Abstract
A vast bibliography on nutrient effects on high-density cultures exists, while it has been overlooked that low densities of starving cells are often the rule in natural environments. By means of a novel sensitive β-galactosidase assay, we examined Escherichia coli transitions to minimal media when the cell concentration was 100 to 10,000 cells per ml. As in high-density cultures, the enzyme activity depended on amino acid availability and was subject to catabolite repression and stringent control. In all conditions tested, despite the presence of other nutrient sources, the relationship between β-galactosidase activity and the l-amino acid pool was hyperbolic. The affinity constant when the amino acid pool was the only nutrient source averaged 14 μM after 90 min and increased up to 222 μM after 4.5 h. While investigating the transition from lag phase to exponential phase, we observed that the cells did not enter into starvation mode in the presence of amino acids, even when the nutrient amount was insufficient to support full survival. Based on these premises, the switch from starvation to hunger was investigated in relation to the amino acid pools. A critical range of concentrations at which E. coli linearly synthesized β-galactosidase despite, at the same time, suffering a large decrease in cell viability was then recognized. Since both β-galactosidase production and the dilution rate were reduced by more than half in the absence of leucine, we examined the contribution of leucine to cell recovery capabilities.
The natural state in which microorganisms conduct their lives oscillates between feast and famine (20). The growth rate in either soil or water is often not measurable, with duplication times in the order of days or weeks (16), because most bacteria are under starving conditions and only bacteria inhabiting “hot spots,” such as the gut, rhizosphere soil, or sites around vegetable debris, have wide access to nutrients (7). Simulating environmental fluctuations of nutrient conditions via controlled nutritional shifts for Escherichia coli showed the activation/deactivation of many different molecular systems coordinating the regulation of the cell metabolism by carbon, nitrogen, and amino acids, like the cyclic AMP (cAMP)-cAMP receptor protein modulon (34), the nitrogen-regulated response (31), and the stringent modulon (4, 9, 24). However, upon the complete exhaustion of an essential nutrient, cells were shown to enter into stationary phase through a general stress response controlled by the rpoS-encoded sigma-S subunit of the RNA polymerase (17). When nutrients were limited yet not completely absent, the cells exhibited the hunger response, which partly overlapped with the stress response (13). It has been thoroughly shown how nutrient downshifts or upshifts also influence the induction of β-galactosidase due to catabolite repression (22), stringent control (30), and stationary-phase control (14). On the other hand, the lack of suitable analytical approaches and the need to uniform experimental conditions directed research mainly towards high bacterial cell densities (around 108 to 109 cells per ml), in continuous or batch cultures (21), despite the fact that numbers of cells of different enteric species in fluid bodies are low (commonly <100 cells per ml) and reach 105 cells per ml only in proximity to a source of contamination.
The aim of this research was to analyze the effect of nutrient downshifts on the bacterial metabolic response at cell densities of 100 to 10,000 cells per ml so as to find the minimum nutrient conditions at which cells resumed growth and/or expressed an adaptive behavior by synthesizing catabolic enzymes. For a model, we used E. coli that was grown in rich medium and diluted in minimal medium and β-galactosidase as an indicator of bacterial metabolism. The amino acid pool proved necessary and sufficient in supporting the cell recovery from the nutritional gap and allowing a faster β-galactosidase expression, in substantial agreement with results at high bacterial densities. We examined in depth the relationship between amino acid concentration and β-galactosidase activity and found it hyperbolic, regardless of other nutrient additions. Then, we compared β-galactosidase and growth kinetics during the cell transition to the exponential phase and observed that the presence of the amino acid pool was sufficient to induce E. coli to synthesize β-galactosidase instead of entering a safer survival-starvation mode, even at the cost of large numbers of individual losses. Finally, we investigated the consequences deriving from single amino acid deficiencies and, particularly, the significant reductions in β-galactosidase synthesis and cell growth caused, in all strains tested, by the removal of leucine from the amino acid pool.
MATERIALS AND METHODS
Bacterial strains and reagents.
Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 13883 were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Citrobacter freundii ATCC 8090 and Escherichia coli K-12 ATCC 23716 were purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). All reagents, unless otherwise specified, were purchased from Sigma-Aldrich, St. Louis, MO.
Cell preparation and counts.
Fresh cultures were prepared by overnight growth in Luria-Bertani medium (LB) (Difco, Sparks, MD) at 37°C. Tests were performed on diluted cell samples after centrifugation (1,800 × g, 4 min) and pellet resuspension in 0.85% NaCl physiological solution. Due to the limited number of cells used in the experiments (500 to 20,000 cells per ml), optical density (turbidity) measurements did not provide significant values. Thus, we performed cell counts by MacConkey agar plating, with 40 h of incubation at 37°C (Oxoid, Basingstoke, United Kingdom).
Induction of β-galactosidase synthesis and the β-galactosidase assay.
The synthesis of β-galactosidase was induced with 0.1 mM of the synthetic inducer IPTG (isopropyl-β-d-thiogalactopyranoside) (6) in phosphate buffer (PB) at pH 7.2, which was made of 47.7 mM Na2HPO4, 22.0 mM KH2PO4, and 0.5 mM MgSO4·7H2O unless otherwise indicated. All tests were performed in 100- by 15-mm glass tubes containing 2.25 ml of PB; they were incubated in a water bath at 37°C. The 20 mM stock solutions of each l-amino acid were prepared in PB and stored frozen. Glassware and media were autoclaved (105 Pa; 121°C) and buffers and solutions were filter sterilized (0.45 μm) before use.
