Abstract
Bacteria swim in liquid environments by means of a complex rotating structure known as the flagellum. Approximately 40 proteins are required for the assembly and functionality of this structure. Rhodobacter sphaeroides has two flagellar systems. One of these systems has been shown to be functional and is required for the synthesis of the well-characterized single subpolar flagellum, while the other was found only after the genome sequence of this bacterium was completed. In this work we found that the second flagellar system of R. sphaeroides can be expressed and produces a functional flagellum. In many bacteria with two flagellar systems, one is required for swimming, while the other allows movement in denser environments by producing a large number of flagella over the entire cell surface. In contrast, the second flagellar system of R. sphaeroides produces polar flagella that are required for swimming. Expression of the second set of flagellar genes seems to be positively regulated under anaerobic growth conditions. Phylogenic analysis suggests that the flagellar system that was initially characterized was in fact acquired by horizontal transfer from a γ-proteobacterium, while the second flagellar system contains the native genes. Interestingly, other α-proteobacteria closely related to R. sphaeroides have also acquired a set of flagellar genes similar to the set found in R. sphaeroides, suggesting that a common ancestor received this gene cluster.
The bacterial flagellum is a complex protein structure which consists of a long helical filament that is connected through a flexible linker known as the hook to an H+- or Na+-driven rotary motor (29). Rotation of the filament produces thrust that allows the cell to swim in liquid or semisolid medium (6). Bacteria perform taxis by controlling the frequency of reorientation, which is commonly regulated by a two-component signal transduction system that senses an environmental stimulus (2, 3, 7). Several reorientation mechanisms have been described for different bacteria (35). In Escherichia coli and Salmonella enterica serovar Typhimurium, in which the flagellar system has been studied more extensively, more than 40 proteins are required for flagellar synthesis and functioning (29). Expression of the flagellar genes is regulated in a hierarchical pattern which results from coordination of flagellar gene expression at the transcriptional or posttranscriptional level with one or more structural checkpoints in flagellum biogenesis (31). This tight regulation has probably evolved to avoid unnecessary synthesis of the large number of flagellar protein subunits required for this structure. The high energetic cost required for the synthesis and functioning of the flagellum is compensated for by the selective advantage conferred by motility. Accordingly, motility seems to be important in several processes, such as colonization, pathogenesis, dispersion, and competition for resources (37). When the growth medium is too dense to allow swimming, many bacteria differentiate into swarming cells. This process consists of enlargement of the cell, hyperflagellation, and secretion of a surfactant. In the peritrichous enterobacteria reviewed by Fraser and Hughes (13) and Rather (46) and in undomesticated Bacillus subtilis (23), the same set of flagellar genes is responsible for both swimming and swarming, while in some monotrichous proteobacterial species (including several Vibrio species [30] and Aeromonas hydrophila [25] in the Gammaproteobacteria and Rhodospirillum centenum [32] and Azospirillum brasilense [36] in the Alphaproteobacteria) a second set of flagellar genes is required for the formation of lateral flagella and swarming. Although no evidence of their functionality has been presented, second flagellar systems have also been found in the ɛ-proteobacterium Chromobacterium violaceum (40, 47), in Yersinia pestis, in Yersinia pseudotuberculosis, and in some E. coli strains (47). In Vibrio parahaemolyticus lateral and polar flagellar systems have diverged sufficiently so that they are unable to compensate for mutations in paralogous genes (8), even though under some conditions these genes are simultaneously expressed (30).
The nonsulfur photosynthetic bacterium Rhodobacter sphaeroides belongs to the α subgroup of the Proteobacteria. Before the release of the genome sequence of this species, several gene duplications were reported to be present in its two chromosomes; these duplications included three rRNA operons (11), two hem genes (hemA and hemT) that encoded the same enzymatic activity but were differentially regulated (38, 39), two sets of genes involved in CO2 fixation for which different metabolic functions have been proposed (15, 16, 22), and several copies of genes encoding different chemotaxis-related proteins (50). The genome sequence of R. sphaeroides 2.4.1 revealed several additional duplications (9), including four functionally specialized copies of the rpoN gene which codes for the σ54 factor (41) and two complete sets of flagellar genes (9).
Prior to the publication of the complete genome of R. sphaeroides, several flagellar genes of the R. sphaeroides WS8 strain (commonly used for chemotaxis and motility studies) had already been described (4, 5, 14, 17-19, 49, 51). In these studies, isolation of nonmotile strains by random or direct mutagenesis always resulted in identification of genes that, with exception of the motAB operon, are part of a gene cluster located in chromosome I; we designated this cluster flagellar cluster 1 (fla1). The fact that several random mutation experiments identified only genes in this cluster indicated that these were the genes required for the synthesis of the previously characterized flagellum and, consequently, responsible for the observed motility of R. sphaeroides. Nonetheless, the presence of a second set of flagellar genes which consists of more than 30 apparently functional cistrons (without any evident mutation), organized mainly in a cluster (the fla2 cluster) present in chromosome I, suggests that these genes are also functional. It has been suggested that the genes in the fla2 cluster could compensate for the loss of a gene in the fla1 cluster, which has been proposed to occur at low frequencies (19, 28). Another possible explanation for the absence of pseudogenes in the second flagellar cluster is a recent duplication event (9), although a horizontal transfer origin for the flagellar genes in the fla1 cluster has also been proposed (20).
