Abstract
In Ralstonia eutropha H16, the nitric oxide (NO)-responsive transcriptional activator NorR controls the expression of a dicistronic operon that encodes a membrane-bound NO reductase, NorB, and a protein of unknown function, NorA. The N-terminal domain (NTD) of NorR is responsible for perception of the signal molecule, nitric oxide. Thirteen out of 29 conserved residues of the NTD were exchanged by site-directed mutagenesis. Replacement of R63, R72, D93, D96, C112, D130, or F137 strongly decreased NorR-dependent promoter activation, while the exchange of Y95 or H110 led to an increase in promoter activity compared to that of the wild type. A purified truncated NorR comprising only the NTD (NorR-NTD) contained one iron atom per molecule and was able to bind NO in the as-isolated state. Based on the iron content of NorR-NTD proteins with single amino acid replacements, residues R72, D93, D96, C112, and D130 are likely candidates for iron ligands. Residues R63, Y95, and H110 appear not to be involved in NO binding but may take part in subsequent steps of the signal transduction mechanism of NorR.
The diatomic radical nitric oxide (NO) is an obligate intermediate of bacterial denitrification. NO is produced from nitrite by respiratory nitrite reductase and is converted to nitrous oxide by nitric oxide reductase (30). Also, nondenitrifiers like Escherichia coli K-12 can produce considerable amounts of NO during growth under hypoxic conditions with nitrate (6). Since NO and derived reactive nitrogen oxides act as toxic and mutagenic agents, defense mechanisms must be induced under conditions of nitrosative stress. NO-responsive transcription factors are found in the Fnr/Crp family (e.g., DnrD [31]), the Rrf2 family (e.g., NsrR [2]), and the NtrC/NifA family (e.g., NorR). The last protein has been identified in the betaproteobacterium Ralstonia eutropha H16 (21) and E. coli (15). However, genomic analysis revealed that NorR orthologs are widely distributed among beta- and gammaproteobacteria (22). Like all proteins of the σ54-dependent NtrC/NifA superfamily (25), NorR shows a typical modular structure, including an N-terminal domain (NTD), a central AAA+ domain, and a helix-turn-helix domain for DNA binding (3, 21). In a recent study, it was shown that the NTD of E. coli NorR harbors a mono-iron center that is able to bind NO as a ferrous nitrosyl (10). Those authors presented a model for the activation of NorR which postulates the existence of an inactive form of NorR, where the ATPase activity of the AAA+ domain is blocked by the NTD. Upon binding of NO, this block is relieved and NorR switches to an active state which is competent to induce promoter activation. This hypothesis is corroborated by the fact that truncated forms of NorR from E. coli and R. eutropha that lack the NTD show signal-independent activation of gene expression (10, 21). To date, neither the iron ligands nor the residues that are involved in the switching mechanism have been identified.
Given the high positional identity (45%) between the amino acid sequences of the NorR-NTD proteins from E. coli and those from R. eutropha, it is likely that both proteins rely on the same mechanism of signal sensing. In this study, amino acid exchanges were introduced into R. eutropha NorR to identify residues that may be involved in iron coordination and/or are critical for the formation of the active state of the protein. Due to its modular structure, NorR is well suited to be targeted by site-directed mutagenesis. Mutations within the NTD most likely do not modify the binding site of the sigma factor (located within the central AAA+ domain) or the helix-turn-helix DNA binding site (located at the C terminus of NorR). Mutated NorR derivatives were investigated with respect to (i) their responses toward an artificial inducer, (ii) their abilities to promote growth by denitrification under physiological conditions, and (iii) the amounts of iron present in the NTD. In combination, the results of this study allow the assignment of candidate ligands of the iron center, as well as residues that appear not to be involved in NO binding but may take part in subsequent steps of the signal transduction mechanism of NorR.
MATERIALS AND METHODS
Media and growth conditions.
E. coli strains were grown in Luria-Bertani broth at 37°C. R. eutropha strains were cultivated at 30°C in mineral salt medium (23) with 0.4% (wt/vol) fructose as the carbon source (FN medium). Solid medium contained 1.5% (wt/vol) agar. Antibiotics were added as follows: for R. eutropha, kanamycin (360 μg/ml) and tetracycline (15 μg/ml); for E. coli, ampicillin (50 μg/ml). Growth curves of denitrifying R. eutropha cells were recorded with a 96-well microtiter plate assay. Cultures were grown in 300 μl FN medium supplemented with 0.1% (wt/vol) sodium nitrate. The plates were sealed with a gastight transparency film, and the increase in turbidity (measured at 436 nm) was followed with an automated optical plate reader (SpectraMAX; Molecular Devices) at 30°C.
Strains and plasmids.
