Abstract
We have developed a gene disruption system in the hyperthermophilic archaeon Thermococcus kodakaraensis using the antibiotic simvastatin and a fusion gene designed to overexpress the 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase gene (hmgTk) with the glutamate dehydrogenase promoter. With this system, we disrupted the T. kodakaraensis amylopullulanase gene (apuTk) or a gene cluster which includes apuTk and genes encoding components of a putative sugar transporter. Disruption plasmids were introduced into wild-type T. kodakaraensis KOD1 cells, and transformants exhibiting resistance to 4 μM simvastatin were isolated. The transformants exhibited growth in the presence of 20 μM simvastatin, and we observed a 30-fold increase in intracellular HMG-CoA reductase activity. The expected gene disruption via double-crossover recombination occurred at the target locus, but we also observed recombination events at the hmgTk locus when the endogenous hmgTk gene was used. This could be avoided by using the corresponding gene from Pyrococcus furiosus (hmgPf) or by linearizing the plasmid prior to transformation. While both gene disruption strains displayed normal growth on amino acids or pyruvate, cells without the sugar transporter genes could not grow on maltooligosaccharides or polysaccharides, indicating that the gene cluster encodes the only sugar transporter involved in the uptake of these compounds. The ΔapuTk strain could not grow on pullulan and displayed only low levels of growth on amylose, suggesting that ApuTk is a major polysaccharide-degrading enzyme in T. kodakaraensis.
Hyperthermophiles are organisms that exhibit optimal growth at temperatures above 80°C (57). The organisms have attracted much attention from an evolutionary viewpoint as they occupy the deepest lineages within the phylogenies of both Archaea and Bacteria based on rRNA sequences (56, 57). Hyperthermophiles are also focused upon as a source of (thermo)stable enzymes that have the potential for application in a broad range of technologies (3, 21). There are now 14 complete genome sequences and many more in progress, providing a wealth of primary structural data from which we can estimate the presence or absence of various metabolic and regulatory mechanisms. However, although biochemical and structural analyses of hyperthermophile proteins are proceeding at a rapid pace, genetic studies to examine gene function in vivo are still limited in number.
In contrast to the hyperthermophilic archaea, a wealth of gene disruption and shuttle vector systems has been developed for the mesophilic archaea. In the halophilic archaea, procedures allowing the uptake of bacteriophage (15), plasmid (14), and genomic DNA (17) have been established with Halobacterium halobium and Halobacterium (now Haloferax) volcanii. Stable shuttle vectors have been developed (29, 37, 45), and homologous recombination has been demonstrated (34, 43). Selection methods include changes in phenotype from auxotrophy to prototrophy (17) and resistance against a variety of antibiotics such as mevinolin (34, 37, 45, 63), novobiocin (29), thiostrepton, and anisomycin (43). Genetic systems have also been developed for Haloarcula strains (16) and other Halobacterium strains including Halobacterium sp. strain NRC-1 (8, 47, 62). In the methanogenic archaea, shuttle vectors and gene disruption systems have been developed in Methanococcus maripaludis based on puromycin and the puromycin N-acetyltransferase gene (pac) (25, 48, 61) or neomycin and aminoglycoside phosphotransferase genes (2). The puromycin-pac system has also been applied for genetic manipulation in Methanococcus voltae (6, 7, 60), as well as in various Methanosarcina species (19, 44). Genetic transformation has also been observed in the thermophilic methanogen Methanobacterium thermoautotrophicum (now Methanothermobacter thermautotrophicus) (64).
In the hyperthermophilic archaea, exchange and recombination of chromosomal markers, as well as homologous recombination of exogenous DNA, have been reported for Sulfolobus acidocaldarius (27, 28, 35, 50). A plasmid based on pNOB8 has been demonstrated to transform Sulfolobus solfataricus (22). An autonomously replicating vector harboring a mutant hygromycin phosphotransferase gene for selection has also been developed for this species (12, 13, 18). In the Euryarchaeota, a shuttle vector has been developed for use in Pyrococcus abyssi (41). In terms of gene disruption, however, only two systems have been reported so far: one for Thermococcus kodakaraensis (52, 53) from the Euryarchaeota and the other for S. solfataricus from the Crenarchaeota (65). Both systems rely on homologous recombination. The former system utilizes various host strains with amino acid/nucleotide auxotrophy and corresponding marker genes that complement the auxotrophy. The latter utilizes a lacS-deficient host strain and a modified but active lacS marker gene with selection based on lactose-dependent growth. The two systems have proved to be powerful tools in examining gene function in the respective strains (5, 30, 54, 55) and can be expected to provide further genetic evidence that will help in understanding the physiological roles of genes in these and closely related organisms.
In this study, we aimed to develop a gene disruption system in hyperthermophiles using antibiotics and a marker gene that would confer resistance to transformant cells. This would relieve the necessity to prepare auxotrophic host cells and also allow selection of transformants in a nutrient-rich medium. Thus, the methodology should not only provide a convenient alternative for gene disruption in T. kodakaraensis but also be helpful in establishing gene disruption systems in other hyperthermophilic archaea. We examined the possibilities of utilizing the mevinolin system established in the halophilic archaea. Mevinolin, along with its analog simvastatin, is a specific inhibitor of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase, an enzyme essential for archaeal membrane lipid biosynthesis (11, 36). HMG-CoA reductases have been extensively examined from a number of archaeal species (9, 10, 31). An overexpression construct of the HMG-CoA reductase gene can be expected to be applicable as a marker gene. Additionally, as the gene is originally present in the hyperthermophile, there is no need for concern about the thermostability of the marker gene product. As all archaeal strains are presumed to require the function of HMG-CoA reductase for lipid and membrane generation, the system described in this study has the potential for application in all hyperthermophilic archaea.
MATERIALS AND METHODS
Strains, media, and plasmids.
T. kodakaraensis KOD1 (4) and the mutant strains were cultivated under anaerobic conditions at 85°C in a nutrient-rich medium, ASW-YT (53), supplemented with various organic substrates or elemental sulfur when appropriate. ASW-YT medium was composed of 0.8× artificial seawater, 5.0 g liter−1 of yeast extract, and 5.0 g liter−1 of tryptone. Resazurin was added at a concentration of 0.8 mg liter−1, and prior to inoculation, Na2S was added to the medium until it became transparent. In the case of plate culture, instead of elemental sulfur and Na2S 9H2O, 2 ml of a polysulfide solution (10 g of Na2S 9H2O and 3 g of sulfur flowers in 15 ml of H2O) per liter and Gelrite (10 g liter−1) were added to solidify the medium. All components were purchased from Wako Pure Chemical Industries (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan). When simvastatin was added to the medium, simvastatin was dissolved in ethanol, and the amount of the solution added was adjusted so that the ethanol concentration in the medium was constant at 0.1% (vol/vol).