In this work we investigated the functionality of the second copy of flagellar genes in R. sphaeroides WS8. Our results show that the fla2 genes are responsible for assembly of a functional flagellum. This flagellum was present under anaerobic growth conditions in liquid or semisolid medium. In contrast to other bacteria with two flagellar systems, in which one system produces lateral flagella, the second flagellar system in R. sphaeroides produces polar flagella. Phylogenetic analysis indicated that the genes in the fla2 cluster are the native R. sphaeroides flagellar genes, while the genes in the fla1 cluster were likely acquired as the result of a horizontal transfer involving a γ-proteobacterium as the possible donor. Interestingly, other α-proteobacteria closely related to R. sphaeroides also have a set of flagellar genes similar to the fla1 cluster, suggesting that the horizontal transfer of these genes occurred before the separation of these bacterial species.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Plasmids and strains of R. sphaeroides and E. coli used in this work are listed in Table 1. R. sphaeroides was grown in Sistrom's minimal medium (54) at 30°C. Photoheterotrophic cultures were grown with continuous illumination and in completely filled screw-cap tubes to generate an anaerobic environment. Heterotrophic cultures were incubated in the dark with strong shaking (250 rpm). Antibiotics were used at the following concentrations: for R. sphaeroides, 50 μg/ml spectinomycin, 25 μg/ml kanamycin, 1 μg/ml tetracycline, and 5 μg/ml gentamicin; and for E. coli, 100 μg/ml spectinomycin, 50 μg/ml kanamycin, 15 μg/ml tetracycline, 30 μg/ml gentamicin, and 100 μg/ml ampicillin. E. coli strains were grown in LB medium at 37°C with shaking at 200 rpm. Swimming assays were carried out in 30-ml, flat-bottom culture vials filled with 20 ml of Sistrom's medium containing the agar concentrations and antibiotics indicated below. The vials were incubated with continuous illumination at 30°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Rhodobacter sphaeroides strains | ||
| WS8 | Wild type | 55 |
| LC1 | flgE1::aadA | 4 |
| SP13 | fleQΔ::kan | 42 |
| SP18 | flgC1::kan | This study |
| SS1 | flhA2::uidA-aadA | This study |
| SS2 | flgC1::kan flhA2::uidA-aadA | This study |
| SS3 | flgE1::aadA flaA::kan | This study |
| SP19 | SP18 fla2+ carrying flgE2::aadA | This study |
| Escherichia coli strains | ||
| LMG174 | Protein expression strain | Invitrogen |
| S17-1 | recA endA thi hsdR RP4-2 Tc::Mu::Tn7, Tpr Smr | 44 |
| TOP10 | Cloning strain | Invitrogen |
| Plasmids | ||
| pRKflgE2 | pRK415 derivative expressing flgE2 | This study |
| pWM5 | Source of the uidA-aadA cassette | 34 |
| pJQ200mp18 | Conjugative suicide plasmid | 45 |
| pUC4Kan | Source of the Kanr cassette | Pharmacia |
| pBADflgE2 | pBAD-His B derivative overexpressing flgE2 | This study |
| pBAD-His B | E. coli expression vector | Invitrogen |
| pRK415 | Conjugative expression vector | 24 |
| pTZ19R | E. coli cloning vector, Apr pUC derivative | Fermentas |
Recombinant DNA techniques.
Strain TOP10 was used for standard recombinant DNA techniques. Plasmid DNA was prepared by using a Mini column plasmid purification kit (QIAGEN Inc., Valencia, CA). Restriction enzymes were used according to the recommendations of the manufacturer. Standard methods were used for transformation, ligation, and other related techniques. To isolate the SS1 and SS2 strains, the corresponding wild-type gene was obtained by PCR using total DNA from WS8 cells and the following nucleotides: flhA2frw (5′-TGACAGCGACATGTCC-3′) and flhA2rev (5′-CGCGGAACAGATCCTGCG-3′). These oligonucleotides were designed using the R. sphaeroides 2.4.1 genomic sequence, and the PCR products were cloned in pTZ19R. A deletion was created by digestion with BamHI, and the resulting product was purified and ligated with the uidA-aadA cassette, which was obtained from pWM5 (34). The fragment carrying the flhA2::uidA-aadA allele was then subcloned in pJQ200mp18 (45) and introduced into the R. sphaeroides WS8 wild-type and SP18 strains. Allelic exchange was confirmed by Southern blotting or PCR. To generate the SP18 strain, an NcoI-PstI fragment obtained from pRS4300 (4) was subcloned, and a Kanr cassette obtained from pUC4kan was cloned in the unique SalI site present in the fragment. The resulting flgC::kan allele was then subcloned in pJQ200mp18 and introduced into the R. sphaeroides WS8 strain by biparental conjugation using the S17-1 strain and donor. To generate the flgE2::aadA allele, we amplified the flgE2 gene with oligonucleotides flgE2F (5′-CCGATGGCGGTGCGAAACAAC-3′) and flgE2R (5′-GGTCGCATTCATCTGCTGCAC-3′) using chromosomal DNA from WS8. The 2,080-bp PCR product was cloned in pTZ19R; the aadA gene was amplified by PCR as described previously (4), and the product was cloned in the SalI site of the flgE2 gene. The resultant allele, flgE2::aadA, was subcloned into pJQ200mp18 and introduced into SP18 fla2+ by conjugation. Allelic exchange was confirmed by PCR.
Proteins and antibodies.
To obtain the FlgE2 protein, the coding region of flgE2 was amplified by PCR and cloned into the pBADHis-B vector (Invitrogen). The resulting plasmid was introduced into the LMG174-1 strain (Invitrogen), and different growth temperatures and arabinose concentrations were tested to determine the optimal conditions for overproduction of this polypeptide. The protein was in the soluble fraction when a culture at an optical density at 600 nm (OD600) of 0.5 was induced with 0.02% arabinose at 30°C for 3 h. The cells were lysed by sonication at 4°C in 10 mM phosphate buffer (pH 7) with 10% glycerol and 10 mM imidazole. The protein was affinity purified by passing the soluble fraction through an Ni-nitrilotriacetic acid agarose (QIAGEN) column. The column was washed with 20 resin volumes of 10 mM phosphate buffer (pH 7) with 10% glycerol and 20 mM imidazole, and the protein was eluted in the same buffer containing 150 mM imidazole. The resulting fraction was then dialyzed three times at 4°C against phosphate-buffered saline (PBS) using 100 times the sample volume.
Mouse anti-FlgE2 antibodies were obtained after three immunizations with approximately 20 μg of protein mixed with incomplete Freund's adjuvant. The immunizations were separated by 4 weeks, and final bleeding occurred 1 week after the third immunization.
The purity of the isolated protein was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis performed using standard techniques. Proteins were transferred to nitrocellulose membranes as described elsewhere. The membranes were probed with the anti-FlgE2 antiserum at a final dilution of 1/100,000. Detection was carried out by chemiluminescence using goat anti-mouse immunoglobulin G-AP(H+L) (Zymed) and the CDP-STAR reagent.
Microscopy.
Swimming cells were observed using a Nikon E600 microscope with a ×40 or ×60 objective and differential interference contrast (DIC) microscopy. Video recording was carried out using a Hamamatsu VE1000 SIT camera and an S-VHS videocassette recorder. To calculate the average swimming speed of bacterial cells during a single linear trajectory, we analyzed the images of a movie of swimming cells recorded at a rate of 26 frames/s. The stacked images of 120 frames were analyzed with the Manual Tracking plug-in of the ImageJ software (NIH, Bethesda, MD). The average velocity was calculated from data for 25 cells. In situ FlgE2 staining was carried out by pelleting 1.5 ml of a photoheterotrophically grown swimming culture (OD600, approximately 0.6) by centrifugation at 2,000 × g and gently resuspending the cells in 500 μl of PBS. The anti-FlgE2 antiserum was added to a final dilution of 1/100. The cells were then incubated at 4°C for 3 h, washed with 1 ml of PBS, and resuspended in 100 μl of PBS with the secondary antibody (Alexa Fluor 488 rabbit anti-mouse immunoglobulin G; Molecular Probes) at the concentration recommended by the manufacturer. The cells were incubated for 30 min at 4°C and observed with a Nikon E600 microscope with epifluorescence. Images were obtained using a Hamamatsu ORCA-ER camera.