E. coli S17-1 (24) served as the donor in conjugative transfers. E. coli XL1-Blue (Stratagene) and E. coli GM2163 (New England Biolabs) were used as hosts for standard cloning procedures. E. coli BL21(DE3) (26) was used for protein overexpression. R. eutropha H16 (ATCC 17699) was used as the wild-type strain, which contains two paralogous norRAB gene regions designated norR1A1B1 and norR2A2B2 (9). While only the norR1A1B1 unit has been studied in detail, mutation analyses have shown that either one of the two is sufficient for denitrification. Throughout Results and Discussion below, designations norR, norA, and norB refer to norR1A1B1. R. eutropha strains HF640 and HF658 are derivatives of the wild-type H16. In HF640 (ΔnorR2A2B2 PnorA1-lacZ), the nor gene region (norR2A2B2) on chromosome 2 was replaced by a PnorA1-lacZ fusion present on plasmid pCH1018 (see below). The NorR-negative strain HF658 (ΔnorR1 ΔnorR2A2B2 PnorA1-lacZ) is a derivative of HF640 and contains an additional in-frame deletion in norR1. This mutation was established with plasmid pCH699 (see below). Mutants were analyzed by Southern blotting to verify the gene replacement and to exclude sequence rearrangements adjacent to the deletion site. Derivatives of the vectors pLO1 and pLO3 were used for gene replacement by homologous recombination (17). Plasmid pCH694 (21) is a derivative of Litmus 28 (New England Biolabs), with a 3.3-kb EcoRI-BamHI insert that contains norR1 and norA1. Plasmid pCH1157 is a pLO3 derivative that contains a 2.3-kb EcoRI-AscI fragment of pCH694.
Selection of targeted residues.
A multiple alignment of NorR proteins was performed with ClustalW (27) and included orthologs from the following organisms: Azotobacter vinelandii AvOP (unfinished genome; Department of Energy, Joint Genome Institute), Burkholderia xenovorans LB400, Burkholderia sp. strain TH2, Erwinia carotovora subsp. atroseptica strain SCRI1043, Escherichia coli CFT073, E. coli K-12, E. coli O157:H7, Idiomarina loihiensis L2TR, Photobacterium profundum SS9, Polaromonas sp. strain JS666, Pseudomonas aeruginosa PAO1, P. aeruginosa UCBPP-PA14, Pseudomonas fluorescens PfO-1, Pseudomonas putida KT2440, R. eutropha H16, R. eutropha JMP134, Ralstonia metallidurans CH34, Ralstonia solanacearum GMI1000, Salmonella enterica subsp. enterica serovar choleraesuis strain SC-B67, Salmonella enterica serovar Typhimurium strain LT2, Shigella flexneri serotype 2a strain 301, Vibrio cholerae, Vibrio fischeri ES114, Vibrio parahaemolyticus RIMD 2210633, Vibrio vulnificus CMCP6, and V. vulnificus YJ016. Both NorR paralogs present in V. fischeri ES114, V. vulnificus CMCP6 and V. vulnificus YJ016 were included.
Construction of plasmids for the generation of norR1 mutant strains.
Plasmid pCH694 served as a template in PCRs for site-directed mutagenesis of norR1. Mutations were generated by the four-primer method (1). Flanking primers carrying unique restriction sites for BstEII and FseI, respectively, were P602 (5′-CGACCTGGTCACCGACCTGCCGCATGCGGTGCGCCTCCAGCGGCTGGTC-3′) and P603 (5′-CGTCGGCCGGCCCTTCACCGTCTGGCTGTTTGCC-3′) (restriction sites are underlined). Mutagenic primers are listed in Table 1. The resulting 499-bp PCR fragments were BstEII-FseI cloned into plasmid pCH1157. All mutations were verified by DNA sequencing. The derivatives of pCH1157 (Table 1) were used for homologous recombination with R. eutropha HF640.
TABLE 1.
Primers and derived plasmids for site-directed mutagenesis
| Amino acid exchange | Primer sequence (5′-3′)a | Directionb | Resulting plasmid |
|---|---|---|---|
| C35S | GCACGCATTTCCGTTcgGGTGCGGTGGCGCTG | f | pCH1174 |
| CAGCGCCACCGCACCcgAACGGAAATGCGTGC | r | ||
| R63K | GCGATACGCTCGGGaaGCGCTTTGCGGTGAG | f | pCH1289 |
| CTCACCGCAAAGCGCttCCCGAGCGTATCGC | r | ||
| H70Q | GCGGTGAGCCTGCAgCCGCGGCTTGCCGCC | f | pCH1326 |
| GGCGGCAAGCCGCGGcTGCAGGCTCACCGC | r | ||
| R72K | TGAGCCTGCACCCGaaGCTTGCCGCCATCC | f | pCH1290 |
| GGATGGCGGCAAGCttCGGGTGCAGGCTCA | r | ||
| C84S | CGGCGCGACGTCACCTcgTTCCATCACGACAGC | f | pCH1176 |
| CTGTCGTGATGGAAcgAGGTGACGTCGCGCCGC | r | ||
| F85L | GACGTCACCTGCcTgCATCACGACAGCATG | f | pCH1291 |
| CATGCTGTCGTGATGcAgGCAGGTGACGTC | r | ||
| D93N | ACAGCATGTTGCCCaACCCCTACGACGGGC | f | pCH1292 |
| GCCCGTCGTAGGGGTtGGGCAACATGCTG | r | ||
| Y95F | GTTGCCCGACCCCTtCGACGGGCTGATCG | f | pCH1149 |
| CGATCAGCCCGTCGaAGGGGTCGGGCAAC | r | ||
| Y95L | GTTGCCCGACCCCctgGACGGGCTGATCG | f | pCH1150 |
| CGATCAGCCCGTCcagGGGGTCGGGCAAC | r | ||
| D96N | TGCCCGACCCCTACaACGGGCTGATCGACG | f | pCH1293 |
| CGTCGATCAGCCCGTtGTAGGGGTCGGGC | r | ||
| H110Q | GCCGCTGCCGGTGCAgGACTGCATGGGCACG | f | pCH1151 |
| CGTGCCCATGCAGTCcTGCACCGGCAGCGGC | r | ||
| C112S | CTGCCGGTGCACGACTcgATGGGCACGAGCCTGC | f | pCH1152 |
| GCAGGCTCGTGCCCATcgAGTCGTGCACCGGCAG | r | ||
| D130N | GCGTGCTGACGCTCaACGCGCTCACGGTCGG | f | pCH1325 |
| CCGACCGTGAGCGCGTtGAGCGTCAGCACGC | r | ||
| F137L | TCACGGTCGGCACGcTgGACGCCGCCGCGC | f | pCH1327 |
| GCGCGGCGGCGTCcAgCGTGCCGACCGTG | r |
Mutated bases are indicated in lowercase.