DNA manipulation and sequence analysis.
Escherichia coli strain DH5α and pUC18/pUC19 (Takara, Kyoto, Japan) were used for DNA manipulation and sequencing. E. coli strains were cultivated in Luria-Bertani medium at 37°C with ampicillin at a concentration of 100 μg ml−1. Restriction and modification enzymes were purchased from Toyobo (Osaka, Japan) and Takara. Plasmid DNA was isolated with a plasmid mini kit from QIAGEN (Hilden, Germany). KOD Plus (Toyobo) was used as a polymerase for PCR, and a GFX PCR DNA and gel band purification kit (GE Life Sciences, Little Chalfont, United Kingdom) was used to recover DNA fragments from agarose gels after electrophoresis. DNA sequencing was performed using a BigDye terminator cycle sequencing kit, version 3.1, and a model 3100 capillary DNA sequencer (Applied Biosystems, Foster City, CA).
Construction of the gene disruption vectors.
Two disruption vectors, pUDapu and pUDmal, were constructed for the targeted disruption of the T. kodakaraensis amylopullulanase gene apu (apuTk) and the sugar transporter gene cluster including apuTk, respectively, via double-crossover homologous recombination. Overexpression cassettes for the HMG-CoA reductase gene from T. kodakaraensis (hmgTk) were constructed by replacing the native promoter with a putative promoter region (−554 to −4) of the glutamate dehydrogenase gene (49). The region −3 to −1 was replaced by 5′-CAT-3′ in order to incorporate an NdeI site for fusion of the promoter to the coding region of hmgTk. Cassettes were designed so that one had SmaI sites at both ends, while another had an XbaI site upstream of the promoter and a BamHI site downstream of hmgTk. The two cassettes were inserted into pUC18 and sequenced. For construction of the apuTk disruption plasmid, a DNA fragment including apuTk along with its flanking regions (about 1,000 bp) was amplified from the genomic DNA of T. kodakaraensis KOD1 with the primer set APU-F1 and APU-TRANS-R1 (5′-AATTCAGAACGGCAAGCTCTACGTAACAGACGGCA-3′ and 5′-GCGTCGTAGATGTCCTCGGGCCTTATGCCGAAGAT-3′, respectively) and inserted into pUC18 at the HincII site. An inverse PCR was then carried out to amplify the flanking regions and pUC18, thereby removing the coding region of gene. The primers used were APU-R2 and APU-F2 (5′-CTTATCACCTCACTCTTTAAGGCCTCCAACAGTGA-3′ and 5′-AGAGGGTGGCGGAATCTGCGGCCCGGCGTTCCTCG-3′, respectively). The DNA fragment was ligated with the hmgTk overexpression cassette excised with SmaI and designated pUDapu. For disruption of the sugar transporter gene cluster, DNA fragments of the 5′ and 3′ flanking regions (about 1,000-bp) of the gene cluster were amplified with the primer pair TRANS-F1 and TRANS-R2 (5′-AGTTCTCAAATCGGACCTTCCGCCGATGGAAAAGT-3′ and 5′-TGTTTATCACCTAGTTATCTCGTTGCATTTGAGTA-3′, respectively) and the pair TRANS-F2 and APU-TRANS-R1 (TRANS-F2, 5′-TCCCCAGGATCCGGCGGTGGTGAAGAGGGTGGCGG-3′). The 5′ flanking region was inserted into pUC19 at the HincII site, followed by insertion of the overexpression cassette in the XbaI and BamHI sites. The 3′ flanking region was then inserted in the BamHI and SmaI sites, resulting in the plasmid pUDmal.
Transformation of T. kodakaraensis.
Transformation procedures were performed as described previously (52, 53), but the host strain used in this study was the wild-type T. kodakaraensis KOD1. After transformation, cells were cultivated in ASW-YT liquid medium supplemented with 0.2% (wt/vol) elemental sulfur (ASW-YT-S0) in the presence of 4 μM simvastatin at 85°C for 12 h. The cells were further grown in ASW-YT-S0 liquid medium with 8 μM simvastatin at 85°C and spread on ASW-YT (polysulfide) plate medium containing 4 μM simvastatin and incubated at 85°C. Genomic DNA was isolated from the transformants and analyzed by PCR and Southern blot analysis.
Southern blot analysis.
A digoxigenin-DNA labeling and detection kit (Roche Diagnostics, Basel, Switzerland) was used according to the manufacturer's instructions. The probes within the coding regions of hmgTk and apuTk were amplified, respectively, with the primer pair HMG-F and HMG-R (5′-TGAGAACATCGGGCACTACTCAATAGATCCCAACC-3′ and 5′-ACCAACGAGGTTCTTGCGGTAGTTCACCTCGGCTA-3′, respectively) and the pair APU-F and APU-R (5′-CTCAACGACAAGACCCTTGAAATCCTAGCGGAGAA-3′ and 5′-GGCTCATCTTATCTTTGTTTTCCATGAGGGCCTTT-3′, respectively). The probe within the coding region of the malE gene of T. kodakaraensis (malETk) was amplified with the primers MalE-F and MalE-R (5′-CACTTCCCCGACCGAGACCACTACTACCTCACCCA-3′ and 5′-CTGCTGGGTGTTGTAGTCGGCAGTCGGGGCCATGT-3′, respectively). The probe within the hmg gene from P. furiosus (hmgPf) was amplified with the primers PfHMG-F and PfHMG-R (5′-AAAGCACATTGGCCACTACTCAATTGATCCAAACG-3′ and 5′-ACCCACTAAGTTCTTTAGGTAGTTTACTTCAGCGA-3′, respectively), and the probe corresponding to the promoter region of the glutamate dehydrogenase gene was amplified with the primers GDHp-F and GDHp-R (5′-ATATCCCACCTCCGATTCCGTTGGTATTTAATCGG-3′ and 5′-TACCACCTCATTTCGGTAATCTGCGAGGTTAACTT-3′, respectively). Genomic DNA from the wild-type and gene disruption mutant strains was digested with PvuII.
Growth properties of T. kodakaraensis and mutant strains.