Electron microscopy images were obtained from negatively stained preparations on carbon-coated grids. The cells in a 1.5-ml sample of an exponential growing culture were concentrated by centrifugation at 2,000 × g for 5 min and resuspended in 100 μl of 10 mM HEPES buffer (pH 7). A grid was placed over a drop of this concentrated sample and left for 5 min. The grid was then placed over a drop of 2% uranyl acetate, left for 2 min, dried at room temperature, and observed with a JEOL 1010 electron microscope.
Phylogenetic analysis.
Orthologous gene families were assembled by using gapped BLASTP and the bidirectional best-hit method, using an E-score cutoff threshold of <10−3 and imposing the limitation that pairs of homologous sequences had to have an overlap of at least 50% of their lengths and >30% overall sequence identity, as described previously (33). Multiple alignment was performed with Muscle 3.51 (12). Best-fit models for each alignment were selected by using the Akaike information criterion (43), as implemented in ProtTest 1.3 (1), and were used to infer maximum likelihood phylogenies with PhyML 2.4.4 (21). Gapped sites were removed from concatenated data sets. Phylogenetic congruence tests with a maximum likelihood framework were performed using the PhyML topologies, and the site likelihood values were computed by using TreePuzzle 5.2 (48) and fed into CONSEL 0.1i (53) to perform the approximately unbiased test. In order to reduce the risk of getting trapped in local maxima, the final tree searches for the concatenated data sets were started in PhyML from 100 random stepwise addition maximum parsimony trees generated with PAUP* 4b10 (57), as well as from single NJBio trees. In-house Perl scripts were used for data manipulation and to process the gene families through the different analysis programs in the pipeline, distributing the jobs to a cluster of 92 Pentium IV processors running Linux Rocks 3.3.0. The genome sequences used in this study were sequences from the following microorganisms: Hahella chejuensis KCTC 2396, Chromohalobacter salexigens DSM 3043, Saccharophagus degradans 2-40, Pseudomonas fluorescens PfO-1, Shewanella oneidensis MR-1, Colwellia psychrerythaea 34H, Photobacterium profundum SS9, Photorhabdus luminescens subsp. laumondii TTO1, E. coli K-12, Nitrosococcus oceani ATCC 19707, Xanthomonas campestris pv. campestris ATCC 33913, Thiobacillus denitrificans ATCC 25259, Methylobacillus flagellatus KT, Nitrosospira multiformis ATCC 25196, Nitrosomonas europaea ATCC 19718, Ralstonia eutropha H16, Burkholderia sp. strain 383, Burkholderia xenovorans LB400, Bordetella bronchiseptica RB50, Chromobacterium violaceum ATCC 12472, Rhodospirillum rubrum ATCC 11170, Zymomonas mobilis subsp. mobilis ZM4, Erythrobacter sp. strain NAP1, Silicibacter pomeroyi DSS-3, Loktanella vestfoldensis SKA53, Rhodobacter sphaeroides 2.4.1, Caulobacter crescentus CB15, Sinorhizobium meliloti 1021, Agrobacterium tumefaciens 58, Mesorhizobium loti MAFF303099, Rhodopseudomonas palustris CGA009, Bradyrhizobium japonicum USDA 110, and Rhizobium leguminosarum bv. viciae 3841.
RESULTS
flgE2 gene does not complement a mutant strain with a lesion in flgE1.
To determine whether the hook protein FlgE1 could be replaced by the FlgE2 protein, we cloned the flgE2 gene in the conjugative plasmid pRK415, so that it was expressed from the plasmid promoter. The resultant plasmid, pRKflgE2, was introduced into the LC1 strain, which has a nonpolar mutation in the flgE1 gene (4). Motility was not observed for the transconjugant cells when they were examined in swarming plates containing 0.25% agar or directly with the light microscope (data not shown). In agreement with previous observations, the motility the LC1 strain was recovered when a plasmid expressing the flgE1 gene was introduced into this strain (4). The FlgE2 protein expressed in the LC1 strain carrying the pRKflgE2 plasmid was detected by Western blotting (Fig. 1A, lane 1) using mouse antibodies raised against purified FlgE2-His protein (see Fig. S1 in the supplemental material). In contrast, no signal was detected in cell extracts of WS8 or LC1 cells (Fig. 1A, lanes 2 and 3). These results show that even though the FlgE2 protein is expressed from the pRKflgE2 plasmid, it cannot replace FlgE1. On the other hand, the fact that no signal was detected in the Western blot for the WS8 strain suggests that the chromosomal flgE2 gene was not expressed in the growth conditions tested, including heterotrophic growth conditions (Fig. 1, lane, 2) and photoheterotrophic growth conditions (Fig. 1, lane 4).
FIG. 1.
Western blot analysis with anti-FlgE2 antibody of cell lysates of various R. sphaeroides strains. (A) Heterotrophic growth conditions. Lane 1, LC1/pRKflgE2; lane 2, WS8; lane 3, LC1. (B) Photoheterotrophic growth conditions. Lane 4, WS8 (OD600, 0.5); lane 5, SP18 (OD600, 0.5); lane 6, SP18 fla2+ (OD600, 0.5); lane 7, SP18 fla2+ (OD600, 1).
Functionality of the fla2 cluster.
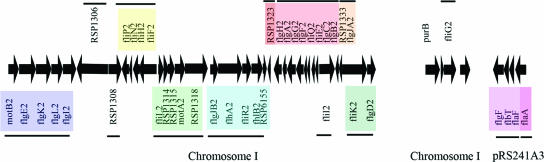
As mentioned above, the fla2 genes are arranged mainly in a single cluster in chromosome I (Fig. 2); the only exceptions are the fliG2 gene, which is located in a different region of the same chromosome, and the flaA gene, which together with its putative regulators is located in plasmid pRS241A3. To determine whether a mutation in a flagellar gene at the fla2 cluster had an effect on the ability of the wild-type strain to swim, the SS1 strain was generated by replacing the wild-type flhA2 gene with the flhA2::uidA-aadA allele. The resulting strain had a wild-type swimming phenotype either when it was tested in soft agar plates or when an aliquot of a culture was observed with the light microscope. The same results were obtained when the cells were grown under heterotrophic and photoheterotrophic conditions (data not shown).
FIG. 2.