f, forward primer; r, reverse primer.
Determination of promoter activities.
A 320-bp region upstream of norA1 containing the promoter for both norA1 (PnorA) and norB1 was PCR amplified using pCH510 (9) as the template and primers 5′-CGGGATCCGCCACCGGCCGCAGGTG-3′ and 5′-TAAAGCTTGTCATGATGTTTCTCCGTCTG-3′ that contain BamHI and HindIII sites (underlined), respectively. The amplified fragment was BamHI-HindIII cloned into the promoter test vector pPHU236 (14) to give plasmid pGE570, containing the PnorA-lacZ fusion. pGE570 was digested with BamHI-SalI, and the resulting 3.5-kb fragment was treated with Klenow polymerase and cloned into the StuI site of pCH696 (21) to give pCH1128. Following the digestion of pCH1128 with KpnI and SpeI and treatment of the 5.9-kb fragment with Klenow polymerase, the fragment was cloned into the PmeI-digested vector pLO1, yielding plasmid pCH1018. The latter plasmid was used for inserting the PnorA-lacZ fusion into chromosome 2 by homologous recombination. Strains containing the PnorA-lacZ fusion were cultivated at 30°C under microaerobic conditions for 4 h to reduce the oxygen tension prior to the addition of 2 mM sodium nitroprusside [SNP; sodium nitrosylpentacyanoferrate(III)-dihydrate] to activate NorR (21). β-Galactosidase activity was assayed as described in reference 21.
Expression of norR in trans.
Since a polyclonal antibody raised against the N-terminal domain of NorR turned out to be unsuitable for full-length NorR, all mutated NorR variants were genetically engineered to contain an N-terminal Strep-tag II affinity peptide (ST) and were detected by Western immunoblot analysis using a commercial anti-ST antibody. The mutated norR genes were expressed in trans under the control of the promoter of the fhp gene (Pfhp). This promoter is induced under low-oxygen tension. The respective plasmids were constructed as follows. The region containing Pfhp was PCR amplified from pGE28 (29) with primers 5′-GCTCTAGACAAGCTTTCGAGCTGTCAGTCCGGCGCCGAGAG-3′ and 5′-GGGTCAGCATATGTCGGTCTCCATGGCGCG-3′ containing sites XbaI, HindIII, and NdeI (underlined), respectively. The resulting 632-bp PCR fragment was NdeI-XbaI cloned into pET22b+ (Novagen) to give pCH1105. To create an NdeI site at the start codon of norR1, the 5′ region of norR1 was PCR amplified from pCH694 with primers 5′-GACCGGTATTTATCACCCATATGCACCATCACCATCACCATACGCCCCTGTATCCCGAGTTGCTGACCGACCTGGTCA-3′ (the NdeI site is underlined) and 5′-CACCAGCCCGTCGACCGCCACCGG-3′. The fragment was NdeI-SalI cloned into pET22b+ to give pCH912. The norR1 sequence was restored by cloning a 1.8-kb SalI-HindIII fragment from pCH694 into pCH912 to give pCH913. A sequence encoding ST (WSHPQFEK) was added 5′ to norR1 by amplifying a fragment from pCH694 with primers 5′-GACCGGTCATATGGCCAGCTGGAGCCACCCGCAGTTCGAGAAGGGCGCCACGCCCCTGTATCCCGAGTTGCTGACCGAC-3′ (the NdeI site and the ST coding sequence are underlined) and 5′-CATCGAGGAACAGGGTGCCGCCGG-3′. This fragment was NdeI-SalI digested and cloned into NdeI-SalI-digested pCH913 to give plasmid pCH1107. The norR1 region was NdeI-BamHI subcloned into pCH1105, yielding pCH1108, which then contained Pfhp upstream of Strep-tag-norR1 gene fusion. Finally, all derivatives of pCH1157 (Table 1) containing mutagenized norR1 genes were BstEII-XhoI digested, and the resulting 1.2-kb fragments were cloned into the broad-host-range vector pCM62 (19). These plasmids were conjugated into R. eutropha HF658. For expression of norR, cells were grown under microaerobic conditions at 30°C for 36 h.