β-Galactosidase activity was determined by the hydrolysis of MUG (methylumbelliferyl-β-d-galactoside) (1). At the end of the induction time, 130 μl of MUG in dimethyl sulfoxide (5 mg/ml), 25 μl of 1% (wt/vol) sodium dodecyl sulfate, and 20 μl of chloroform were added to the cultures; after vortexing, the hydrolysis mixture was incubated for 60 min at 37°C. The hydrolysis was stopped by adding 100 μl of 2 N NaOH. After the solution was gently mixed, 2.21 ml of the solution, containing the fluorescent product of β-galactosidase hydrolysis 4-methylumbelliferone, was transferred to a 10- by 10-mm quartz cuvette by avoiding the sampling of chloroform phase. The heating block of the spectrofluorimeter, a PerkinElmer LS50B (Wellesley, MA), was set at 37°C, the excitation wavelength at 362 nm, and the emission wavelength at 445 nm, with both slit widths at 2.5 nm. β-Galactosidase was expressed as standard enzymatic units. One unit will hydrolyze 1.0 μmol of o-nitrophenyl β-d-galactoside to o-nitrophenol and d-galactose per min at pH 7.3 and 37°C.
Preparation of MOPS medium.
MOPS (potassium morpholinepropane sulfonate) medium was prepared according to the method of Neidhardt et al. (27) and comprised 40 mM MOPS, 4 mM Tricine, 10 μM FeSO4·7H2O, 1.32 mM K2HPO4, 9.52 mM NH4Cl, 276 μM K2SO4, 500 nM CaCl2·2H2O, 523 μM MgCl2, 50 mM NaCl, 3 nM (NH4)6Mo7O24·4H2O, 400 nM H3BO3, 30 nM CoCl2, 10 nM CuSO4, 80 nM MnCl2, and 10 nM ZnSO4.
Induction of β-galactosidase synthesis and β-galactosidase assay in the absence of a single amino acid.
Overnight cultures in LB (0.5 ml) were centrifuged, diluted to 1,000 to 5,000 (E. coli) or 20,000 (C. freundii) cells per ml in 0.1 mM IPTG in PB, and aliquots of 2.25 ml were incubated at 37°C in the presence of different amino acid pools. After 90 min, β-galactosidase was measured as described above. The amino acid pool, designated AA, contained all 20 natural l-amino acids in equimolar quantities, each at 13.3 μM (1/20 each; total concentration, 266 μM). Certain amino acid mixture AA-x contained 19 natural l-amino acids in equimolar quantities, each at 14.0 μM (total concentration, 266 μM), without the indicated amino acid (x).
RESULTS
Dependence on the presence of amino acids of β-galactosidase kinetics in the lag phase.
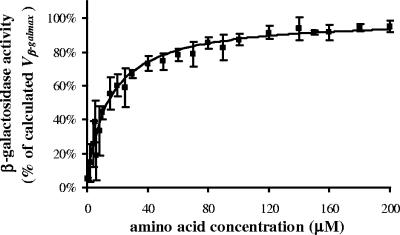
We subjected Escherichia coli ATCC 25922 to a remarkable nutritional downshift, from the highly nutritional LB to poor minimal medium PB, and we assessed the metabolic performances of E. coli by monitoring the synthesis of the inducible enzyme β-galactosidase in environments with low cell densities (1,000 to 5,000 cells per ml) under different nutrient conditions. A short incubation time (90 min) was selected so as to evaluate the cell behavior during the lag phase. The presence of amino acids supported the synthesis of β-galactosidase (Fig. 1) already before bacterial growth. In the absence of external amino acids, β-galactosidase activity was very low (<5%), while it increased to 75% of maximum activity at the given conditions, when the amino acid concentration was increased from 10 to 50 μM. The induced β-galactosidase activity was plotted against the total amino acid concentration ranging between 0 and 200 μM (0 to 10 μM each). The Hill sigmoidal curve was selected at first to interpolate experimental data. Since the best-fitting calculated n value (measure of the degree of sigmoidicity) was close to 1 (0.98), the study was redirected to the hyperbolic function, which proved more suitable for the application. Both Km (Table 1) and Vmax constants were calculated by setting the β-galactosidase activity measured in the absence of amino acids (β-galAA=0) to 0 units to be used as a blank reference. Second, we took into account the real values of β-galAA=0, in order to calculate KAA, which represents the amount of external amino acids required to induce 50% of the Vβ-galmax (Vmax plus β- galAA=0), the hypothetical maximum β-galactosidase activity. The difference between Km and KAA was termed IntAA, a value for the intracellular supply of amino acids and representing the concentration of external amino acids that the cell would require in order to induce an enzymatic activity equivalent to a β-galAA=0.
FIG. 1.