Cluster and operon arrangement of fla2 genes of R. sphaeroides. The arrows represent genes and their directions of transcription. When possible, gene designations were assigned on the basis of similarity with a gene having a known function. The lines and colors show the possible operon organization, using the criterion of a maximal distance of 30 bp between stop and initiation codons of two contiguous genes. Chromosome and plasmid localizations of the fla2 genes are indicated at the bottom.
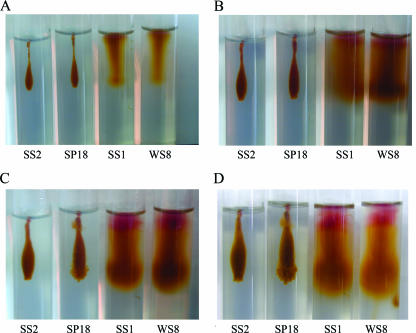
To rule out the possibility that the phenotype of the SS1 strain (flhA2::uidA-aadA) was concealed by the strong swimming produced by the fla1 flagellum, we compared the abilities of the WS8 wild-type, SP18 (flgC1::kan), SS1 (flhA2::uidA-aadA), and SS2 (flgC1::kan, flhA2::uidA-aadA) strains to swim. The phenotypes of these strains were determined in stab tubes under photoheterotrophic growth conditions, since swimming of Rhodobacter capsulatus is observed better by this method (27). As in the swimming plate assay, swimming in stab tubes is observed as spreading from the inoculation point, and the only difference is that the stab tubes are inoculated by stabbing the agar with a straightened loop, resulting in a long stripe of growth that the cells move away from. Motility of the WS8 wild-type and SS1 (flhA2::uidA-aadA) strains was observed in 0.25% agar as soon as 36 h after inoculation (Fig. 3A). Longer incubation times resulted in a homogeneous spreading pattern that completely saturated the tube (Fig. 3B, C, and D). Interestingly, motility of the SP18 (flgC1::kan) strain was observed under these conditions after 1 week of growth (Fig. 3C). In contrast with the homogeneous dispersion pattern of the wild-type strain, the SP18 strain dispersed as several independent flares, resulting in a broken cloud appearance (Fig. 3C and D). Motile cells that produced a pattern in agar tubes similar to the pattern described for SP18 (flgC1::kan) were also observed for the LC1 (flgE1::aadA) and SP13 (fleQΔ::kan) strains (data not shown), indicating that this behavior is not specifically associated with the flgC1::kan allele. The SS2 strain carrying the flhA2::uidA-aadA and flgC1::kan alleles was unable to form a dispersion pattern similar to that formed by either the wild-type or SP18 strain (Fig. 3). This suggests that the motility of the SP18 strain that was observed was dependent on the fla2 genes. In agreement with this suggestion, no swimming cells were observed when the fla2 mutant allele flaA2::kan was introduced into the LC1 strain (flgE1::aadA) and the resulting strain was tested in agar stab tubes (data not shown). Additionally, we tested the abilities of the WS8, SP18, SS1, and SS2 strains to swim when the media contained different agar concentrations. Swimming of the wild-type and SS1 (flhA2::uidA-aadA) strains diminished progressively as the agar concentration increased from 0.25 to 0.41%, and when the medium contained 0.5% agar, no motility was observed (data not shown). Swimming of the SP18 strain was as sensitive to the agar concentration as swimming of the wild-type and SS1 strains (data not shown).
FIG. 3.
Swimming patterns of various R. sphaeroides strains in stab tubes with 0.25% agar. The tubes were incubated with constant illumination for 36 h (A), 3 days (B), 7 days (C), and 10 days (D). The strains used were WS8 (wild type), SS1 (flhA2::uidA-aadA), SP18 (flgC1::kan), and SS2 (flgC1::kan flhA2::uidA-aadA).
Examination with the light microscope of a culture sample obtained from one of the growing flares of the SP18 strain inoculated into a stab tube revealed that only a few cells in the population (approximately 0.1% of the population) were actively swimming, possibly explaining the nonhomogeneous dispersal pattern of this strain. To determine whether the swimming cells were the result of a compensatory mutation or of some adaptation mechanism induced by the growth conditions in the agar tubes, the swimming cells that appeared in an SP18 culture growing in a stab tube were purified. This was carried out by streaking the cells that moved away from the inoculation strip and testing individual colonies for swimming in 0.25% agar tubes. This procedure was repeated three times, after which all the colonies tested showed the same motility pattern, which consisted of a uniformly dispersed cloud that formed around the inoculation strip (see Fig. S2 in the supplemental material). In the stab tubes, swimming of the purified strain was clearly visible after only 72 h of growth, in contrast with the 7 days that was required to observe swimming of the original SP18 strain (compare Fig. 4B with Fig. S2 in the supplemental material). Swimming cells could be easily detected in an anaerobic exponentially growing liquid culture of the purified strain (approximately 10% of the population was actively swimming [see Video S1 in the supplemental material]). These results suggested that the swimming cells that appeared during incubation of a nonswimming culture of the SP18 strain had acquired a mutation that allowed swimming that was dependent on the fla2 flagellum. Interestingly, no swimming cells were detected when the purified SP18 strain was grown aerobically (see Discussion). The mean swimming velocities of SP18 cells during and after the purification steps were the same (i.e., 18 ± 5 μm/s), eliminating the possibility that the differences in the time required to observe swimming was due to an increase in the swimming velocity. In this paper we refer to the purified SP18 strain as SP18 fla2+. In contrast to the results obtained with either the WS8 wild-type or SP18 strain, the FlgE2 protein could be easily detected by Western blotting in cell extracts from the SP18 fla2+ strain (Fig. 1B, compare lanes 4 and 5 with lanes 6 and 7), further supporting the role of fla2 genes in the swimming phenotype of the SP18 fla2+ strain.
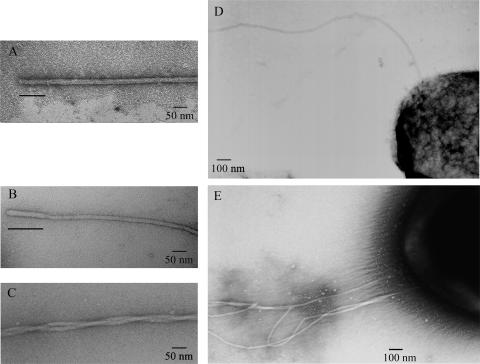
FIG. 4.
Electron microscopy images of the two different types of flagella assembled by R. sphaeroides. (A) Representative hook-filament structure found in the WS8 strain. (B) Representative hook-filament structure found in the SP18 fla2+ strain. (C) Intertwined filaments frequently found in preparations of the SP18 fla2+ strain. (D) Representative strain WS8 cell showing a subpolar flagellum. (E) Representative strain SP18 fla2+ cell showing several polar flagella. The bars in panels A and B indicate the hook.
