. [DOI] [PubMed] [Google Scholar]
- 6.Aucken, H. M., S. G. Wilkinson, and T. L. Pitt. 1997. Identification of capsular antigens in Serratia marcescens. J. Clin. Microbiol. 35:59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ausubel, F. A., R. Brent, R. F. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
- 8.Bigger, J. F., G. Meltzer, A. Mandell, and R. M. Burde. 1971. Serratia marcescens endophthalmitis. Am. J. Ophthalmol. 72:1102-1105. [DOI] [PubMed] [Google Scholar]
- 9.Bruton, B. D., J. Fletcher, S. D. Pair, M. Shaw, and H. Sittertz-Bhatkar. 1998. Association of a phloem-limited bacterium with yellow vine disease in cucurbits. Plant Dis. 82:512-520. [DOI] [PubMed] [Google Scholar]
- 10.Carbonell, G. V., H. H. M. Della Colleta, T. Yano, A. L. C. Darini, C. E. Levy, and B. A. L. Fonseca. 2000. Clinical relevance and virulence factors of pigmented Serratia marcescens. FEMS Immunol. Med. Microbiol. 28:143-149. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, R., and I. Constable. 1977. Infective keratitis in soft contact lens wearers. Br. J. Ophthalmol. 61:250-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 13.Dunlap, P. V., and E. P. Greenberg. 1988. Control of Vibrio fischeri lux gene transcription by a cyclic AMP receptor protein-LuxR protein regulatory circuit. J. Bacteriol. 170:4040-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberl, L., G. Christiansen, S. Molin, and M. Givskov. 1996. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J. Bacteriol. 178:554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberl, L., M. K. Winson, C. Sternberg, G. S. A. B. Stewart, G. Christiansen, S. R. Chhabra, B. Bycroft, P. Williams, S. Molin, and M. Givskov. 1996. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 20:127-136. [DOI] [PubMed] [Google Scholar]
- 16.Elkins, J. G., D. J. Hassett, P. S. Stewart, H. P. Schweizer, and T. R. McDermott. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ. Microbiol. 65:4594-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleiszig, S. M. J., T. S. Zaidi, M. J. Preston, M. Grout, D. J. Evans, and G. B. Pier. 1996. Relationship between cytotoxicity and corneal epithelial cell invasion by clinical isolates of Pseudomonas aeruginosa. Infect. Immun. 64:2288-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fletcher, M. 1996. Bacterial attachment in aquatic environments: a diversity of surfaces and adhesion strategies, p. 1-24. In M. Fletcher (ed.), Bacterial adhesion: molecular and ecological diversity, vol. 1. Wiley-Liss, New York. NY. [Google Scholar]
- 19.Gandhi, P. A., A. D. Sawant, L. A. Wilson, and D. G. Ahearn. 1993. Adaptation and growth of Serratia marcescens in contact lens disinfectant solutions containing chlorhexidine gluconate. Appl. Environ. Microbiol. 59:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Givskov, M., R. de Nys, M. Manefield, L. Gram, R. Maximilien, L. Eberl, S. Molin, P. D. Steinberg, and S. Kjelleberg. 1996. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol. 178:6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Givskov, M., L. Olsen, and S. Molin. 1988. Cloning and expression in Escherichia coli of the gene for extracellular phospholipase A1 from Serratia liquefaciens. J. Bacteriol. 170:5855-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Givskov, M., J. Östling, L. Eberl, P. W. Lindum, A. B. Christensen, G. Christiansen, S. Molin, and S. Kjelleberg. 1998. Two separate regulatory systems participate in control of swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:742-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guasch, G. F., S. Ferrer, J. Enfedaque, M. B. Viejo, and M. Regue. 1995. A 17 kDa outer-membrane protein (Omp4) from Serratia marcescens confers partial resistance to bacteriocin 28b when expressed in Escherichia coli. Microbiology 141:2535-2542. [DOI] [PubMed] [Google Scholar]
- 24.Hansen, L. H., S. J. Sorensen, and L. B. Jensen. 1997. Chromosomal insertion of the entire Escherichia coli lactose operon, into two strains of Pseudomonas, using a modified mini-Tn5 delivery system. Gene 186:167-173. [DOI] [PubMed] [Google Scholar]
- 25.Hejazi, A., and F. R. Falkiner. 1997. Serratia marcescens. J. Med. Microbiol. 46:903-912. [DOI] [PubMed] [Google Scholar]
- 26.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Anderson, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Hoiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87-102. [DOI] [PubMed] [Google Scholar]
- 27.Hertle, R., and H. Schwartz. 2004. Serratia marcescens internalization and replication in human bladder epithelial cells. BMC Infect. Dis. 4:16-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hild, E., K. Takayama, R.-M. Olsson, and S. Kjelleberg. 2000. Evidence for a role of rpoE in stressed and unstressed cells of marine Vibrio angustum S14. J. Bacteriol. 182:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoiby, N., K. Johansen, C. Moser, Z. J. Song, O. Ciofu, and A. Kharazmi. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect. 3:23-35. [DOI] [PubMed] [Google Scholar]
- 30.Hume, E. B. H., and M. D. P. Willcox. 1997. Adhesion and growth of Serratia marcescens on artificial closed eye tears soaked hydrogel contact lenses. Aust. N. Z. J. Ophthalmol. 25(Suppl. 1):S39-S41. [DOI] [PubMed] [Google Scholar]
- 31.Hume, E. B. H., M. D. P. Willcox, D. F. Sweeney, and B. A. Holden. 1996. An examination of the clonal variants of Serratia marcescens that infect the eye during contact lens wear. J. Med. Microbiol. 45:127-132. [DOI] [PubMed] [Google Scholar]
- 32.Klemm, P., and M. A. Schembri. 2000. Bacterial adhesins: function and structure. Int. J. Med. Microbiol. 290:27-35. [DOI] [PubMed] [Google Scholar]
- 33.Koutsoudis, M. D., D. Tsaltas, T. D. Minogue, and S. B. von Bodman. 2006. Quorum sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. USA 103:5983-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labbate, M., S. Y. Queck, K. S. Koh, S. A. Rice, M. Givskov, and S. Kjelleberg. 2004. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 186:692-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai, H.-C., P.-C. Soo, J.-R. Wei, W.-C. Yi, S.-J. Liaw, Y.-T. Horng, S.-M. Lin, S.-W. Ho, S. Swift, and P. Williams. 2005. The RssAB two-component signal transduction system in Serratia marcescens regulates swarming motility and cell envelope architecture in response to exogenous saturated fatty acids. J. Bacteriol. 187:3407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY.