T. kodakaraensis KOD1 and the mutant strains were grown in ASW-YT-S0 medium at 85°C for 12 h and inoculated into 15 ml of ASW-YT-S0 or ASW-YT medium supplemented with 0.5% (wt/vol) sodium pyruvate, a 0.5% (wt/vol) concentration of a specific maltooligosaccharide (3, 4, 5, 6, or 7 glucose units), 0.5% (wt/vol) amylose (polysaccharide consisting of glucose connected solely by α-1,4-glycosidic bonds), or 0.5% (wt/vol) pullulan (polysaccharide consisting of maltotriose units connected by α-1,6-glycosidic bonds). Cell densities (optical density at 660 nm) were measured at appropriate intervals with a UV spectrometer mini photo 518R (Taitec, Koshigaya, Japan). In order to estimate resistance toward simvastatin, cells were cultivated in 15 ml of ASW-YT-S0 supplemented with 1, 5, 10, or 20 μM simvastatin.
Measurements of HMG-CoA reductase activity.
Activity measurements were performed at 60°C in a final volume of 1 ml containing cell extracts, 200 μM NADPH, and 0.5 mM HMG-CoA (Sigma, St. Louis, MO) in 50 mM potassium phosphate buffer (pH 7.0). The consumption of NADPH was monitored at 340 nm by a UV-visible light spectrophotometer (UV-1600PC; Shimadzu, Kyoto, Japan). Cell extracts were prepared as follows. T. kodakaraensis and the disruptants were cultivated in ASW-YT-S0 medium at 85°C for approximately 8 h. Cells were collected and sonicated on ice, and the supernatant after centrifugation (20,000 × g for 30 min at 4°C) was used as the cell extract. Protein concentrations were determined with a protein assay kit (Bio-Rad, Hercules, CA) using bovine serum albumin as a standard.
RESULTS
Effect of various concentrations of simvastatin on the growth of T. kodakaraensis.
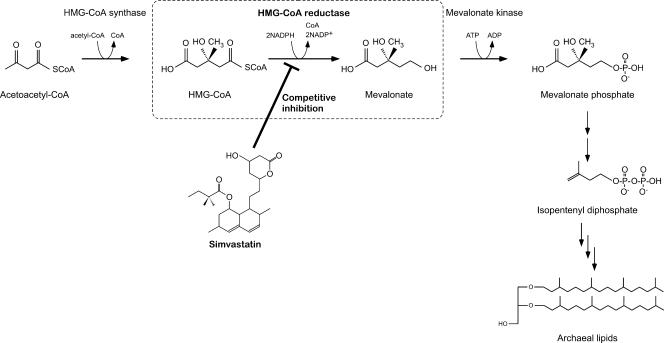
As isopentenyl diphosphate is the major precursor for archaeal lipid membranes, we supposed that inhibition of HMG-CoA reductase would have severe effects on the growth of T. kodakaraensis (Fig. 1). Our main concerns were whether the uptake and inhibitory effects of simvastatin were sufficient to allow use of the antibiotic at realistic concentrations and whether the compound was stable enough at temperatures of >80°C to inhibit growth for several days, which is necessary for the formation of colonies.
FIG. 1.
The mevalonate pathway for isoprenoid lipid biosynthesis in Archaea. The reaction catalyzed by HMG-CoA reductase is boxed with dotted lines. It should be noted that the two reactions converting mevalonate phosphate to isopentenyl diphosphate are distinct from the reactions in the classical mevalonate pathway (26).
We examined the growth of T. kodakaraensis KOD1 in the presence of various concentrations of simvastatin in the nutrient-rich medium ASW-YT-S0. Simvastatin was dissolved in ethanol, and the amount of ethanol added to the medium was constant at 0.1% (vol/vol). In ASW-YT-S0 medium, T. kodakaraensis KOD1 cells reach the stationary phase within 24 h. No effect on growth was observed with the addition of ethanol alone. In the presence of 1 or 2 μM simvastatin, growth was observed only after 24 h, while 48 h was necessary for growth with 3 μM simvastatin. At concentrations of 4 or 5 μM simvastatin, growth was not observed for at least 5 days, indicating that these concentrations would be suitable for selecting transformants with resistance against simvastatin. We also confirmed that these concentrations were sufficient to prevent colony formation of T. kodakaraensis KOD1 on nutrient-rich plate medium.
A cassette for the overexpression of the HMG-CoA reductase gene.
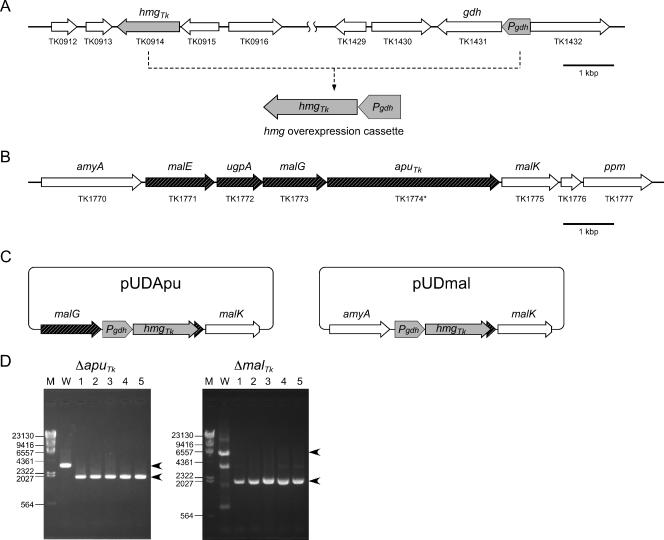
As simvastatin is a competitive inhibitor of HMG-CoA reductase, we expected that overexpression of its gene from T. kodakaraensis (hmgTk) would reduce the inhibitory effects of simvastatin on cell growth. Previous studies have indicated that the enzyme glutamate dehydrogenase is abundant in T. kodakaraensis cells grown in various media (49), suggesting that the gene (gdhTk) is under the control of a strong promoter. We therefore utilized a 551-bp intergenic region between the coding regions of gdhTk (TK1431) and the adjacent gene TK1432 and fused the region upstream of hmgTk (see Materials and Methods). This overexpression cassette (Pgdh-hmg) was used as the marker gene for construction of disruption plasmids (Fig. 2A).
FIG. 2.