Comparison of the two types of R. sphaeroides flagella by electron microscopy.
To further characterize the flagellum synthesized by the genes in fla2, we examined the WS8 wild-type, SS1, SP18 fla2+, and SS2 strains with the electron microscope. Flagella similar to the flagella characterized previously for R. sphaeroides WS8 (26, 58) were observed in the wild-type and SS1 (flhA2::uidA-aadA) strains (Fig. 4A and D). When the SP18 fla2+ strain was examined with the electron microscope, flagella that had a thinner filament and a more compact subunit arrangement than the flagella present in preparations of the WS8 or SS1 strains were observed (Fig. 4B). No flagella were detected in preparations of the SS2 strain (data not shown). The different widths observed for the two types of flagella (20 and 10 nm for the filaments of WS8 and SP18 fla2+, respectively) could be due to the difference in the molecular masses of the flagellins used to assemble the filaments (50,009-Da FliC and 28,288-Da FlaA for the fla1 and fla2 filaments, respectively). The hook lengths were also different for the two types of flagella (approximately 100 nm for the fla1 flagella and 130 nm for the fla2 flagella) (Fig. 4A and B). The number of flagella assembled by SP18 fla2+ cells was variable, but cells with two or three filaments were common (Fig. 4E). In addition, flagella in these preparations were frequently found to be intertwined with each other (Fig. 4C). Although many of the filaments observed for SP18 fla2+ cells were broken or detached from the cell body, cells like the one shown in Fig. 4E and intertwined filaments like the filaments shown in Fig. 4C were frequently observed, suggesting that several polar flagella are assembled in the cells expressing the fla2 genes.
Localization of flagella assembled by the products of the fla2 genes.
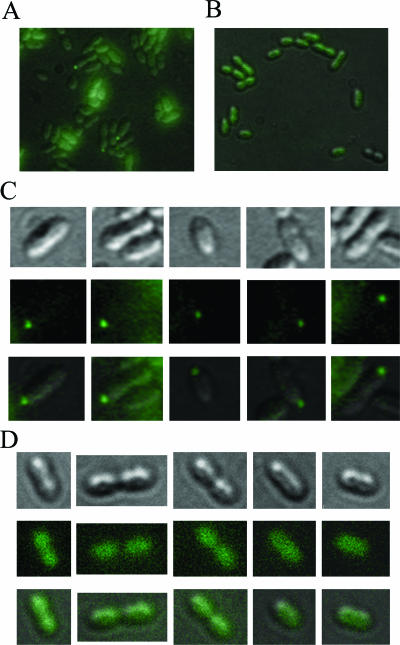
To reliably determine the localization pattern of the fla2 flagella of R. sphaeroides, we labeled the assembled hook with anti-FlgE2 antibody coupled to a secondary fluorescent anti-mouse antibody, as described in Materials and Methods. The images obtained for the SP18 fla2+ strain showed that there was a single polar fluorescent focus in each cell (Fig. 5A and C) . Localized fluorescence was detected in a small proportion of the cells in the field, possibly due to differences in the focal points between the cells and, as mentioned above, to the low number of swimming cells in the population. No localized fluorescence was detected when SP18 fla2+ cells were incubated with only the secondary antibody (Fig. 5B and D) or when SS1 (flhA2::uidA-aadA) cells were incubated with the primary and secondary antibodies (data not shown). Additionally, we introduced the flgE2::aadA allele into SP18 fla2+; the resultant strain (SP19) was not able to swim in 0.25% agar tubes (data not shown) and had a growth pattern similar to that observed for the SS2 (flhA2::uidA-aadA flgC1::kan) strain in these conditions (Fig. 3). As expected, SP19 cells did not exhibit fluorescent foci when they were incubated with both antibodies (data not shown).
FIG. 5.
In vivo fluorescent immunodetection of the hook protein (FlgE2). (A and C) Strain SP18 fla2+ incubated with anti-FlgE2 and fluorescent anti-mouse antibodies. (B and D) SP18 fla2+ cells incubated with only the fluorescent anti-mouse antibody. (A and B) Sections of the fields of view from composite images of fluorescent and DIC images. (C and D) Representative cells from (from top to bottom) the DIC image, the fluorescent image, and the composite of the DIC and fluorescent images.
Phylogenetic relationship between fla1 and fla2 genes.
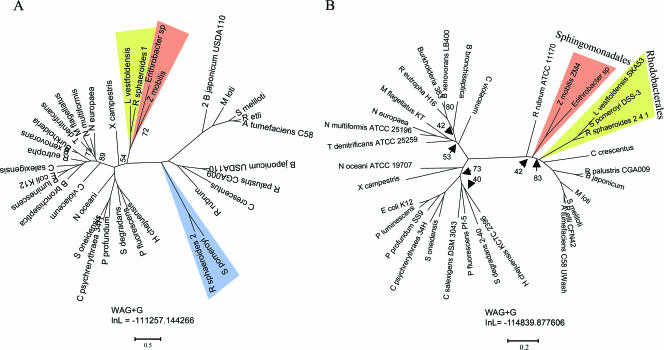
A phylogenetic analysis of several well-conserved flagellar proteins (FliG, FlhA, FlhB, FliP, and FliF) was carried out in order to determine if the R. sphaeroides fla2 cluster originated through duplication of the previously characterized flagellar cluster or was the result of a lateral gene transfer event. The individual protein alignments were concatenated, and the resulting supermatrix was used to infer a flagellar gene phylogeny using the maximum likelihood criterion. The resulting phylogram (Fig. 6A) was compared with the topology of a highly resolved maximum likelihood species tree inferred from a supermatrix generated by concatenation of 10 orthologous, single-copy core protein alignments (Fig. 6B). The latter alignments were selected because of their high phylogenetic information content as assessed by likelihood mapping (56); because of their low composition heterogeneity, and because they yielded congruent topologies as judged by the approximately unbiased test (52) (see Materials and Methods and the legend to Fig. 6 for details). An exploratory analysis that included the sequences of all flagellated bacteria for which fully sequenced genomes are available in the GenBank database indicated that only the α-, β-, and γ-proteobacteria were required to determine the phylogenetic relationship between the genes in the R. sphaeroides fla1 and fla2 clusters. Species belonging to these subgroups were selected to represent the sequenced genome lineages as comprehensively as possible. A comparison of the topologies and bootstrap values for the species and flagellar protein trees provided strong evidence indicating that the fla1 cluster of R. sphaeroides originated as the result of a horizontal gene transfer event involving a γ-proteobacterium as the likely donor (Fig. 6), as previously suggested (20). The α-proteobacteria Z. mobilis, Erithrobacter sp., and L. vestfoldensis seem to have acquired their flagellar genes from the same γ-proteobacterial donor lineage as R. sphaeroides. Surprisingly, the fla2 genes of R. sphaeroides are grouped with the flagellar genes of the α-proteobacteria, indicating that the fla2 cluster contains the native flagellar genes of R. sphaeroides. These results are supported by the gene arrangements observed for the fla1 cluster of R. sphaeroides and the flagellar clusters of Z. mobilis, Erithrobacter sp., and L. vestfoldensis, which are similar to the flagellar gene arrangement found in Pseudomonas aeruginosa (see Fig. S3 in the supplemental material) (10). Accordingly, the fla2 cluster of R. sphaeroides shows evident synteny with the flagellar cluster of the α-proteobacterium S. pomeroyi (see Fig. S3 in the supplemental material). It is noteworthy that of the different α-proteobacteria that have received the γ-proteobacterial flagellar gene cluster, only R. sphaeroides has maintained the two systems, while in Z. mobilis, L. vestfoldensis, and Erythrobacter sp. the native flagellar genes have been lost.