Overproduction and purification of NorR-NTD.
The NTD of NorR was modified to contain a C-terminal six-histidine coding sequence (His tag) and was overproduced with E. coli BL21(DE3) with a pET expression system (Stratagene). Plasmid pCH1088 is a pET22b+ derivative containing a 535-bp NdeI-SacI fragment that encodes the first 169 amino acids of NorR fused 3′ to a sequence coding six additional histidines (His tag). This fragment was generated by PCR with pCH694 as a template and primers 5′-GACCGGTATTTATCACCCATATGACGCCCATG-3′ and 5′-CGAGCTCCAAGCTTCTAATGGTGATGGTGATGGTGCTGCAGGGCGCGGATCTCGGCCTCCAGCCGGGTGGTG-3′ (restriction sites and sequences for the His tag are underlined). All derivatives of pCH1157 (Table 1) containing mutagenized norR1 genes were SexAI-NcoI or SexAI-PstI digested and cloned into pCH1088, thereby replacing the native norR1 sequence.
For overproduction of NorR-NTD, strains were cultivated overnight at 30°C under microaerobic conditions. IPTG (isopropyl-β-d-thiogalactopyranoside; 0.1 mM) was added after 3 h of preincubation. NO-treated cultures were grown aerobically at 30°C with shaking (120 rpm) for 1.5 h after induction. Cells were spun down at 5,000 × g and resuspended in 140 ml of prewarmed LB medium. The culture was transferred into a sealed flask (150 ml) and incubated for 15 min at 30°C without aeration to achieve anaerobic conditions. Nitric oxide-saturated solution was then added to a final concentration of about 5 μM. After 5 min of incubation, cells were harvested at 5,000 × g for 15 min.
To purify NorR-NTD, cell pellets were resuspended in lysis buffer (300 mM NaCl, 10 mM imidazole, 50 mM phosphate buffer [pH 8], including a protease inhibitor cocktail [Sigma]), and cells were disrupted by sonication. Subsequent steps were carried out in a glove box. The extract was centrifuged at 18,000 × g for 30 min at 10°C and subjected to affinity chromatography using Ni-nitrilotriacetic acid spin columns (QIAGEN). This procedure yielded pure NorR-NTD protein according to in-gel staining with Coomassie brilliant blue. Protein concentrations were determined according to the method described by Lowry et al. (17a). Iron was determined using the ferene method (12). UV-visible spectra were recorded on a Cary 300 SCAN UV-Vis spectrophotometer.
RESULTS
Site-directed mutagenesis.
Based on a multiple alignment of NorR orthologs (not shown), a number of residues within the NTD of NorR were selected for site-directed mutagenesis (Fig. 1). These included C35, R63, H70, R72, D93, D96, H110, C112, D130, and F137, which are conserved in most or all NorR orthologs and may serve as putative iron ligands. In a distinct subgroup of NorR orthologs, Y95 (using R. eutropha numbering) is replaced by phenylalanine in the native sequence. Members of this subgroup are encoded predominantly in Vibrio species and also differ from other NorR proteins with respect to a conserved motif in the helix-turn-helix domain (MGKKLG instead of the common LAKRLG sequence). To investigate whether the Y- to-F conversion is mechanistically important, Y95 of R. eutropha NorR was replaced by phenylalanine and leucine. In addition, the nonconserved cysteine C84 was targeted. Cysteine exchange mutations were performed since these residues are potential targets of S-nitrosylation and hence may take part in the activation of NorR by NO. Amino acid exchanges in NorR were established by mutation of the norR gene in cis to avoid potential titration effects that might arise from a multicopy in trans system.
FIG. 1.
Conserved residues in NorR-NTD and amino acid exchanges. Boxed residues are strictly conserved in NorR orthologs; encircled residues are conserved in the majority of orthologs. Sequence positions of marked residues are indicated. Examples of alternative residues are given below the sequence along with the corresponding organism: Bxv, Burkholderia xenovorans LB400; Ppr, Photobacterium profundum SS9; Avn, Azotobacter vinelandii AvOP; Prm, Polaromonas sp. strain JS666. A conserved tyrosine that is replaced by phenylalanine in a distinct subset of NorR orthologs (see text for details) is marked by a diamond-shaped outline (middle sequence row). Arrows denote exchanges constructed in this study.
Activation of the norA promoter.