Disruption of the apuTk and malTk loci of T. kodakaraensis. (A) Design of the hmgTk overexpression cassette using the 5′ upstream flanking region of gdhTk. (B) Gene organization of the putative maltooligosaccharide transporter of T. kodakaraensis. TK1774* represents the correct sequence of the apuTk gene (see text). Black arrows indicate the gene(s) disrupted in this study. (C) The two plasmids constructed for the disruption of the apuTk and malTk loci via double-crossover recombination. (D) PCR analyses of the apuTk and malTk loci confirming gene disruption. Primers were designed in the 5′ and 3′ flanking regions of the gene(s) to be disrupted. DNA size markers were run in lane M, and their sizes (bp) are indicated to the left of the gels. The results of PCR with wild-type T. kodakaraensis KOD1 and five individual transformants are indicated in lane W and lanes 1 to 5, respectively. The arrowheads to the right of the gels indicate the amplified fragments expected before and after recombination. The decreases in lengths of the amplified fragments reflect the differences in length between apuTk (∼3,500 bp) and Pgdh-hmg (∼2,000 bp) and between malTk (∼7,000 bp) and Pgdh-hmg. Nonspecific amplifications of DNA fragments observed for the wild-type malTk locus were due to the prolonged reaction time necessary to amplify the entire locus.
Design and construction of the gene disruption plasmids.
The genes disrupted in this study were a putative amylopullulanase gene (apuTk, or TK1774) and a gene cluster including apuTk and three additional genes encoding the components of a sugar transporter (TK1771 to TK1773) of T. kodakaraensis (Fig. 2B) (24). In the latter stages of this study, we discovered an error in the original genome sequence of TK1774 (an excess A at position 1,581,978 of the genome). The correct sequence leads to a protein with a change and elongation in sequence in the C-terminal region from residue Asn1070 (see Discussion). In this report we will refer to the corrected apuTk gene as TK1774* and, for simplicity, to the four-gene cluster (TK1771 to TK1774*) as malTk.
Amylopullulanases, or type II pullulanases, exhibit both α-amylase and pullulanase activities and can therefore cleave both α-1,4- and α-1,6-glucosidic bonds (20). There are a number of other homologs on the genome (24), some with putative signal sequences for secretion, expected to harbor the ability to degrade α-linked polysaccharides. In particular, the TK1884 protein has been experimentally confirmed to exhibit α-amylase activity (59). On the other hand, in contrast to the two sugar transporters present in Pyrococcus furiosus (Mal-I, PF1739 to PF1744, and Mal-II, PF1933 to PF1938) (33), only one putative gene cluster is found on the T. kodakaraensis genome (TK1771 to TK1775) (24). Based on primary structure similarity, the transporter from T. kodakaraensis corresponds to Mal-II, suggesting that it is specific to maltooligosaccharides with three or more glucose units. By disrupting apuTk, we expected to gain insight into the actual degree of influence apuTk has, among the multiple amylase homologs of T. kodakaraensis, on the degradation of various extracellular polysaccharides. Growth characteristics of the malTk disruptant were expected to clarify the presence or absence of other sugar transporters as well as to provide information on the substrate specificity of the MalTk transporter in vivo.
Similar to the design of gene disruption plasmids in a previously described system using pyrF or trpE as selectable markers (52), Pgdh-hmg was inserted between the 5′ and 3′ flanking regions (∼1,000 bp) of the target gene(s) (Fig. 2C). The plasmids pUDapu and pUDmal were used to transform wild-type T. kodakaraensis KOD1, and transformants were selected based on their resistance toward simvastatin.
Isolation of the gene disruption strains ΔapuTk and ΔmalTk.
After transformation, cells were grown in ASW-YT-S0 liquid medium in the presence of 4 μM simvastatin. Growth was observed with cells transformed with pUDapu and pUDmal but not for cells treated without plasmid. Cells were further inoculated in the same liquid medium with 8 μM simvastatin and then spread on plate medium with 4 μM simvastatin. Five colonies were selected for each gene disruption and grown in ASW-YT-S0 medium. We examined the apuTk and malTk loci by PCR (Fig. 2D). As expected, we observed shorter amplified fragments from the transformants than from the wild-type strain, corresponding to the decrease in length brought about by the replacement of apuTk and malTk by Pgdh-hmg.
Extent of simvastatin resistance of the transformants.
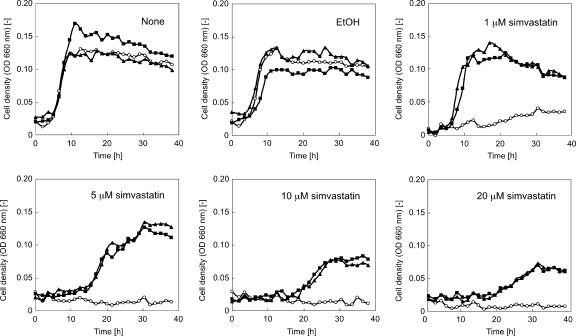
As the transformants harbored Pgdh-hmg on their genomes, we examined their resistance against various concentrations of simvastatin (Fig. 3). One ΔapuTk and one ΔmalTk transformant were grown in the presence of 1, 5, 10, and 20 μM simvastatin, and their growth characteristics were compared with those observed in medium without simvastatin. Although the wild-type strain could not grow at all with 5 μM simvastatin, specific growth rates and cell yields of the transformants were still comparable to those in the absence of simvastatin. The degree of inhibition became prominent at higher concentrations, but we found that concentrations over 20 μM were necessary to completely inhibit growth of the transformants. We further examined the levels of HMG-CoA reductase activity in the wild-type and transformant cells. Specific activity in the cell extracts of wild-type cells was approximately 25 nmol min−1 mg−1. In contrast, the level observed in the extracts of the ΔapuTk strain was 760 nmol min−1 mg−1, indicating an increase in activity of over 30-fold. The resistance against simvastatin and the increase in HMG-CoA reductase activity in the transformants are consistent with the presumption that simvastatin inhibits growth of T. kodakaraensis KOD1 by specifically inhibiting the activity of HMG-CoA reductase.
FIG. 3.
Growth of wild-type T. kodakaraensis KOD1 and ΔapuTk and ΔmalTk mutant strains in the presence of various concentrations of simvastatin. Open circle, wild-type strain; filled square, ΔapuTk strain; filled triangle, ΔmalTk strain; OD, optical density.
Phenotype analyses of the gene disruption strains.