- 37.Lazachek, G. W., G. L. Boyle, A. L. Schwartz, and I. H. Leopold. 1971. Serratia marcescens, an ocular pathogen: new considerations. Arch. Ophthalmol. 86:599-603. [DOI] [PubMed] [Google Scholar]
- 38.Leranoz, S., P. Orús, F. Berlanga, F. Dalet, and M. Viñas. 1997. New fimbrial adhesins of Serratia marcescens isolated from urinary tract infections: description and properties. J. Urol. 157:694-698. [PubMed] [Google Scholar]
- 39.Lindum, P. W., U. Anthoni, C. Christophersen, L. Eberl, S. Molin, and M. Givskov. 1998. N-acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynch, M. J., S. Swift, D. F. Kirke, C. W. Keevil, C. E. R. Dodd, and P. Williams. 2002. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4:18-28. [DOI] [PubMed] [Google Scholar]
- 41.Matz, C., T. Bergfeld, S. A. Rice, and S. Kjelleberg. 2004. Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ. Microbiol. 6:218-226. [DOI] [PubMed] [Google Scholar]
- 42.McGrath, S., D. S. Wade, and E. C. Pesci. 2004. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 230:27-34. [DOI] [PubMed] [Google Scholar]
- 43.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, V. L., and J. J. Mekalanos. 1984. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc. Natl. Acad. Sci. USA 81:3471-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuno, T., and S. Mizushima. 1990. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol. Microbiol. 4:1077-1082. [DOI] [PubMed] [Google Scholar]
- 46.Nichols, W. A., S. Clegg, and M. R. Brown. 1990. Characterization of the type I fimbrial subunit gene (fimA) of Serratia marcescens. Mol. Microbiol. 4:2119-2126. [DOI] [PubMed] [Google Scholar]
- 47.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 48.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in Gram-negative bacteria: a signalling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Queck, S. Y., M. Weitere, A. M. Moreno, S. A. Rice, and S. Kjelleberg. 2006. The role of quorum sensing mediated development traits in the resistance of Serratia marcescens biofilms against protozoan grazing. Environ. Microbiol. 8:1017-1025. [DOI] [PubMed] [Google Scholar]
- 50.Redfield, R. J. 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10:365-370. [DOI] [PubMed] [Google Scholar]
- 51.Rice, S. A., K. S. Koh, S. Y. Queck, M. Labbate, K. W. Lam, and S. Kjelleberg. 2005. Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J. Bacteriol. 187:3477-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riedel, K., T. Ohnesorg, K. A. Krogfelt, T. S. Hansen, K. Omori, M. Givskov, and L. Eberl. 2001. N-acyl-l-homoserine lactone-mediated regulation of the Lip secretion system in Serratia liquefaciens MG1. J. Bacteriol. 183:1805-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riegman, N., R. Kusters, H. van Veggel, H. Bergmans, P. van Bergen En Henegouwen, J. Hacker, and I. van Die. 1990. F1C fimbriae of a uropathogenic Escherichia coli strain: genetic and functional organization of the foc gene cluster and identification of minor subunits. J. Bacteriol. 172:1114-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 55.Schooling, S. R., P. Gilbert, and D. G. Allison. 2001. Is there a role for homoserine lactones in the biofilm phenotype? p. 201-210. In P. Gilbert, D. Allison, M. Brading, J. Verran, and J. Walker (ed.), Biofilm community interactions: chance or necessity? BioLine, Cardiff, United Kingdom.
- 56.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 57.Siebert, P. D., A. Chenchik, D. E. Kellogg, K. A. Lukyanov, and S. A. Lukyanov. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 59.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 60.Steidle, A., K. Sigl, R. Schuhegger, A. Ihring, M. Schmid, S. Gantner, M. Stoffels, K. Riedel, M. Givskov, A. Hartmann, C. Langebartels, and L. Eberl. 2001. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67:5761-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thuruthyil, S. J., H. Zhu, and M. D. P. Willcox. 2001. Serotype and adhesion of Pseudomonas aeruginosa isolated from contact lens wearers. Clin. Exp. Ophthalmol. 29:147-149. [DOI] [PubMed] [Google Scholar]
- 62.Tillett, D. 2000. Microcystin: genetics, biosynthesis and evolution. Ph.D. thesis. University of New South Wales, Sydney, Australia.
- 63.Tillett, D., and B. A. Neilan. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 36:251-258. [Google Scholar]
- 64.Vica Pacheco, S., O. G. González, and G. L. P. Contreras. 1997. The lom gene of bacteriophage λ is involved in Escherichia coli K12 adhesion to human buccal epithelial cells. FEMS Microbiol. Lett. 156:129-132. [DOI] [PubMed] [Google Scholar]
- 65.Watnick, P. I., K. J. Fullner, and R. Kolter. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae E1 Tor. J. Bacteriol. 181:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]