β-Galactosidase activity affected by amino acid concentration. The final curve was the result of 20 different experiments conducted in an 8-month period to reduce effects due to climatic influence. The experimental points (▪) are expressed as mean percentages of β-galactosidase activity versus amino acid concentrations ± standard errors (SE) (error bars) of 3 to 20 tests. At first, the absolute value of β-galAA=0 was calculated in every test and subtracted from the experimental points, in order to evaluate β-galactosidase activity only due to added amino acids. The upper limit of the β-galactosidase/cell production Vmax was then determined in every test by nonlinear least-squares analysis (Microcal Origin version 5.0 software) in the following hyperbole equation: y = (Vmax × x)/(Km + x). After that, the following equation was calculated: Vβ-galmax = Vmax + β-galAA=0. In order to recalculate, in every test, β-galAA=0 and all experimental points as percentages of Vβ-galmax, we assumed the limiting value Vβ-galmax to be the highest hypothetical β-galactosidase activity per cell (100%) inducible at the test conditions. Finally, by introducing the mean β-galAA=0 as a constant value in the hyperbole equation y = β-galAA=0 + [(Vmax × x)/(Km + x)], we generated a final curve which was calculated from the means of normalized experimental points.
TABLE 1.
Kinetic parameters with amino acid as the only nutrient sourcea
| Parameter | Variable for indicated strain with noted incubation time
|
||
|---|---|---|---|
|
E. coli ATCC 25922
|
C. freundii ATCC 8090 (1.5 h) | ||
| 1.5 h | 4.5 h | ||
| Km (μM) | 15.0 ± 7 | 223.6 ± 51.8 | 14 ± 2 |
| KAA (μM) | 13.5 ± 7 | 221.8 ± 49.7 | 10 ± 2 |
| IntAA (μM) | 1.5 ± 1.2 | 1.8 ± 0.5 | 3.8 ± 3 |
| β-galAA=0 (% of Vβ-galmax) | 5 ± 3 | 0.37 ± 0.32 | 14 ± 1 |
| Vβ-galmax (10−8× U/cell)b | 1.0 ± 0.4 | 40.3 ± 7.0 | |
| μmax (h−1) | 0.94 ± 0.08 | ||
| KAA-μc (μM) | 122.3 ± 33.2 | ||
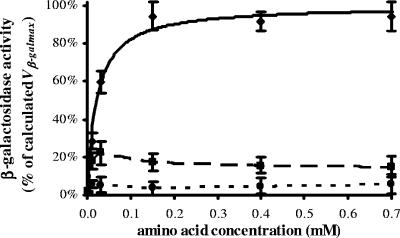
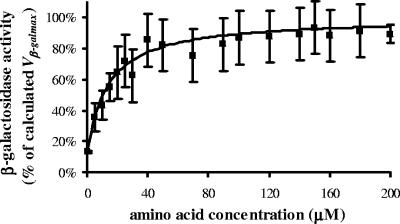
The study was extended to E. coli K-12 with results similar to those of studies with E. coli ATCC 25922 (Fig. 2). Also, Citrobacter freundii (Fig. 3) was taken into consideration to verify similarities/differences of enteric non-E. coli strains. A close hyperbole-fashioned relationship was recorded at low amino acid concentrations, despite the fact that C. freundii had a 10-fold-lower ability to produce the β-galactosidase enzyme, and consequently, a 10-fold-higher number of cells was required to equalize E. coli test sensitivity.
FIG. 2.
β-Galactosidase activity of E. coli K-12 affected by amino acid concentration. See Materials and Methods for the procedure. The AA curve (for all 20 amino acids; solid line) was determined as described in the legend to Fig. 1. Microcal Origin version 5.0 software was used to plot the effects of AA-leucine (dashed line) and AA-isoleucine (dotted line) by interpolation. The experimental points (⧫, ▪, and •) are expressed as mean percentages ± SE (error bars) of three to six tests.
FIG. 3.
β-Galactosidase activity affected by amino acid concentration. The procedure used is as described in the legend to Fig. 1 except for the use of Citrobacter freundii ATCC 8090 at 17,000 to 19,000 cells per ml. The experimental points (▪) are expressed as mean percentages ± standard deviations (SD) (error bars) of two or three tests.
Retaining the hyperbolic relationship between the amino acids and the synthesis of β-galactosidase in the presence of other carbon or nitrogen sources.
We improved the nutritional quality of the minimal medium PB by adding several carbon and nitrogen sources singularly and in combination. However, when amino acids were lacking, the synthesis of β-galactosidase in E. coli was severely restricted (β-galAA=0 < 5%), except when the complete MOPS minimal medium (27) was used (β-galAA=0 < 20%). The presence of amino acids stimulated, at different degrees, the enzyme production, depending on the presence of other nutrients for absolute yields (Table 2) . Despite the variability of medium composition, the correlation with the amino acid concentration was unaffected. As expected, the addition of glucose induced severe catabolite repression, impairing 60% of the β-galactosidase production, partly reactivated by cyclic AMP, which also relieved catabolite repression caused by amino acids as the only carbon sources (10 to 20%). The value of KAA-n, i.e., KAA in the presence of other nutrients (n), was reduced by 80% by the simultaneous presence of succinate and ammonium chloride. Also, maltose and lactate reduced KAA-n values (between 40 and 60%), while other nutrients were noneffective. The use of the widely recognized MOPS allowed us to investigate a less severe nutritional downshift due to the presence of micronutrients (see Materials and Methods); for this reason, the cell production of β-galactosidase was nearly 20% even in the absence of amino acids, but the presence of external amino acids was still required to stimulate 80% of the β-galactosidase production both in the presence and absence of glycerol.