FIG. 6.
Maximum likelihood species and flagellar gene phylogenies. (A) Phylogram inferred from the concatenated FliG, FliP, FlhA, FlhB, and FliF flagellar protein alignments for 13 α-proteobacteria, 9 β-proteobacteria, and 11 γ-proteobacteria, resulting in a supermatrix of 2,122 residues after removal of gapped sites (for FlhA, residues 1 to 688; for FlhB, residues 689 to 1044; for FliF, residues 1045 to 1553; for FliG, residues 1554 to 1883; for FliP, residues 1884 to 2122). (B) Hypothetical phylogenetic relationships of species inferred from concatenation of 10 core proteins (for CarB, residues 1 to 284; for FtsH, residues 285 to 876; for UvrA, residues 877 to 1808; for NusA, residues 1809 to 2293; for DnaB, residues 2294 to 2737; for RplC, residues 2738 to 2935; for RplF, residues 2936 to 3108; for Lgt, residues 3109 to 3348; for PhoB, residues 3349 to 3572; for ClpP, residues 3573 to 3766) that resulted in a supermatrix of 3,766 aligned residues after removal of gapped sites. Most bipartitions in these phylogenies had >95% bootstrap support (based on 100 pseudoreplicates); the exceptions were the bipartitions whose bootstrap values are indicated.
DISCUSSION
Swimming and chemotaxis in the photosynthetic bacterium R. sphaeroides have been intensively studied. Although analysis of the complete genome sequence of this bacterium revealed the presence of two flagellar systems, previous motility screening and phenotypic analyses of strains generated by random or directed mutagenesis allowed characterization of only genes that belong to the fla1 flagellar system. In this work we describe swimming of R. sphaeroides mediated by flagella synthesized by the genes of the second flagellar system (fla2) of this bacterium.
Detection of fla2-dependent swimming required an initial incubation for 1 week. After this, swimming of purified cells was observed after 3 days of incubation. This suggests that the swimming cells accumulated a mutation. The identity of the gene or genes responsible for this phenotypic change is unknown. Nevertheless, since no expression of the flgE2 gene was detected before selection of the SP18 fla2+ strain, we hypothesized that the mutation involves a regulatory gene. The swimming phenotype of the SP18 fla2+ strain could be the result of reversion of an undetected mutation commonly present in laboratory strains of R. sphaeroides. Alternatively, the phenotypic change of the SP18 fla2+ strain could be the result of a mutation in a transcriptional regulator that usually allows expression of the fla2 genes only under particular conditions that have not been determined yet.
The fact that no swimming cells were obtained when fla1-fla2 double mutants were generated (strains SS2 [flgC1::kan flhA2::uidA-aadA], SS3 [flgE1::aadA flaA::kan], and SP19 [flgC1::kan flgE2::aadA]) supports the notion that the swimming detected for the fla1 mutants is mediated by the flagella synthesized by the fla2 genes. These results also suggest that the two flagellar systems of R. sphaeroides are independent, i.e., that the fla1 genes cannot replace the fla2 genes and vice versa. However, it remains to be determined if other proteins with a higher degree of similarity could be functional in both flagellar structures.
Observations made with the electron microscope further supported the functionality of the fla2 system by revealing different morphological characteristics of the fla1 and fla2 hook-filament structures. The flagellar filament formed by the fla1 flagellin is thicker than that formed by the flagellin of the fla2 system. Additionally, the fla2 hook is longer than the hook present in the fla1 flagellum; it should be noted that in both cases a straight hook was present.
In several species of proteobacteria, dual flagellar systems have been associated with swarming motility mediated by lateral flagella (30). However, this functional specialization does not occur in R. sphaeroides, since fla2 flagella are polarly localized and motility dependent on the fla2 system was observed in liquid medium. Anaerobic growth conditions seem to be the main requirement for induction of swimming of the SP18 fla2+ strain since no swimming cells were observed in aerobic cultures of this strain. However, it should be emphasized that anaerobic conditions per se are not sufficient to induce expression of fla2 cluster genes in wild-type R. sphaeroides, since in accordance with previous reports, no swimming cells were detected when different fla1 mutant strains were grown under these conditions (41). In line with these results, the FlgE2 protein was not present in cell extracts obtained from the wild-type WS8 strain grown under anaerobic conditions (Fig. 1, lane 4). Therefore, as mentioned above, the first requirement for expression of the fla2 genes seems to be acquisition of a mutation in a putative regulatory gene. After this event, fla2-dependent motility is observed only if the cells are cultured under anaerobic conditions. The factor(s) involved in this regulation remains to be investigated.
The facts that no mutations have accumulated in the fla2 genes of the R. sphaeroides 2.4.1. strain and that other R. sphaeroides strains (R. sphaeroides ATCC 17025 and ATCC 17029) have the two flagellar clusters in their chromosomes suggest that even though the fla1 flagellum allows strong swimming of this bacterium, swimming mediated by the fla2 flagellum could be advantageous under some conditions. When liquid cultures of the SP18 fla2+ strain were observed with the light microscope, approximately 10% of the population was motile. This could reflect heterogeneity in the population in spite of the three purification steps that were carried out; alternatively, it is possible that the optimal conditions required for expression of the fla2 genes have not been identified yet.