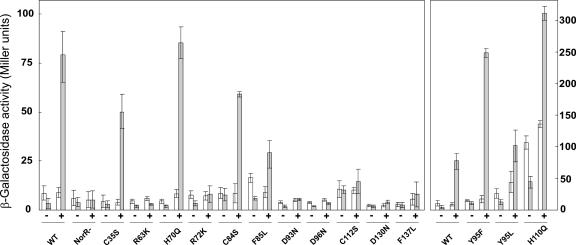
To date, the only genes known to be regulated by NorR in R. eutropha are norA and norB. These genes form a dicistronic operon and are expressed from PnorA. While norB encodes a membrane-bound NO reductase, the physiological function of the norA gene product is as yet unclear. Preliminary data indicate that NorA is involved in NO metabolism (8). Promoter activation by NorR was monitored using a transcriptional fusion of PnorA with the lacZ gene of E. coli. The PnorA-lacZ fusion was integrated into chromosome 2 of R. eutropha to yield a single-copy reporter system. To follow the activation of PnorA under controlled conditions, the nitrosylating agent SNP was added as an artificial inducer to nondenitrifying cell cultures that were depleted of oxygen. A PnorA response to SNP under these conditions has been shown previously (21). β-Galactosidase activity was determined at 4 h and 20 h after the addition of SNP. As a control, β-galactosidase activity in uninduced cells was also determined. The results of this experiment are shown in Fig. 2. The promoter activity in strains containing NorR with the C35S, H70Q, C84S, or Y95L exchange deviated from that for wild-type NorR (strain HF640) by less than 40%. Taking into account the standard deviations shown in Fig. 2, these residues are considered not essential for signal perception. Surprisingly, exchanges Y95F and H110Q resulted in a significant increase in promoter activity. The most prominent effect was observed with H110Q. Although the activity in the H110Q exchange mutant was increased even in the absence of SNP, it was still SNP dependent rather than constitutive. About 20 to 25% of activity was left in strains containing a F85L or a C112S exchange. All other exchanges resulted in activity levels in a range equal to that of the NorR-negative mutant. Particularly low activities were observed with exchanges R63K, R72K, D93N, D96N, and D130N. Western immunoblot analysis of strains containing Strep-tag-carrying derivatives of either of the mutated NorR proteins confirmed that none of the amino acid exchanges led to a significantly reduced level of NorR (data not shown).
FIG. 2.
Transcriptional activation of the norA promoter by NorR and mutated derivatives of NorR. Promoter activation by NorR was induced by the addition of 2 mM SNP (+) and compared to uninduced cells (−). Activities (expressed in Miller units) were determined at 4 h (white bars) and 20 h (gray bars) after induction. The wild type (WT) and NorR mutants are indicated at the bottom. Standard deviations (error bars) were estimated from three to six independent experiments. The panel at the right is presented at a different scale to fit mutants with high activities.
Physiological impact of amino acid exchanges in NorR.
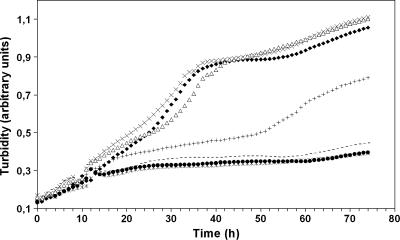
It was demonstrated previously that a mutant lacking the NO reductase NorB is conditionally lethal under denitrifying growth conditions (9). Therefore, reduced activation of PnorA (and hence a decreased level of norB expression) should impair growth by denitrification. Growth of denitrifying cells was monitored with a microtiter plate assay with an automated plate reader device to obtain highly resolved data sets. Curves of a representative subset of mutant strains are illustrated in Fig. 3. In general, strains showing wild-type-like promoter activity were almost unimpaired in growth, while strains exhibiting basal levels of promoter activity showed virtually no growth by denitrification. Namely, the R63K, R72K, D93N, D96N, C112S, D130N, and F137L strains belonged to the latter group. Intermediate growth phenotypes were observed for F85L and C84S strains. The enhanced promoter activity in the Y95F and H110Q mutants did not result in a major improvement of growth. This result indicates that the expression of norB beyond the wild-type level is not beneficial for denitrification, perhaps because the concentration of NorB in wild-type cells is high enough not to be limiting for the reduction of NO.
FIG. 3.
Growth of mutant strains by denitrification as shown in a microtiter plate assay. Each curve represents the mean of four to six individual cultures. Turbidity was measured at 436 nm. Δ, wild type; •, NorR-negative mutant; +, NorR-F85L; *, NorR-D96N; ⧫, NorR-Y95F; ×, NorR-H110Q; -, NorR-C112S. The inflection point of the curves around 12 h reflects a lag phase of cell growth during adaptation from aerobic growth to denitrification, which was due to residual oxygen in the culture wells.
Iron content and NO-binding behavior of NorR-NTD.