TABLE 2.
Vβ-galmax-n and KAA-n under different nutrient conditions
| Amino acid pool AA (20 mM) plus: | Mean % of calculated valuea
|
|
|---|---|---|
| Vβ-galmax-n | KAA-n | |
| Maltose (20 mM) | 80 | 60 |
| Glucose | ||
| 0.18% | 40 | 110 |
| 0.18% plus cAMP (1 mM) | 80 | 110 |
| Mannitol (20 mM) | 80 | 70 |
| Glycerol (0.2%) | 100 | 100 |
| Succinate | ||
| 0.2% | 90 | 90 |
| 0.2% plus ammonium chloride (0.1%) | 90 | 20 |
| Lactate (40%) | 100 | 40 |
| Acetate (0.25%) | 60 | 100 |
| Citrate (20 mM) | 90 | 90 |
| Ammonium chloride (0.02%) | 90 | 70 |
| Ammonium sulfate (0.1%) | 90 | 80 |
| Ammonium phosphate (0.2%) | 90 | 110 |
| cAMP (1 mM) | 110 | 140 |
| MOPS | 120 | 70 |
| MOPS plus glycerol (0.2%) | 110 | 40 |
Mean percentages of calculated Vβ-galmax or KAA with AA as the only source. See the legend to Fig. 1 for the procedure and text for the description of parameters (three to six tests). PB was enriched with different carbon/nitrogen sources in the presence of growing concentrations of amino acids (0, 10, 30, and 150 μM).
Longer-term influence of amino acid concentration on growth rate and β-galactosidase activity.
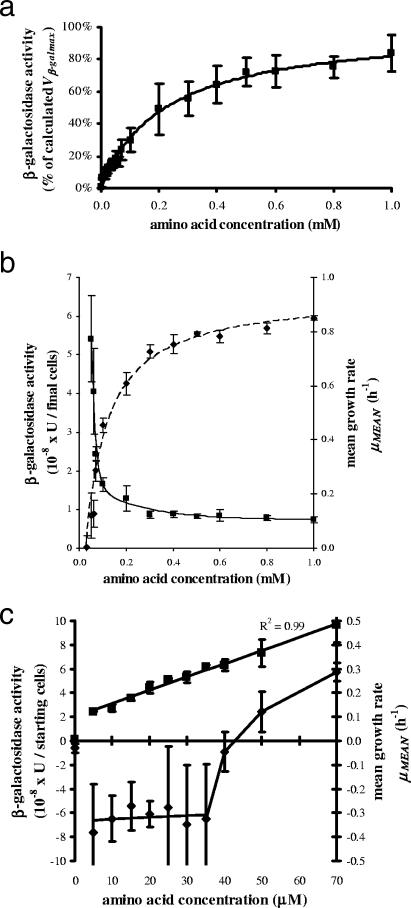
We investigated the effect of amino acids on β-galactosidase production and the dilution rate after a 4.5-hour incubation (Fig. 4a to c). In the absence of amino acids, β-galactosidase activity was 300 times lower than the Vβ-galmax, with a mean value of 1.5 × 10−9 U/cell, and cell growth approached zero, with a −0.03 h−1 mean growth rate {μmean = tx−1 × 2.303 × [log10 (cells at tx) − log10 (cells at t0)]} (2) corresponding to a 10% decrease in the number of culturable cells (Fig. 4c). At a 5 μM amino acid concentration, β-galactosidase activity increased by 17-fold (2.5 × 10−8 U/cell). In the range of 5 to 70 μM, the enzyme synthesis was linearly correlated with the amino acid concentration, whereas the cell duplication was not sufficiently supported when the concentration was lower than 40 μM, such that after 270 min, the number of culturable cells suffered a significant 60 to 80% decrease in all experiments involving amino acid concentrations higher than 0 μM but lower than 40 μM (Fig. 4c). It was only at amino acid concentrations higher than 40 μM that a positive μmean was measured and the cells reached the exponential stage. When the amino acid concentration was raised to 1 mM, the behavior of β-galactosidase was similar to that obtained after 90 min of incubation (Fig. 4a), and the μmean was also fitted with a hyperbolic function, while the β-galactosidase activity per cell showed an exponential decay in correspondence with the beginning of cell growth (Fig. 4b). Because of longer incubation time and consequent cell proliferation, the KAA constant, indicative of the need for amino acids, increased from 14 to 222 μM. Since the IntAA values were equivalent in the lag and exponential phases, we assumed that this parameter was dependent only on inoculum history, confirming its reliability as a parameter for measuring the internal amino acid supplies (Table 1).
FIG. 4.