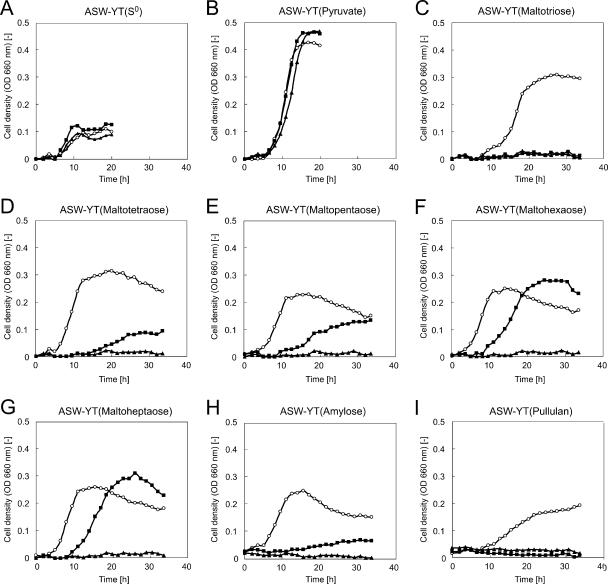
We examined the growth characteristics of the ΔapuTk and ΔmalTk strains in various media and compared them with those of the wild-type strain (Fig. 4). No change in phenotype was observed when the three strains were grown on amino acids (ASW-YT-S0) or amino acids and pyruvate (ASW-YT-pyruvate) as carbon sources. However, disruption of apuTk and malTk brought about dramatic changes in phenotype when the strains were grown on various sugars. Disruption of the MalTk transporter abolished growth on all sugars examined. Although the ΔapuTk strain displayed growth on a number of maltooligosaccharides, the strain could not grow on pullulan (Fig. 4I). Interestingly, while several α-amylase homologs (including TK1884) are present on the genome, we found that disruption of apuTk led to a significant decrease in growth rates when strains were grown on amylose, a maltopolysaccharide consisting of only α-1,4-linkages (Fig. 4H). Another intriguing finding was that the disruption of apuTk had a greater detrimental effect on growth with shorter maltooligosaccharides, which was rather surprising as amylopullulanases are presumed to function in the breakdown of poly- or oligosaccharides. In contrast to the wild-type strain, no growth was observed for ΔapuTk in the medium supplemented with maltotriose (Fig. 4C).
FIG. 4.
Growth of wild-type T. kodakaraensis KOD1 and ΔapuTk and ΔmalTk mutant strains on various carbon sources. The carbon sources examined are indicated above each panel. Glucose and maltose were not examined, as the wild-type strain cannot utilize these sugars. Open circle, wild-type strain; filled square, ΔapuTk strain; filled triangle, ΔmalTk strain; OD, optical density.
Recombination at the hmgTk locus.
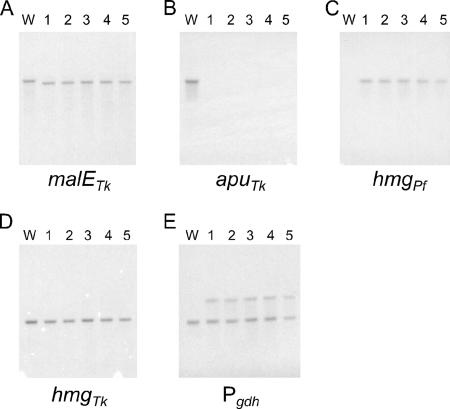
As the plasmid used in this study harbors the endogenous hmgTk and Pgdh, there will always be a possibility of these regions recombining with the corresponding native loci present on the genome. We have actually applied this property in developing a single-crossover insertion/pop-out recombination system using the pyrF gene as a selective marker (52). We therefore examined whether recombination events had occurred at the hmgTk and/or Pgdh glutamate dehydrogenase locus. We observed that in some strains the native hmgTk locus had been disturbed (data not shown). As the locus of the target gene is stable in a disrupted form via double-crossover recombination, the recombination at the hmgTk locus does not directly pose a problem. Nevertheless, we examined possibilities to prevent or decrease the frequencies of unintended recombination. Linearizing the plasmids prior to introducing them into the cells would prevent single-crossover recombination. Utilizing an hmg gene from a heterologous host would also decrease the possibilities of single-crossover recombination as well as further recombination due to the presence of two identical regions on the genome. We found that both methods could be used for gene disruption. In examining the latter possibility, we used the hmg gene from the closely related P. furiosus and constructed plasmids to disrupt the apuTk gene. In five randomly chosen transformants with resistance against simvastatin, we clearly observed that double-crossover recombination had occurred at the apuTk locus (Fig. 5A and B) along with the appearance of a single copy of the hmgPf gene (Fig. 5C) and an additional copy of the gdh promoter (Fig. 5E), whereas neither the native hmgTk locus nor the gdh promoter regions were disturbed (Fig. 5D and E). We further confirmed that the strains obtained using hmgPf as a marker gene (i) were able to grow in the presence of 20 μM simvastatin, (ii) exhibited significant HMG-CoA reductase activities in the cell extracts (720 nmol min−1 mg−1), and (iii) displayed the same growth characteristics toward various carbon sources as those shown in Fig. 4 (data not shown).
FIG. 5.
Southern blot analyses on ΔapuTk strains obtained with the hmgPf gene as a selectable marker. Genomic DNA from five selected ΔapuTk strains and from wild-type T. kodakaraensis KOD1 (W) was subjected to Southern blot analyses using probes within the regions indicated below each membrane.
DISCUSSION
In this study, we have developed in T. kodakaraensis a gene disruption system based on resistance against antibiotics using simvastatin and an overexpression cassette of hmgTk. The system has many advantages for initiating gene disruption studies in hyperthermophilic archaea. First, one does not need to construct a host strain with a particular defect or auxotrophy toward an amino acid. There is also no need for selection to be carried out in minimal medium. Positive selection of mutant strains is possible in nutrient-rich medium. If the genome sequence is available, the possibilities to disrupt genes with this system can be examined immediately. It should be noted that we have not examined the stability of simvastatin under acidic conditions, which should be examined prior to application of the method on strains such as Sulfolobus. When the genome sequence is not available, it may be possible to use the genes of a closely related strain whose genome has been sequenced. As demonstrated in this study, the heterologous hmgPf gene was applicable for gene disruption in T. kodakaraensis. In order to avoid initial single-crossover recombination and possible recombination events afterwards due to the presence of two identical regions on the same chromosome, a heterologous marker may be more advantageous than an endogenous marker. We hope this methodology will promote gene disruption studies in a broader range of hyperthermophilic archaea.