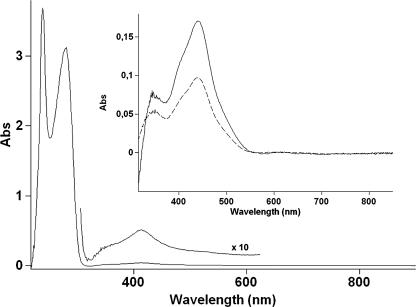
Attempts to overproduce and purify full-length NorR neither resulted in sufficiently high yields of soluble protein nor allowed reproducible quantification of iron. Alternatively, the iron content of NorR was assessed from the isolated signaling domain of NorR. This approach was considered to be feasible since (i) the modular structure of NorR implies that the N-terminal portion behaves as a functional domain and (ii) the N-terminal domain of E. coli NorR binds NO and is indistinguishable from full-length NorR according to its electron spin resonance spectrum (10). A truncated protein (NorR-NTD) was constructed that contains the N-terminal 169 residues of R. eutropha NorR and 6 additional histidines (His tag) at the C terminus. NorR-NTD was overproduced in E. coli cells and purified under anaerobic conditions by a single-step procedure. The iron content determination of various preparations yielded a ratio of 0.79 to 0.87 mol of iron per mol of NorR-NTD. Although a detailed spectroscopic investigation was beyond the scope of this study, we sought to prove that purified NorR-NTD was functional in terms of NO binding. The reaction of NorR-NTD with NO gas was monitored by UV-visible spectroscopy. NorR-NTD in the as-isolated state showed a maximum of absorption at 415 nm and a shoulder at around 350 nm in the visible spectrum (Fig. 4). The addition of stoichiometric amounts of NO to this preparation resulted in a change of absorption, showing a new signal at 440 nm and shoulders at around 350 and 410 nm. Signals in the 400- to 500-nm region are indicative of a ferrous nitrosyl; similar features have been reported for aquated iron-NO (28) and the nitrosyl adducts of several ferrous nonheme mono-iron and di-iron proteins (4, 5, 7, 18). While as-isolated NorR-NTD was very sensitive to traces of oxygen, leading to an immediate loss of iron and a precipitation of protein, the NO-adduct of NorR-NTD was quite stable in air. After 2 h of incubation at 30°C, about one-third of the signal at 440 nm was still visible. Stability toward oxygen was also reported for NorR-NO from E. coli (10). To show that R. eutropha NorR-NTD binds NO in vivo, the protein was purified from E. coli cultures treated with NO. The optical spectrum, recorded immediately after anaerobic purification of NorR-NTD from these cells, yielded essentially the same spectrum as that for the NO-adduct prepared in vitro (Fig. 4). NorR-NTD purified in the presence of oxygen showed no conspicuous signals in optical spectroscopy (not shown) and contained virtually no iron. This result excludes a possible contribution of the His tag to the iron binding properties of NorR-NTD.
FIG. 4.
Optical spectra of NorR-NTD (200 μM). The main panel shows NorR-NTD in the as-isolated state. The inset shows difference spectra of NorR-NTD incubated with NO in vitro and in vivo, respectively: dashed line, NorR-NTD incubated with NO (200 μM) minus as-isolated NorR-NTD; solid line, NorR-NTD purified from cultures amended with NO minus NorR-NTD purified from cultures without the addition of NO. Abs, absorption.
Iron content and spectroscopic investigation of mutant proteins.
All exchanges introduced into full-length NorR were also established in a truncated NorR-NTD protein. Each of the mutated proteins was overproduced in E. coli cells, and the iron content was determined from the purified proteins. As shown in Table 2, the iron content of the C35S, R63K, H70Q, C84S, F85L, and H110Q variants was comparable to that of NorR-NTD. The Y95F and F137L variants showed about half of the iron content of NorR-NTD. Only up to 24% iron was detected in the D93N, Y95L, D96N, R72K, C112S, and D130N variants. Given that the Y95L and Y95F exchange mutants were still susceptible to SNP induction (and thus should contain NorR with an intact iron center), we suppose that the low iron content of the respective NorR-NTD mutant proteins is due to a loss of iron during the purification procedure. In contrast, the relatively high iron content of NorR-NTD (F137L) and NorR-NTD (R63K) did not correlate with weak promoter induction and growth observed with the NorR (F137L) and NorR (R63K) mutant strains. To investigate whether this effect was caused by the inability of these proteins to bind NO, NorR-NTD (F137L) and NorR-NTD (R63K) were examined by UV-visible spectroscopy (data not shown). As-isolated NorR-NTD (F137L) did not show an increase at 440 nm upon the addition of NO, indicating that the F137L exchange altered the properties of the iron center and hence prevented binding of NO. NO-exposed NorR-NTD (R63K) protein yielded essentially the same spectrum as NO-exposed NorR-NTD. Therefore, R63 is not important for NO binding but is probably involved in a subsequent step of intramolecular signal transduction.
TABLE 2.
Iron content of mutated NorR-NTD variants
| Amino acid exchange | % of iron in NorR-NTD variants vs wild-type NorR-NTDa |
|---|---|
| C35S | 73 |
| R63K | 100 |
| H70Q | 80 |
| R72K | 7 |
| C84S | 78 |
| F85L | 96 |
| D93N | 10 |
| Y95L | 20 |
| Y95F | 60 |
| D96N | 15 |
| H110Q | 100 |
| C112S | 3 |
| D130N | 15 |
| F137L | 50 |
Values are given as the percentage of the iron content of wild-type NorR-NTD (0.8 mol of iron per mol of protein).
DISCUSSION
In this study we have analyzed mutant NorR proteins with single amino acid replacements at 13 residues, 8 of which belong to a set of 22 residues strictly conserved in the NTD of NorR orthologs. Strictly conserved residues that have not been considered are nonpolar leucines and alanines (L12, L25, A52, L56, and L91), a single threonine (T128), and the helix breakers glycine and proline (G55, G62, P71, P92, P94, G97, G114, and G125). Although these residues cannot be excluded as ligands since backbone nitrogen and oxygen atoms can be involved in metal coordination (13), preferred ligands of iron include imidazole nitrogens, carboxylates, sulfur, and carbonyl oxygens (16). Hence, all residues conserved in the NTD of NorR that are commonly found as side chain ligands in mono-iron proteins were targeted by site-directed mutagenesis in this study. Given that exchange of C35, H70, C84, Y95, and H110 did not abolish SNP-dependent promoter activation in vivo, it is unlikely that these residues participate in the coordination of iron. Likewise, F85 is clearly not essential, albeit the F85L exchange in NorR led to a decreased promoter activity and retarded growth of this mutant. In contrast, residues R63, R72, D93, D96, C112, D130, and F137 are of obvious functional importance since replacements at these positions resulted in virtually inactive NorR. Of these, all three conserved aspartates, R72, and C112 are essential for the coordination of the iron center.