(a) β-Galactosidase activity affected by amino acid concentration. Overnight cultures in LB (0.5 ml) were centrifuged and diluted to 50 to 300 cells per ml in 0.1 mM IPTG in PB, and then aliquots of 2.25 ml were incubated at 37°C in the presence of growing concentrations of amino acids. After 270 min, β-galactosidase activity was measured as described in Materials and Methods. The experimental points (▪) are expressed as mean percentages ± SD (error bars) of five to seven tests. The curve was determined as described in the legend to Fig. 1. (b) β-Galactosidase activity and mean growth rate affected by amino acid concentration. See the description for Fig. 4a for the procedure. β-Galactosidase activity (▪) was expressed as enzymatic units per the final number of cells. Cell counts were performed as described in Materials and Methods. β-Galactosidase activity versus amino acid concentration was fitted as an exponential decay of the third order via Microcal Origin version 5.0 software. Mean growth rate (μmean; ⧫) versus amino acid concentration was determined by nonlinear least-squares analysis via Microcal Origin version 5.0 software with the following hyperbole equation: y = μAA=0 + [(μmax × x)/(Km + x)]. Since the hyperbolic relationship was valid only for μ values of >0, μAA=0 is a fictitious value which was calculated only for regression purposes. For μmean values of ≤0, see Fig. 4c. The experimental points (▪ and ⧫) are expressed as means ± SE (error bars) of five to seven tests. (c) β-Galactosidase activity and mean growth rate affected by amino acid concentration. See the description for Fig. 4a for the procedure. The curve representing β-galactosidase activity (▪) was determined by linear least-squares analysis. μmean (⧫) was calculated as described for Fig. 4b. The experimental points (▪ and ⧫) are expressed as means ± SE (error bars) of five to seven tests.
On a daily basis, both 90-min and 4.5-hour incubation tests were significantly repeatable, whereas we observed large day-to-day variations for Vβ-galmax (±40%) and KAA (±50%), determinations with both constants not being statistically correlated with each other, with the number of cells, or with external climate conditions. We believed that these discrepancies could be ascribed to the fact that, even under identical circumstances, every cell culture expressed a different and exclusive behavior. However, the high number of experiments performed (20 90-min experiments and 7 4.5-hour experiments) allowed the calculation of mean values.
Variation in β-galactosidase production caused by the absence of a single amino acid.
We evaluated the levels of β-galactosidase synthesis of different strains affected by a single amino acid deficiency in a low-cell-density environment (Table 3). In E. coli ATCC 25922, most amino acid deficiencies were not influential; however, in a few circumstances, the absence of a single amino acid accounted for a major decrease, depending on the final amino acid pool concentration. Up to an 80% loss in the ability of this strain to synthesize β-galactosidase was caused by leucine or tryptophan deficiency. For instance, E. coli K-12 was extremely sensitive to single deficiencies of amino acids of the ilv pathway, while C. freundii was scarcely receptive to the need for a particular amino acid, except for leucine. Eventually, the absence of leucine yielded a more-than-50% reduction of β-galactosidase activity and growth rate in all strains tested (E. coli K-12 and ATCC 25922, C. freundii ATCC 8090, and K. pneumoniae ATCC 13883).
TABLE 3.
Effect of a missing amino acid on the synthesis of β-galactosidase
| Missing amino acid in AA-x | β-Galactosidase activitya
|
||
|---|---|---|---|
| E. coli ATCC 25922 | E. coli ATCC 23716 | C. freundii ATCC 8090 | |
| Alanine | 80.9 ± 12.6 | 92.6 ± 3.2 | 71.3 ± 7.5 |
| Cysteine | 102.7 ± 0.9 | 86.5 ± 6.4 | 88.1 ± 6.5 |
| Aspartate | 92.2 ± 0.4 | 85.8 ± 6.4 | 80.8 ± 4.2 |
| Glutamate | 93.0 ± 1.2 | 87.3 ± 4.1 | 75.7 ± 7.8 |
| Phenylalanine | 72.1 ± 10.3 | 89.3 ± 4.0 | 83.3 ± 0.8 |
| Glycine | 89.9 ± 4.6 | 39.2 ± 0.9 | 83.8 ± 1.7 |
| Histidine | 79.5 ± 3.6 | 80.0 ± 5.3 | 78.2 ± 0.3 |
| Isoleucine | 57.5 ± 5.6 | 3.0 ± 1.3 | 78.2 ± 3.6 |
| Lysine | 81.8 ± 11.2 | 91.2 ± 3.8 | 82.1 ± 4.8 |
| Leucine | 19.9 ± 4.5 | 12.5 ± 1.7 | 47.3 ± 15.8 |
| Methionine | 59.6 ± 4.8 | 77.1 ± 5.6 | 81.3 ± 5.9 |
| Asparagine | 93.9 ± 0.3 | 88.1 ± 4.4 | 79.7 ± 8.9 |
| Proline | 88.9 ± 2.9 | 80.4 ± 5.4 | 82.3 ± 7.3 |
| Glutamine | 74.9 ± 1.4 | 85.6 ± 1.9 | 81.9 ± 10.3 |
| Arginine | 68.2 ± 4.1 | 80.4 ± 3.3 | 76.9 ± 9.7 |
| Serine | 77.0 ± 11.1 | 86.2 ± 4.4 | 85.1 ± 10.1 |
| Threonine | 54.9 ± 6.9 | 85.6 ± 7.5 | 93.7 ± 9.7 |
| Valine | 48.1 ± 12.2 | 35.4 ± 3.0 | 107.5 ± 5.3 |
| Tryptophan | 35.7 ± 11.0 | 75.3 ± 7.5 | 95.1 ± 14.2 |
| Tyrosine | 66.4 ± 6.4 | 87.3 ± 0.5 | 93.8 ± 7.3 |
See Materials and Methods for the procedure. The experimental points are expressed as mean percentages ± SE of three to six tests, by using AA as a reference (100%).