Phylogenetic and synteny analyses of the two flagellar clusters showed that the fla1 genes originated from a horizontal transfer event, while fla2 contains the native flagellar genes of R. sphaeroides. Flagellar genes are part of a highly integrated system that involves several interactions for each protein, making integration of a newly acquired protein into a particular flagellar system difficult. It is possible that for this reason examples of horizontally acquired independent flagellar genes are infrequent. An exception to this is the MotA/B proteins which form a proton channel that has limited interactions with the flagellar protein FliG (6). It has been shown that in R. centenum the motAB operon required for functioning of the polar flagella of this bacterium was acquired by horizontal transfer (32). In the case of the fla1 genes of R. sphaeroides, only the motAB operon is located outside the flagellar cluster, suggesting that the motAB genes were not acquired simultaneously with the rest of the fla1 cluster. Alternatively, it is possible that after acquisition of the motAB genes together with the fla1 cluster, a chromosomal rearrangement changed the position of this operon.
The flagellar genes of at least three other α-proteobacteria were also laterally acquired. However, in contrast to R. sphaeroides, the original flagellar set is not present in these microorganisms, suggesting that the conservation of the native flagellar cluster of R. sphaeroides is the result of positive selection pressure.
Even though the bacterial species with flagellar systems acquired by horizontal transfer belong to two different subgroups of α-proteobacteria (Rhodobacterales [R. sphaeroides and L. vestfoldensis] and Sphingomonadales [Erythrobacter sp. and Z. mobilis]), the four sets of acquired flagellar genes group in a single clade, indicating that there was a single donor or similar donors (Fig. 6). The inner topology of the clade that contains the horizontally transferred flagellar systems is similar to that of the clade formed by these microorganisms in the species tree (compare Fig. 6A and B). This suggests that the horizontal transfer of the flagellar cluster may have occurred between a γ-proteobacterium and the common ancestor of the Rhodobacterales and Sphingomonadales. Supporting this idea, the flagellar genes present in the genomes of two other sphingomonads (Sphingomonas sp. strain SKA58 and Sphingopyxis alaskensis RB2256) were apparently acquired from the same γ-proteobacterium that the R. sphaeroides fla1 system was acquired from.
Supplementary Material
Acknowledgments
We thank Teresa Ballado, Javier de la Mora, Alejandro Marmolejo, and Georgina Díaz Herrera for their technical assistance.
This work was funded in part by CONACyT grant P42600-Q and DGAPA IN222103-3.
Footnotes
Published ahead of print on 9 February 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abascal, F., R. Zardoya, and D. Posada. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104-2105. [DOI] [PubMed] [Google Scholar]
- 2.Adler, J. 1966. Chemotaxis in bacteria. Science 153:708-716. [DOI] [PubMed] [Google Scholar]
- 3.Baker, M. D., P. M. Wolanin, and J. B. Stock. 2006. Systems biology of bacterial chemotaxis. Curr. Opin. Microbiol. 9:187-192. [DOI] [PubMed] [Google Scholar]
- 4.Ballado, T., L. Camarena, B. González-Pedrajo, E. Silva-Herzog, and G. Dreyfus. 2001. The hook gene (flgE) is expressed from the flgBCDEF operon in Rhodobacter sphaeroides: study of an flgE mutant. J. Bacteriol. 183:1680-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballado, T., A. Campos, L. Camarena, and G. Dreyfus. 1996. Flagellar genes from Rhodobacter sphaeroides are homologous to genes of the fliF operon of Salmonella typhimurium and to the type-III secretion system. Gene 170:69-72. [DOI] [PubMed] [Google Scholar]
- 6.Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19-54. [DOI] [PubMed] [Google Scholar]
- 7.Blair, D. F. 1995. How bacteria sense and swim. Annu. Rev. Microbiol. 49:489-522. [DOI] [PubMed] [Google Scholar]
- 8.Boles, B. R., and L. L. McCarter. 2000. Insertional inactivation of genes encoding components of the sodium-type flagellar motor and switch of Vibrio parahaemolyticus. J. Bacteriol. 182:1035-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choudhary, M., Y. X. Fu, C. Mackenzie, and S. Kaplan. 2004. DNA sequence duplication in Rhodobacter sphaeroides 2.4.1: evidence of an ancient partnership between chromosomes I and II. J. Bacteriol. 186:2019-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809-824. [DOI] [PubMed] [Google Scholar]
- 11.Dryden, S. C., and S. Kaplan. 1990. Localization and structural analysis of the ribosomal RNA operons of Rhodobacter sphaeroides. Nucleic Acids Res. 18:7267-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser, G. M., and C. Hughes. 1999. Swarming motility. Curr. Opin. Microbiol. 2:630-635. [DOI] [PubMed] [Google Scholar]
- 14.García, N., A. Campos, A. Osorio, S. Poggio, B. González-Pedrajo, L. Camarena, and G. Dreyfus. 1998. The flagellar switch genes fliM and fliN of Rhodobacter sphaeroides are contained in a large flagellar gene cluster. J. Bacteriol. 180:3978-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson, J. L., and F. R. Tabita. 1987. Organization of phosphoribulokinase and ribulose bisphosphate carboxylase/oxygenase genes in Rhodopseudomonas (Rhodobacter) sphaeroides. J. Bacteriol. 169:3685-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson, J. L., and F. R. Tabita. 1988. Localization and mapping of CO2 fixation genes within two gene clusters in Rhodobacter sphaeroides. J. Bacteriol. 170:2153-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González-Pedrajo, B., T. Ballado, A. Campos, R. E. Sockett, L. Camarena, and G. Dreyfus. 1997. Structural and genetic analysis of a mutant of Rhodobacter sphaeroides WS8 deficient in hook length control. J. Bacteriol. 179:6581-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González-Pedrajo, B., J. de la Mora, T. Ballado, L. Camarena, and G. Dreyfus. 2002. Characterization of the flgG operon of Rhodobacter sphaeroides WS8 and its role in flagellum biosynthesis. Biochim. Biophys. Acta 1579:55-63. [DOI] [PubMed] [Google Scholar]
- 19.Goodfellow, I. G., C. E. Pollitt, and R. E. Sockett. 1996. Cloning of the fliI gene from Rhodobacter sphaeroides WS8 by analysis of a transposon mutant with impaired motility. FEMS Microbiol. Lett. 142:111-116. [DOI] [PubMed] [Google Scholar]
- 20.Gophna, U., E. Z. Ron, and D. Graur. 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312:151-163. [DOI] [PubMed] [Google Scholar]