The phenotypes of the ΔapuTk and ΔmalTk strains in various media not only provide valuable information on the physiological roles of the disrupted genes themselves but also allow us to estimate the contribution of other genes on the genome. The growth characteristics of the ΔmalTk strain indicated that it is the only transporter in T. kodakaraensis involved in the uptake of the poly- and oligosaccharides examined in this study. As the ΔapuTk strain could not grow at all on pullulan, there is a possibility that ApuTk is the only relevant enzyme responsible for the initial hydrolysis of pullulan, meaning that TK0977, annotated as a type II pullulan hydrolase, is not directly involved in the degradation of extracellular pullulan. Another possibility is that ApuTk and the TK0977 product display activity specified toward pullulan substrates of different lengths and are unable to complement each other. A third possibility would be that the lack of growth is due to the fact that the ΔapuTk strain is defective in the uptake of maltotriose (Fig. 4C), a major product of pullulan hydrolysis. From the results shown in Fig. 4H, we also found that ApuTk plays a much greater role than expected in the cleavage of α-1,4-glycosidic linkages, suggesting that the experimentally verified α-amylase TK1884 protein (59) may not be the major amylose-degrading enzyme in T. kodakaraensis. This agrees well with the results of a transcriptome analysis of P. furiosus grown on starch, which revealed that PF1935*, the homolog of TK1774*, is the protein most up-regulated in the presence of starch (39).
An intriguing change in phenotype was observed in the ΔapuTk strain grown on maltooligosaccharides. As the amylopullulanase was presumed to function in the degradation of poly- or oligosaccharides, we expected that gene disruption would have little effect on growth with maltooligosaccharides. Even if phenotypic changes were to be observed, the effects brought about by disrupting apuTk were expected to be greater with longer substrates. However, the results obtained with ΔapuTk were just the opposite. This may be the result of polar effects brought about by insertion of the hmgTk overexpression cassette. With our disruption strategy, the downstream genes, in particular, malKTk (TK1775), would be under the control of the gdh promoter and would thereby disturb the stoichiometric expression of the transporter subunits. Another possibility is that the up-regulation of transporter expression was disturbed by the absence of apuTk. Further, ApuTk itself may also be a component of the sugar transporter complex, resulting in a decrease in stability or efficiency of the complex when ApuTk is absent. This is consistent with the fact that the apuTk gene is clustered within the subunit genes of the transporter itself. The possibility that amylopullulanase resides on the cell surface has been proposed in closely related hyperthermophilic archaea (1, 23, 32). The enzyme from Thermococcus hydrothermalis has been reported to harbor a large C-terminal extension with three domains in addition to the central catalytic domain (23). One of the domains consists of two sequence repeats, each containing motifs with similarity to the S-layer homology signature (42), which is considered responsible for the anchoring of proteins to the cell surface in several bacteria (40, 42, 46, 51). The second region is extremely rich in threonine residues and is followed by the third, putative transmembrane domain. This architecture resembles the C-terminal regions of S-layer proteins of the haloarchaea, in which the Thr-rich regions are targets for O-linked glycosylation and the transmembrane domain serves as either a transmembrane anchor or hydrophobic cell wall anchor (38, 58). Moreover, as pointed out by Albers et al. (1), the two regions are also found in a number of proteins of the Thermococcales annotated as periplasmic components of ABC-type dipeptide transport systems, further supporting the involvement of these domains in cell surface attachment (Fig. 6). As amylopullulanases from P. furiosus, P. abyssi, and T. kodakaraensis also harbor these domains, it can be presumed that the amylopullulanases from the Thermococcales are attached to the cell surface. Further biochemical examination will be necessary to clarify whether these amylopullulanases have any additional function besides their roles in poly- and oligosaccharide hydrolysis.
FIG. 6.
Carboxy-terminal regions of various amylopullulanase proteins from the Thermococcales, along with the corresponding regions of periplasmic components of two putative ABC-type dipeptide transport systems of T. kodakaraensis. All of these proteins harbor a threonine (or serine)-rich region, followed by a putative transmembrane domain and a stretch of basic residues (indicated by circles) at the extreme C terminus. The subscripts of the amylopullulanase proteins identify the source organism as follows (accession number): ApuTl, Thermococcus litoralis (BAC10983); ApuTh, Thermococcus hydrothermalis (AAD28552); ApuPf, P. furiosus (ABA33719); ApuPa, P. abyssi (CAB49104). Accession numbers for TK1760 and TK1804 are BAD85949 and BAD85993, respectively.
Acknowledgments
This study was supported by a Grant-in-Aid for Scientific Research to T.I. (no. 14103011), a Grant-in-Aid for Scientific Research on Priority Areas, “Applied Genomics,” to H.A. (no. 18018026), and a Grant-in-Aid for Scientific Research to R.M. (no. 18608002), from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 26 January 2007.