The molecular mechanism of signal transduction by NorR is still unclear. We have shown that a stable NO adduct is formed from stoichiometric amounts of NorR-NTD and NO. This result opens the question of how the active form of NorR is deactivated. While the initial event in the activation of NorR is probably the formation of an iron-mononitrosyl, we considered the possibility that subsequent steps involve an S-nitrosylation reaction. However, our results clearly showed that only one of the three cysteines (C112) in the NTD is important for the function of NorR. Since NorR-NTD (C112S) is virtually devoid of iron, C112 is probably an iron ligand rather than a direct target of nitrosylation. A promising candidate for a residue involved in a later step of signal transduction is R63. Although an R63K exchange affected neither the iron content of NorR-NTD nor its ability to bind NO, full-length NorR (R63K) did not respond to induction by SNP and led to poor growth by denitrification. Thus, we conclude that the R63K mutation obstructs the activation mechanism of NorR, leading to an “always-locked” state of the regulator. Two further residues, Y95 and H110, appear to be involved in a subsequent step of signal transduction. The high promoter activity of the Y95F and H110Q mutants compared to that of the wild-type NorR suggest that, in the inactive form of NorR, these residues take part in the interaction of the NTD with the central domain. This domain-domain interaction is essential for the postulated switching mechanism as it blocks the ATPase activity of NorR (10). The exchange of Y95 or H110 may weaken the locked state, resulting in a partly activated NorR even in the absence of SNP. In the presence of SNP, about threefold-higher promoter activity was observed with NorR (H110Q) than with wild-type NorR. This superactivity is most likely the result of a cumulative effect, because induction by SNP-induced NorR adds to a high basal level of induction. While PnorA activity was increased in the presence of NorR (Y95F), only a marginal effect was observed for NorR (Y95L). This difference may be explained by a more stable iron center in NorR (Y95F) than in NorR (Y95L), as observed for purified NorR-NTD (Y95F) versus NorR-NTD (Y95L). On the basis of the present data, it is not possible to decide if the presence of a phenylalanine at this position in the native sequence of some NorR orthologs is of any physiological relevance. However, it is interesting to note that in most cases, the putative target genes of these NorR proteins encoded a hybrid cluster protein and its cognate reductase. Since hybrid cluster proteins are assumed to be involved in the detoxification of hydroxylamine, the presence of phenylalanine instead of tyrosine in NorR might reflect the adaptation to a signal molecule other than NO.
The spectroscopic features of E. coli NorR-NO suggest a five- or six-coordinate mono-iron structure with NO being the sixth ligand in the active state (10). The exchange of D99 (which corresponds to D96 in R. eutropha NorR) with alanine resulted in iron-free protein that was unable to activate gene expression. Our results agree with these data and suggest that in R. eutropha NorR, the iron is coordinated by five residues: R72, D93, D96, C112, and D130. The stability of the iron center is also affected by F137. Moreover, this residue is essential for NO binding. Notably, none of the histidine residues of the NTD appeared to be involved in iron coordination. To date, only a few proteins containing mono-iron centers without any histidine ligand are known. Prominent examples are Fe-(Cys)4 centers of rubredoxins (11) and the unusual mono-iron center of iron nitrile hydratases (20). However, in the latter case at least three cysteine residues are metal ligands, whereas only one cysteine is essential for iron coordination in NorR. In conclusion, the iron in NorR is coordinated by an unprecedented ligand environment. Future work will include a comprehensive spectroscopic investigation of wild-type NorR and the mutant proteins generated in this study, in order to get further insight into the metal coordination site and the NO binding behavior of NorR.