Tests of cells in the lag phase evidenced that when the amino acid concentration was further increased (>1 mM), β-galactosidase activity approached closely the asymptotical maximum Vβ-galmax at first but then started to decrease (data not shown). This tendency was amplified in the absence of leucine (Fig. 2), when the enzyme activity suffered a remarkable reduction after the peak corresponding with an amino acid concentration of 10 μM. Finally, mixtures with an overall amino acid concentration of 200 μM, including 1 to 20 amino acids (from 200 to 10 μM each), were tested sequentially. As expected, β-galactosidase activity increased progressively with the increase in the number of different amino acids present in the solution (data not shown).
DISCUSSION
Three main findings emerged from the present study involving nutritional downshifts in environments in which E. coli has low cell density: (i) the recovery of β-galactosidase activity and the concentration of the 20 natural amino acids were hyperbolically related, and the shape of their relation was not affected by the simultaneous presence of other carbon/nitrogen sources; (ii) a 4.5-hour absence of nutrients induced a starvation behavior characterized by high rates of cell survival but no measurable β-galactosidase activity, whereas low concentrations of amino acids (5 μM) were sufficient to promote a transition to an adaptive mode, in which E. coli resumed enzyme synthesis while simultaneously enduring a population drop by between 60 and 80 percent; and (iii) β-galactosidase activity and growth rate were negatively affected by leucine deficiency in all conditions tested.
β-Galactosidase activity in environments populated at low cell densities after nutritional downshift.
Earlier experiments carried out with high-density cultures proved that when E. coli is transferred to a nutrient-limited medium, the overall synthesis of protein (38), including β-galactosidase (39), is reduced. Kuroda et al. (23) substantiated the dependence of β-galactosidase activity on the presence of amino acids after nutrient down-transition via Northern blot analysis at high cell densities and indicated translation as the critical step; the resulting β-galactosidase activity was found to be closely related to amino acid concentration despite the fact that the measured amount of IPTG-induced lacZ mRNA was not influenced whatsoever. We achieved similar results with low cell densities by using an experimental procedure for quantitatively monitoring β-galactosidase activity, and additionally, we assessed the correlation between the enzyme activity and the amino acid concentration. We observed a hyperbolic relationship and defined the cell affinity for the amino acid pool under conditions for the maximum expression of scavenging capabilities, due to both glucose and ammonium shortage, via a Michaelian constant (KAA). In order to calculate β-galactosidase production, we replaced the Monod parameter μmax, normally expressing maximum growth rate, with Vβ-galmax, which represented the hypothetical maximum achievable rate of β-galactosidase synthesis.
Other factors affected the kinetics of enzymatic production, with different outcomes for Vβ-galmax-n and KAA-n, as evidenced by multiple-nutrient experiments (Table 2). As in high-density environments, catabolite repression strongly reduced Vβ-galmax-n, in view of the fact that β-galactosidase was repressed in cAMP-deficient glucose medium and the Vβ-galmax-n of cells in MOPS was higher than that of cells in glycerol-supplemented MOPS. Since the addition of cAMP to the glucose medium left KAA-n unchanged while KAA-n was unexpectedly increased in cAMP-supplemented media having amino acids as the only carbon source, the effect of cAMP on KAA-n could not be significantly defined. On the other hand, KAA-n was distinctly subject to cell dilution effects. The remarkable KAA-n reductions in nutrient media with nitrogen and carbon sources in a balanced ratio and in glycerol-supplemented MOPS were ascribable to a shorter lag phase (data not shown), with an earlier resumption of cell growth and consequent faster, but not greater, β-galactosidase production.
Downshift responses triggered by the presence of amino acids: starvation or hunger?
After 4.5 h of incubation, β-galactosidase activity and cell growth showed conflicting behaviors at amino acid concentrations between 5 and 35 μM, since the enzyme activity grew linearly while the chances of bacterial survival were significantly more reduced during the whole interval than under conditions in which nutrient was absent (Fig. 4c). To find an interpretation of these results, we investigated the cell responses before and after this interval. In the absence of amino acids, β-galactosidase activity was extremely low, cell growth ceased, and the number of culturable cells endured only a 10% decrease. We deduced that the cells switched to a maintenance metabolism characterized by improved resistance to stress conditions and consistent with the rpoS-mediated survival response (15, 17). On the other hand, cells growing at suboptimal dilution rates, like at amino acid concentrations higher than 35 μM, express the “in between feast and famine” hunger response (13), which is elicited by carbon or nitrogen limitations (18) and characterized by improved scavenging skills, due to the overexpression of transport and assimilation systems (12). In view of the fact that the activation of the rpoS-mediated survival response causes inhibition of these genes, hunger and starvation responses are in conflict (29). Since the expression of RpoS is inversely correlated with specific growth rate (19) and, above all, attenuated by the influence of amino acids on the leucine responsive regulatory protein (Lrp) regulon (5), a central system of regulation of feast-to-famine transitions (35), we have associated this amino acid-mediated cell growth to the exponential phase with the rising expression of a hunger response.