REFERENCES
- 1.Albers, S.-V., S. M. Koning, W. N. Konings, and A. J. M. Driessen. 2004. Insights into ABC transport in archaea. J. Bioenerg. Biomembr. 36:5-15. [DOI] [PubMed] [Google Scholar]
- 2.Argyle, J. L., D. L. Tumbula, and J. A. Leigh. 1996. Neomycin resistance as a selectable marker in Methanococcus maripaludis. Appl. Environ. Microbiol. 62:4233-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atomi, H. 2005. Recent progress towards the application of hyperthermophiles and their enzymes. Curr. Opin. Chem. Biol. 9:166-173. [DOI] [PubMed] [Google Scholar]
- 4.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atomi, H., R. Matsumi, and T. Imanaka. 2004. Reverse gyrase is not a prerequisite for hyperthermophilic life. J. Bacteriol. 186:4829-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardy, S. L., and K. F. Jarrell. 2003. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol. Microbiol. 50:1339-1347. [DOI] [PubMed] [Google Scholar]
- 7.Berghöfer, Y., and A. Klein. 1995. Insertional mutations in the hydrogenase vhc and frc operons encoding selenium-free hydrogenases in Methanococcus voltae. Appl. Environ. Microbiol. 61:1770-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berquist, B. R., and S. DasSarma. 2003. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J. Bacteriol. 185:5959-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bischoff, K. M., and V. W. Rodwell. 1996. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase from Haloferax volcanii: purification, characterization, and expression in Escherichia coli. J. Bacteriol. 178:19-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bochar, D. A., J. R. Brown, W. F. Doolittle, H.-P. Klenk, W. Lam, M. E. Schenk, C. V. Stauffacher, and V. W. Rodwell. 1997. 3-Hydroxy-3-methylglutaryl coenzyme A reductase of Sulfolobus solfataricus: DNA sequence, phylogeny, expression in Escherichia coli of the hmgA gene, and purification and kinetic characterization of the gene product. J. Bacteriol. 179:3632-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabrera, J. A., J. Bolds, P. E. Shields, C. M. Havel, and J. A. Watson. 1986. Isoprenoid synthesis in Halobacterium halobium. Modulation of 3-hydroxy-3-methylglutaryl coenzyme A concentration in response to mevalonate availability. J. Biol. Chem. 261:3578-3583. [PubMed] [Google Scholar]
- 12.Cannio, R., P. Contursi, M. Rossi, and S. Bartolucci. 1998. An autonomously replicating transforming vector for Sulfolobus solfataricus. J. Bacteriol. 180:3237-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannio, R., P. Contursi, M. Rossi, and S. Bartolucci. 2001. Thermoadaptation of a mesophilic hygromycin B phosphotransferase by directed evolution in hyperthermophilic Archaea: selection of a stable genetic marker for DNA transfer into Sulfolobus solfataricus. Extremophiles 5:153-159. [DOI] [PubMed] [Google Scholar]
- 14.Charlebois, R. L., W. L. Lam, S. W. Cline, and W. F. Doolittle. 1987. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc. Natl. Acad. Sci. USA 84:8530-8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cline, S. W., and W. F. Doolittle. 1987. Efficient transfection of the archaebacterium Halobacterium halobium. J. Bacteriol. 169:1341-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cline, S. W., and W. F. Doolittle. 1992. Transformation of members of the genus Haloarcula with shuttle vectors based on Halobacterium halobium and Haloferax volcanii plasmid replicons. J. Bacteriol. 174:1076-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cline, S. W., L. C. Schalkwyk, and W. F. Doolittle. 1989. Transformation of the archaebacterium Halobacterium volcanii with genomic DNA. J. Bacteriol. 171:4987-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Contursi, P., F. M. Pisani, A. Grigoriev, R. Cannio, S. Bartolucci, and M. Rossi. 2004. Identification and autonomous replication capability of a chromosomal replication origin from the archaeon Sulfolobus solfataricus. Extremophiles 8:385-391. [DOI] [PubMed] [Google Scholar]
- 19.de Macario, E. C., M. Guerrini, C. B. Dugan, and A. J. L. Macario. 1996. Integration of foreign DNA in an intergenic region of the archaeon Methanosarcina mazei without effect on transcription of adjacent genes. J. Mol. Biol. 262:12-20. [DOI] [PubMed] [Google Scholar]
- 20.Doman-Pytka, M., and J. Bardowski. 2004. Pullulan degrading enzymes of bacterial origin. Crit. Rev. Microbiol. 30:107-121. [DOI] [PubMed] [Google Scholar]
- 21.Egorova, K., and G. Antranikian. 2005. Industrial relevance of thermophilic Archaea. Curr. Opin. Microbiol. 8:649-655. [DOI] [PubMed] [Google Scholar]
- 22.Elferink, M. G. L., C. Schleper, and W. Zillig. 1996. Transformation of the extremely thermoacidophilic archaeon Sulfolobus solfataricus via a self-spreading vector. FEMS Microbiol. Lett. 137:31-35. [DOI] [PubMed] [Google Scholar]
- 23.Erra-Pujada, M., P. Debeire, F. Duchiron, and M. J. O'Donohue. 1999. The type II pullulanase of Thermococcus hydrothermalis: molecular characterization of the gene and expression of the catalytic domain. J. Bacteriol. 181:3284-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukui, T., H. Atomi, T. Kanai, R. Matsumi, S. Fujiwara, and T. Imanaka. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner, W. L., and W. B. Whitman. 1999. Expression vectors for Methanococcus maripaludis: overexpression of acetohydroxyacid synthase and β-galactosidase. Genetics 152:1439-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grochowski, L. L., H. Xu, and R. H. White. 2006. Methanocaldococcus jannaschii uses a modified mevalonate pathway for biosynthesis of isopentenyl diphosphate. J. Bacteriol. 188:3192-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grogan, D. W. 1996. Exchange of genetic markers at extremely high temperatures in the archaeon Sulfolobus acidocaldarius. J. Bacteriol. 178:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen, J. E., A. C. Dill, and D. W. Grogan. 2005. Conjugational genetic exchange in the hyperthermophilic archaeon Sulfolobus acidocaldarius: intragenic recombination with minimal dependence on marker separation. J. Bacteriol. 187:805-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmes, M. L., and M. L. Dyall-Smith. 1990. A plasmid vector with a selectable marker for halophilic archaebacteria. J. Bacteriol. 172:756-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imanaka, H., A. Yamatsu, T. Fukui, H. Atomi, and T. Imanaka. 2006. Phosphoenolpyruvate synthase plays an essential role for glycolysis in the modified Embden-Meyerhof pathway in Thermococcus kodakarensis. Mol. Microbiol. 61:898-909. [DOI] [PubMed] [Google Scholar]
- 31.Kim, D.-Y., C. V. Stauffacher, and V. W. Rodwell. 2000. Dual coenzyme specificity of Archaeoglobus fulgidus HMG-CoA reductase. Protein Sci. 9:1226-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koning, S. M., S.-V. Albers, W. N. Konings, and A. J. M. Driessen. 2002. Sugar transport in (hyper)thermophilic archaea. Res. Microbiol. 153:61-67. [DOI] [PubMed] [Google Scholar]