- 21.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 22.Hallenbeck, P. L., R. Lerchen, P. Hessler, and S. Kaplan. 1990. Phosphoribulokinase activity and regulation of CO2 fixation critical for photosynthetic growth of Rhodobacter sphaeroides. J. Bacteriol. 172:1749-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581-590. [DOI] [PubMed] [Google Scholar]
- 24.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 25.Kirov, S. M., B. C. Tassell, A. B. Semmler, L. A. O'Donovan, A. A. Rabaan, and J. G. Shaw. 2002. Lateral flagella and swarming motility in Aeromonas species. J. Bacteriol. 184:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi, K., T. Saitoh, D. S. Shah, K. Ohnishi, I. G. Goodfellow, R. E. Sockett, and S. I. Aizawa. 2003. Purification and characterization of the flagellar basal body of Rhodobacter sphaeroides. J. Bacteriol. 185:5295-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang, A. S., and J. T. Beatty. 2002. A bacterial signal transduction system controls genetic exchange and motility. J. Bacteriol. 184:913-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackenzie, C., M. Choudhary, F. W. Larimer, P. F. Predki, S. Stilwagen, J. P. Armitage, R. D. Barber, T. J. Donohue, J. P. Hosler, J. E. Newman, J. P. Shapleigh, R. E. Sockett, J. Zeilstra-Ryalls, and S. Kaplan. 2001. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth. Res. 70:19-41. [DOI] [PubMed] [Google Scholar]
- 29.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 30.McCarter, L. L. 2004. Dual flagellar systems enable motility under different circumstances. J. Mol. Microbiol. Biotechnol. 7:18-29. [DOI] [PubMed] [Google Scholar]
- 31.McCarter, L. L. 2006. Regulation of flagella. Curr. Opin. Microbiol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 32.McClain, J., D. R. Rollo, B. G. Rushing, and C. E. Bauer. 2002. Rhodospirillum centenum utilizes separate motor and switch components to control lateral and polar flagellum rotation. J. Bacteriol. 184:2429-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medrano-Soto, A., G. Moreno-Hagelsieb, P. Vinuesa, J. A. Christen, and J. Collado-Vides. 2004. Successful lateral transfer requires codon usage compatibility between foreign genes and recipient genomes. Mol. Biol. Evol. 21:1884-1894. [DOI] [PubMed] [Google Scholar]
- 34.Metcalf, W. W., and B. L. Wanner. 1993. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene 129:17-25. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell, J. G., and K. Kogure. 2006. Bacterial motility: links to the environment and a driving force for microbial physics. FEMS Microbiol. Ecol. 55:3-16. [DOI] [PubMed] [Google Scholar]
- 36.Moens, S., K. Michiels, V. Keijers, F. Van Leuven, and J. Vanderleyden. 1995. Cloning, sequencing, and phenotypic analysis of laf1, encoding the flagellin of the lateral flagella of Azospirillum brasilense Sp7. J. Bacteriol. 177:5419-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moens, S., and J. Vanderleyden. 1996. Functions of bacterial flagella. Crit. Rev. Microbiol. 22:67-100. [DOI] [PubMed] [Google Scholar]
- 38.Neidle, E. L., and S. Kaplan. 1993. 5-Aminolevulinic acid availability and control of spectral complex formation in hemA and hemT mutants of Rhodobacter sphaeroides. J. Bacteriol. 175:2304-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neidle, E. L., and S. Kaplan. 1993. Expression of the Rhodobacter sphaeroides hemA and hemT genes, encoding two 5-aminolevulinic acid synthase isozymes. J. Bacteriol. 175:2292-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pereira, M., J. A. Parente, L. A. Bataus, D. D. Cardoso, R. B. Soares, and C. M. Soares. 2004. Chemotaxis and flagellar genes of Chromobacterium violaceum. Genet. Mol. Res. 3:92-101. [PubMed] [Google Scholar]
- 41.Poggio, S., A. Osorio, G. Dreyfus, and L. Camarena. 2002. The four different σ54 factors of Rhodobacter sphaeroides are not functionally interchangeable. Mol. Microbiol. 46:75-85. [DOI] [PubMed] [Google Scholar]
- 42.Poggio, S., A. Osorio, G. Dreyfus, and L. Camarena. 2005. The flagellar hierarchy of Rhodobacter sphaeroides is controlled by the concerted action of two enhancer-binding proteins. Mol. Microbiol. 58:969-983. [DOI] [PubMed] [Google Scholar]
- 43.Posada, D., and T. R. Buckley. 2004. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst. Biol. 53:793-808. [DOI] [PubMed] [Google Scholar]
- 44.Priefer, U. B., R. Simon, and A. Pühler. 1985. Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J. Bacteriol. 163:324-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 46.Rather, P. N. 2005. Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 7:1065-1073. [DOI] [PubMed] [Google Scholar]
- 47.Ren, C. P., S. A. Beatson, J. Parkhill, and M. J. Pallen. 2005. The Flag-2 locus, an ancestral gene cluster, is potentially associated with a novel flagellar system from Escherichia coli. J. Bacteriol. 187:1430-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 49.Shah, D. S., J. P. Armitage, and R. E. Sockett. 1995. Rhodobacter sphaeroides WS8 expresses a polypeptide that is similar to MotB of Escherichia coli. J. Bacteriol. 177:2929-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah, D. S., S. L. Porter, D. C. Harris, G. H. Wadhams, P. A. Hamblin, and J. P. Armitage. 2000. Identification of a fourth cheY gene in Rhodobacter sphaeroides and interspecies interaction within the bacterial chemotaxis signal transduction pathway. Mol. Microbiol. 35:101-112. [DOI] [PubMed] [Google Scholar]
- 51.Shah, D. S., and R. E. Sockett. 1995. Analysis of the motA flagellar motor gene from Rhodobacter sphaeroides, a bacterium with a unidirectional, stop-start flagellum. Mol. Microbiol. 17:961-969. [DOI] [PubMed] [Google Scholar]
- 52.Shimodaira, H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51:492-508. [DOI] [PubMed] [Google Scholar]
- 53.Shimodaira, H., and M. Hasegawa. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246-1247. [DOI] [PubMed] [Google Scholar]
- 54.Sistrom, W. R. 1962. The kinetics of the synthesis of photopigments in Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 28:607-616. [DOI] [PubMed] [Google Scholar]
- 55.Sockett, R. E., J. C. A. Foster, and J. P. Armitage. 1990. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp. 53:473-479. [Google Scholar]
- 56.Strimmer, K., and A. von Haeseler. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA 94:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swofford, D. L. PAUP*: phylogenetic analysis using parsimony and other methods. Sinauer Associates, Sunderland, MA.
- 58.West, M. A., and G. Dreyfus. 1997. Isolation and ultrastructural study of the flagellar basal body complex from Rhodobacter sphaeroides WS8 (wild-type) and a polyhook mutant PG. Biochem. Biophys. Res. Commun. 238:733-737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.