In the transition from 0 to 5 μM, the lack of the starvation response for an adaptive response caused a fall in the survival rate from 90% to between 20 and 40%. Since an effective hunger response involves specific growth rates from 0.1 to 0.9 h−1 (13), we deduced that the hunger response was not expressed with homogeneous efficacy. Only cells fitted with advanced capabilities for the transport of amino acids and their use as both carbon (26) and nitrogen sources (32) were able to survive in the absence of ammonium and sugars without expressing a starvation stress response. Cell viability stabilized during the 5 μM-to-35 μM interval, and the cells that survived synthesized β-galactosidase linearly with respect to the amino acid pool concentrations. These observations agree with the reported slow shut-off of RpoS transcription in concurrence with the reactivation of lacZ after diauxie (14). We have argued whether, instead of having simply died, the nonculturable cells would have entered a viable but not culturable state; however, we discarded this possibility when, by using tests in the absence of nutrients as an experimental model for the maximum degree of starvation achievable in such time at those conditions, no more than 10% reduction due to starvation was observed in culturability. The minimum amino acid nutritional limit for allowing full survival at those cell densities was thus identified to be between 40 and 50 μM.
Amino acid diversity as crucial as amino acid quantity: the importance of leucine.
Another aspect that was investigated concerned the importance of the single amino acids. Despite the fact that the same level of nutrient concentration was maintained, the β-galactosidase rate decreased dramatically when a few components were not present, confirming that amino acid diversity played a critical role. Although most of the reductions in β-galactosidase activity, due to single or multiple deficiencies, were ascribed to the logical effort of the cell to biosynthesize the required building blocks (4), some amino acids produced distinctive strain-dependent inhibition of E. coli, like at higher cell densities (33). We verified that a single isoleucine deficiency inhibited the synthesis of β-galactosidase in K-12 (Fig. 2), since the presence of valine impairs the expression of genes responsible for the synthesis of enzymes of the common biosynthetic pathway, inducing a stringent response due to amino acid starvation (8).
Leucine was the only amino acid significantly affecting enzyme synthesis and growth in all strains tested. Interestingly, these reductions of β-galactosidase activity and dilution rate in leucine-deficient environments agree favorably with observations reported in literature covering leucine-dependent effects on the Lrp (3), which is activated in minimal medium, due to carbon or ammonia limitation (28). Above all, leucine was shown to counteract the negative regulation of Lrp on alanine, serine, and threonine catabolism (25, 37) and neutralize the 70% reduction in β-galactosidase synthesis measured in lrp mutants (36). Our results support the assumption that E. coli interprets leucine as an indication of nutrient availability, and thus, in the presence of leucine, the starvation response is slowed down (10).
In summary, we developed a novel sensitive tool for calculating β-galactosidase activity, which could also be extensively used for regular analyses of gene expression based on genetic fusions with the lac operon. By means of this method, we investigated low-cell-density environments and recognized effects due to catabolite repression and stringent control, in agreement with what is common knowledge regarding environments with high bacterial density. While doing so, we observed that the presence of low concentrations of amino acids was actually sufficient for E. coli to switch from survival to adaptation when moving to adverse environments.
Cells subject to the above-mentioned feast-to-famine transitions find themselves in a nutrient-deprived regimen, similar to that characterizing cells in stationary phase after nutrient exhaustion. For this reason, our results, which pointed out the key role played by amino acids, particularly of leucine, in the cell responses to nutrient disadvantage, are also consistent with the findings that cells in stationary phase tend to escape starvation by optimizing tools for amino acid scavenging and catabolism (41) via a genetic development of lrp mutants (40), which exhibit a growth advantage in stationary phase phenotype (11).
Future research will address the investigation of longer-term low-cell-density nutritional shortages in the presence or absence of amino acids with the purpose of exploring the borders between starvation and hunger.
Acknowledgments
This work was supported by the Italian Ministry of University and Research (MIUR), via L.488/92, grant N. S634-P, and also by the Italian private foundation Fondazione Cassa di Risparmio di Terni e Narni.