- 33.Koning, S. M., W. N. Konings, and A. J. M. Driessen. 2002. Biochemical evidence for the presence of two α-glucoside ABC-transport systems in the hyperthermophilic archaeon Pyrococcus furiosus. Archaea 1:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krebs, M. P., R. Mollaaghababa, and H. G. Khorana. 1993. Gene replacement in Halobacterium halobium and expression of bacteriorhodopsin mutants. Proc. Natl. Acad. Sci. USA 90:1987-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurosawa, N., and D. W. Grogan. 2005. Homologous recombination of exogenous DNA with the Sulfolobus acidocaldarius genome: properties and uses. FEMS Microbiol. Lett. 253:141-149. [DOI] [PubMed] [Google Scholar]
- 36.Lam, W. L., and W. F. Doolittle. 1992. Mevinolin-resistant mutations identify a promoter and the gene for a eukaryote-like 3-hydroxy-3-methylglutaryl-coenzyme A reductase in the archaebacterium Haloferax volcanii. J. Biol. Chem. 267:5829-5834. [PubMed] [Google Scholar]
- 37.Lam, W. L., and W. F. Doolittle. 1989. Shuttle vectors for the archaebacterium Halobacterium volcanii. Proc. Natl. Acad. Sci. USA 86:5478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lechner, J., and M. Sumper. 1987. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J. Biol. Chem. 262:9724-9729. [PubMed] [Google Scholar]
- 39.Lee, H.-S., K. R. Shockley, G. J. Schut, S. B. Conners, C. I. Montero, M. R. Johnson, C.-J. Chou, S. L. Bridger, N. Wigner, S. D. Brehm, F. E. Jenney, Jr., D. A. Comfort, R. M. Kelly, and M. W. W. Adams. 2006. Transcriptional and biochemical analysis of starch metabolism in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 188:2115-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lemaire, M., H. Ohayon, P. Gounon, T. Fujino, and P. Béguin. 1995. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J. Bacteriol. 177:2451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas, S., L. Toffin, Y. Zivanovic, D. Charlier, H. Moussard, P. Forterre, D. Prieur, and G. Erauso. 2002. Construction of a shuttle vector for, and spheroplast transformation of, the hyperthermophilic archaeon Pyrococcus abyssi. Appl. Environ. Microbiol. 68:5528-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lupas, A., H. Engelhardt, J. Peters, U. Santarius, S. Volker, and W. Baumeister. 1994. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J. Bacteriol. 176:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mankin, A. S., I. M. Zyrianova, V. K. Kagramanova, and R. A. Garrett. 1992. Introducing mutations into the single-copy chromosomal 23S rRNA gene of the archaeon Halobacterium halobium by using an rRNA operon-based transformation system. Proc. Natl. Acad. Sci. USA 89:6535-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metcalf, W. W., J. K. Zhang, E. Apolinario, K. R. Sowers, and R. S. Wolfe. 1997. A genetic system for archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 94:2626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nieuwlandt, D. T., and C. J. Daniels. 1990. An expression vector for the archaebacterium Haloferax volcanii. J. Bacteriol. 172:7104-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olabarría, G., J. L. Carrascosa, M. A. de Pedro, and J. Berenguer. 1996. A conserved motif in S-layer proteins is involved in peptidoglycan binding in Thermus thermophilus. J. Bacteriol. 178:4765-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peck, R. F., S. DasSarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 48.Porat, I., W. Kim, E. L. Hendrickson, Q. Xia, Y. Zhang, T. Wang, F. Taub, B. C. Moore, I. J. Anderson, M. Hackett, J. A. Leigh, and W. B. Whitman. 2006. Disruption of the operon encoding Ehb hydrogenase limits anabolic CO2 assimilation in the archaeon Methanococcus maripaludis. J. Bacteriol. 188:1373-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahman, R. N. Z. A., S. Fujiwara, M. Takagi, and T. Imanaka. 1998. Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and comparison of the enzymatic characteristics of native and recombinant GDHs. Mol. Gen. Genet. 257:338-347. [DOI] [PubMed] [Google Scholar]
- 50.Reilly, M. S., and D. W. Grogan. 2001. Characterization of intragenic recombination in a hyperthermophilic archaeon via conjugational DNA exchange. J. Bacteriol. 183:2943-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sára, M., E. M. Egelseer, C. Dekitsch, and U. B. Sleytr. 1998. Identification of two binding domains, one for peptidoglycan and another for a secondary cell wall polymer, on the N-terminal part of the S-layer protein SbsB from Bacillus stearothermophilus PV72/p2. J. Bacteriol. 180:6780-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71:3889-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sato, T., H. Imanaka, N. Rashid, T. Fukui, H. Atomi, and T. Imanaka. 2004. Genetic evidence identifying the true gluconeogenic fructose-1,6-bisphosphatase in Thermococcus kodakaraensis and other hyperthermophiles. J. Bacteriol. 186:5799-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schelert, J., V. Dixit, V. Hoang, J. Simbahan, M. Drozda, and P. Blum. 2004. Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J. Bacteriol. 186:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stetter, K. O. 1999. Extremophiles and their adaptation to hot environments. FEBS Lett. 452:22-25. [DOI] [PubMed] [Google Scholar]
- 57.Stetter, K. O. 1996. Hyperthermophilic procaryotes. FEMS Microbiol. Rev. 18:149-158. [Google Scholar]
- 58.Sumper, M., E. Berg, R. Mengele, and I. Strobel. 1990. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J. Bacteriol. 172:7111-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tachibana, Y., M. M. Leclere, S. Fujiwara, M. Takagi, and T. Imanaka. 1996. Cloning and expression of the α-amylase gene from the hyperthermophilic archaeon Pyrococcus sp. KOD1, and characterization of the enzyme. J. Ferment. Bioeng. 82:224-232. [Google Scholar]
- 60.Thomas, N. A., S. Mueller, A. Klein, and K. F. Jarrell. 2002. Mutants in flaI and flaJ of the archaeon Methanococcus voltae are deficient in flagellum assembly. Mol. Microbiol. 46:879-887. [DOI] [PubMed] [Google Scholar]
- 61.Tumbula, D. L., T. L. Bowen, and W. B. Whitman. 1997. Characterization of pURB500 from the archaeon Methanococcus maripaludis and construction of a shuttle vector. J. Bacteriol. 179:2976-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, G., S. P. Kennedy, S. Fasiludeen, C. Rensing, and S. DasSarma. 2004. Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. J. Bacteriol. 186:3187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wendoloski, D., C. Ferrer, and M. L. Dyall-Smith. 2001. A new simvastatin (mevinolin)-resistance marker from Haloarcula hispanica and a new Haloferax volcanii strain cured of plasmid pHV2. Microbiology 147:959-964. [DOI] [PubMed] [Google Scholar]
- 64.Worrell, V. E., D. P. Nagle, Jr., D. McCarthy, and A. Eisenbraun. 1988. Genetic transformation system in the archaebacterium Methanobacterium thermoautotrophicum Marburg. J. Bacteriol. 170:653-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Worthington, P., V. Hoang, F. Perez-Pomares, and P. Blum. 2003. Targeted disruption of the α-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 185:482-488. [DOI] [PMC free article] [PubMed] [Google Scholar]