Acknowledgments
We thank Anne Pohlmann for providing plasmids pCH1088 and pCH1108 and Edward Schwartz for critically reading the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Allemandou, F., J. Nussberger, H. R. Brunner, and N. Brakch. 2003. Rapid site-directed mutagenesis using two-PCR-generated DNA fragments reproducing the plasmid template. J. Biomed. Biotechnol. 2003:202-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodenmiller, D. M., and S. Spiro. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188:874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Büsch, A., A. Pohlmann, B. Friedrich, and R. Cramm. 2004. A DNA region recognized by the nitric oxide-responsive transcriptional activator NorR is conserved in β- and γ-proteobacteria. J. Bacteriol. 186:7980-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, V. J., A. M. Orville, M. R. Harpel, C. A. Frolik, K. K. Surerus, E. Munck, and J. D. Lipscomb. 1989. Spectroscopic studies of isopenicillin N synthase. A mononuclear nonheme Fe2+ oxidase with metal coordination sites for small molecules and substrate. J. Biol. Chem. 264:21677-21681. [PubMed] [Google Scholar]
- 5.Clay, M. D., C. A. Cosper, F. E. Jenney, Jr., M. W. Adams, and M. K. Johnson. 2003. Nitric oxide binding at the mononuclear active site of reduced Pyrococcus furiosus superoxide reductase. Proc. Natl. Acad. Sci. USA 100:3796-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corker, H., and R. K. Poole. 2003. Nitric oxide formation by Escherichia coli. Dependence on nitrite reductase, the NO-sensing regulator Fnr, and flavohemoglobin Hmp. J. Biol. Chem. 278:31584-31592. [DOI] [PubMed] [Google Scholar]
- 7.Coufal, D. E., P. Tavares, A. S. Pereira, B. H. Hyunh, and S. J. Lippard. 1999. Reactions of nitric oxide with the reduced non-heme diiron center of the soluble methane monooxygenase hydroxylase. Biochemistry 38:4504-4513. [DOI] [PubMed] [Google Scholar]
- 8.Cramm, R., A. Büsch, and K. Strube. 2006. NO-dependent transcriptional activation of gene expression in Ralstonia eutropha H16. Biochem. Soc. Trans. 34:182-184. [DOI] [PubMed] [Google Scholar]
- 9.Cramm, R., R. A. Siddiqui, and B. Friedrich. 1997. Two isofunctional nitric oxide reductases in Alcaligenes eutrophus H16. J. Bacteriol. 179:6769-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Autreaux, B., N. P. Tucker, R. Dixon, and S. Spiro. 2005. A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature 437:769-772. [DOI] [PubMed] [Google Scholar]
- 11.Frey, M., L. Sieker, F. Payan, R. Haser, M. Bruschi, G. Pepe, and J. LeGall. 1987. Rubredoxin from Desulfovibrio gigas. A molecular model of the oxidized form at 1.4 Å resolution. J. Mol. Biol. 197:525-541. [DOI] [PubMed] [Google Scholar]
- 12.Hennessy, D. J., G. R. Reid, F. E. Smith, and S. L. Thompson. 1984. Ferene: a new spectrophotometric reagent for iron. Can. J. Chem. 62:721-724. [Google Scholar]
- 13.Holm, R. H., P. Kennepohl, and E. I. Solomon. 1996. Structural and functional aspects of metal sites in biology. Chem. Rev. 96:2239-2314. [DOI] [PubMed] [Google Scholar]
- 14.Hubner, P., J. C. Willison, P. M. Vignais, and T. A. Bickle. 1991. Expression of regulatory nif genes in Rhodobacter capsulatus. J. Bacteriol. 173:2993-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutchings, M. I., N. Mandhana, and S. Spiro. 2002. The NorR protein of Escherichia coli activates expression of the flavorubredoxin gene norV in response to reactive nitrogen species. J. Bacteriol. 184:4640-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlin, S., Z. Y. Zhu, and K. D. Karlin. 1997. The extended environment of mononuclear metal centers in protein structures. Proc. Natl. Acad. Sci. USA 94:14225-14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenz, O., E. Schwartz, J. Dernedde, M. Eitinger, and B. Friedrich. 1994. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J. Bacteriol. 176:4385-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 18.Lu, S., E. Libby, L. Saleh, G. Xing, J. M. Bollinger, Jr., and P. Moenne-Loccoz. 2004. Characterization of NO adducts of the diiron center in protein R2 of Escherichia coli ribonucleotide reductase and site-directed variants; implications for the O2 activation mechanism. J. Biol. Inorg. Chem. 9:818-827. [DOI] [PubMed] [Google Scholar]
- 19.Marx, C. J., and M. E. Lidström. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 20.Nagashima, S., M. Nakasako, N. Dohmae, M. Tsujimura, K. Takio, M. Odaka, M. Yohda, N. Kamiya, and I. Endo. 1998. Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms. Nat. Struct. Biol. 5:347-351. [DOI] [PubMed] [Google Scholar]
- 21.Pohlmann, A., R. Cramm, K. Schmelz, and B. Friedrich. 2000. A novel NO-responding regulator controls the reduction of nitric oxide in Ralstonia eutropha. Mol. Microbiol. 38:626-638. [DOI] [PubMed] [Google Scholar]
- 22.Rodionov, D. A., I. L. Dubchak, A. P. Arkin, E. J. Alm, and M. S. Gelfand. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 1:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies. Arch. Mikrobiol. 38:209-222. [PubMed] [Google Scholar]
- 24.Simon, R., U. B. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 25.Studholme, D. J., and R. Dixon. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185:1757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wanat, A., T. Schneppensieper, G. Stochel, R. van Eldik, E. Bill, and K. Wieghardt. 2002. Kinetics, mechanism, and spectroscopy of the reversible binding of nitric oxide to aquated iron(II). An undergraduate text book reaction revisited. Inorg. Chem. 41:4-10. [DOI] [PubMed] [Google Scholar]
- 29.Weihs, V., K. Schmidt, B. Schneider, and B. Friedrich. 1989. The formation of an oxygen-binding flavohemoprotein from Alcaligenes eutrophus is plasmid-determined. Arch. Microbiol. 151:546-550. [Google Scholar]
- 30.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zumft, W. G. 2002. Nitric oxide signaling and NO dependent transcriptional control in bacterial denitrification by members of the FNR-CRP regulator family. J. Mol. Microbiol. Biotechnol. 4:277-286. [PubMed] [Google Scholar]