We thank Paolo Nannipieri for the unrivalled support in the revision process, Mark Falvo for the English revision and Centro Servizi Bibliotecari, Universitá di Perugia, Sezione Terni for providing references.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Berg, J. D., and L. Fiksdal. 1988. Rapid detection of total and fecal coliforms in water by enzymatic hydrolysis of 4-methylumbelliferone-β-d-galactoside. Appl. Environ. Microbiol. 54:2118-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Button, D. K. 1985. Kinetics of nutrient-limited transport and microbial growth. Microbiol. Rev. 49:270-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvo, J. M., and R. G. Matthews. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 5.Chen, C. F., J. Lan, M. Korovine, Z. Q. Shao, L. Tao, J. Zhang, and E. B. Newman. 1997. Metabolic regulation of lrp gene expression in Escherichia coli K-12. Microbiology 143:2079-2084. [DOI] [PubMed] [Google Scholar]
- 6.Cho, S., S. Scharpf, M. Franko, and C. W. Vermeulen. 1985. Effect of iso-propyl-thio-beta-d-galactoside concentration on the level of lac-operon induction in steady state Escherichia coli. Biochem. Biophys. Res. Commun. 128:1268-1273. [DOI] [PubMed] [Google Scholar]
- 7.Davison, J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73-91. [DOI] [PubMed] [Google Scholar]
- 8.De Felice, M., M. Levinthal, M. Iaccarino, and J. Guardiola. 1979. Growth inhibition as a consequence of antagonism between related amino acids: effect of valine in Escherichia coli K-12. Microbiol. Rev. 43:42-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dennis, P. P., M. Ehrenberg, and H. Bremer. 2004. Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol. Mol. Biol. Rev. 68:639-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernsting, B. R., M. R. Atkinson, A. J. Ninfa, and R. G. Matthews. 1992. Characterization of the regulon controlled by the leucine-responsive regulatory protein in Escherichia coli. J. Bacteriol. 174:1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell, M. J., and S. E. Finkel. 2003. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J. Bacteriol. 185:7044-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferenci, T. 1999. Regulation by nutrient limitation. Curr. Opin. Microbiol. 2:208-213. [DOI] [PubMed] [Google Scholar]
- 13.Ferenci, T. 2001. Hungry bacteria—definition and properties of a nutritional state. Environ. Microbiol. 3:605-611. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, D., A. Teich, P. Neubauer, and R. Hengge-Aronis. 1998. The general stress sigma factor σS of Escherichia coli is induced during diauxic shift from glucose to lactose. J. Bacteriol. 180:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentry, D. R., V. J. Hernandez, L. H. Nguyen, D. B. Jensen, and M. Cashel. 1993. Synthesis of the stationary-phase sigma factor σS is positively regulated by ppGpp. J. Bacteriol. 175:7982-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, T. R. G., and S. T. Williams. 1971. Microbial productivity in soil. Symp. Soc. Gen. Microbiol. 21:255-286. [Google Scholar]
- 17.Hengge-Aronis, R. 1996. Regulation of gene expression during entry into stationary phase, p. 1497-1512. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 18.Hua, Q., C. Yang, T. Oshima, H. Mori, and K. Shimizu. 2004. Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl. Environ. Microbiol. 70:2354-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ihssen, J., and T. Egli. 2004. Specific growth rate and not cell density controls the general stress response in Escherichia coli. Microbiology 150:1637-1648. [DOI] [PubMed] [Google Scholar]
- 20.Koch, A. L. 1971. The adaptive responses of Escherichia coli to a feast and famine existence. Adv. Microb. Physiol. 6:147-217. [DOI] [PubMed] [Google Scholar]
- 21.Kovárová-Kovar, K., and T. Egli. 1998. Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol. Mol. Biol. Rev. 62:646-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuo, J.-T., Y.-J. Chang, and C.-P. Tseng. 2003. Growth rate regulation of lac operon expression in Escherichia coli is cyclic AMP dependent. FEBS Lett. 553:397-402. [DOI] [PubMed] [Google Scholar]
- 23.Kuroda, A., S. Tanaka, T. Ikeda, J. Kato, N. Takiguchi, and H. Ohtake. 1999. Inorganic polyphosphate kinase is required to stimulate protein degradation and for adaptation to amino acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. USA 96:14264-14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magnusson, L. U., A. Farewell, and T. Nyström. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13:236-242. [DOI] [PubMed] [Google Scholar]
- 25.Mathew, E., J. Zhi, and M. Freundlich. 1996. Lrp is a direct repressor of the dad operon in Escherichia coli. J. Bacteriol. 178:7234-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McFall, E., and E. B. Newman. 1996. Amino acids as carbon sources, p. 358-379. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 27.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman, E. B., R. T. Lin, and R. D'Ari. 1996. The leucine/Lrp regulon, p. 1513-1525. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 29.Notley-McRobb, L., T. King, and T. Ferenci. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184:806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Primakoff, P. 1981. In vivo role of the relA+ gene in regulation of the lac operon. J. Bacteriol. 145:410-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reitzer, L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57:155-176. [DOI] [PubMed] [Google Scholar]
- 32.Reitzer, L. J. 1996. Sources of nitrogen and their utilization, p. 380-390. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 33.Rowley, D. 1953. Inhibition of E. coli strains by amino-acids. Nature 171:80-81. [DOI] [PubMed] [Google Scholar]
- 34.Saier, M. H., Jr., T. M. Ramseier, and J. Reizer. 1996. Regulation of carbon utilization, p. 1325-1343. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 35.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tchetina, E., and E. B. Newman. 1995. Identification of Lrp-regulated genes by inverse PCR and sequencing: regulation of two mal operons of Escherichia coli by leucine-responsive regulatory protein. J. Bacteriol. 177:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuan, L. R., R. D'Ari, and E. B. Newman. 1990. The leucine regulon of Escherichia coli K-12: a mutation in rblA alters expression of l-leucine-dependent metabolic operons. J. Bacteriol. 172:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westover, K. C., and L. A. Jacobson. 1974. Control of protein synthesis in Escherichia coli. I. Translation and functional inactivation of messenger ribonucleic acid after energy source shift-down. J. Biol. Chem. 249:6272-6279. [PubMed] [Google Scholar]
- 39.Westover, K. C., and L. A. Jacobson. 1974. Control of protein synthesis in Escherichia coli. II. Translation and degradation of lactose operon messenger ribonucleic acid after energy source shift-down. J. Biol. Chem. 249:6280-6287. [PubMed] [Google Scholar]
- 40.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]
- 41.Zinser, E. S., and R. Kolter. 1999. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 181:5800-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]