Abstract
Upon tryptophan induction of tna operon expression in Escherichia coli, the leader peptidyl-tRNA, TnaC-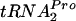 , resists cleavage, resulting in ribosome stalling at the tnaC stop codon. This stalled ribosome blocks Rho factor binding and action, preventing transcription termination in the tna operon's leader region. Plasmid-mediated overexpression of tnaC was previously shown to inhibit cell growth by reducing uncharged
, resists cleavage, resulting in ribosome stalling at the tnaC stop codon. This stalled ribosome blocks Rho factor binding and action, preventing transcription termination in the tna operon's leader region. Plasmid-mediated overexpression of tnaC was previously shown to inhibit cell growth by reducing uncharged 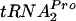 availability. Which factors relieve ribosome stalling, facilitate TnaC-
availability. Which factors relieve ribosome stalling, facilitate TnaC- cleavage, and relieve growth inhibition were addressed in the current study. In strains containing the chromosomal tna operon and lacking a tnaC plasmid, the overproduction of ribosome recycling factor (RRF) and release factor 3 (RF3) reduced tna operon expression. Their overproduction in vivo also increased the rate of cleavage of TnaC-
cleavage, and relieve growth inhibition were addressed in the current study. In strains containing the chromosomal tna operon and lacking a tnaC plasmid, the overproduction of ribosome recycling factor (RRF) and release factor 3 (RF3) reduced tna operon expression. Their overproduction in vivo also increased the rate of cleavage of TnaC-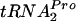 , relieving the growth inhibition associated with plasmid-mediated tnaC overexpression. The overproduction of elongation factor G or initiation factor 3 did not have comparable effects, and tmRNA was incapable of attacking TnaC-
, relieving the growth inhibition associated with plasmid-mediated tnaC overexpression. The overproduction of elongation factor G or initiation factor 3 did not have comparable effects, and tmRNA was incapable of attacking TnaC-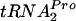 in stalled ribosome complexes. The stability of TnaC-
in stalled ribosome complexes. The stability of TnaC- was increased appreciably in strains deficient in RRF and RF3 or deficient in peptidyl-tRNA hydrolase. These findings reveal the existence of a natural mechanism whereby an amino acid, tryptophan, binds to ribosomes that have just completed the synthesis of TnaC-
was increased appreciably in strains deficient in RRF and RF3 or deficient in peptidyl-tRNA hydrolase. These findings reveal the existence of a natural mechanism whereby an amino acid, tryptophan, binds to ribosomes that have just completed the synthesis of TnaC-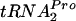 . Bound tryptophan inhibits RF2-mediated cleavage of TnaC-
. Bound tryptophan inhibits RF2-mediated cleavage of TnaC- , resulting in the stalling of the ribosome translating tnaC mRNA. This stalling results in increased transcription of the structural genes of the tna operon. RRF and RF3 then bind to this stalled ribosome complex and slowly release TnaC-
, resulting in the stalling of the ribosome translating tnaC mRNA. This stalling results in increased transcription of the structural genes of the tna operon. RRF and RF3 then bind to this stalled ribosome complex and slowly release TnaC-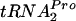 . This release allows ribosome recycling and permits the cleavage of TnaC-
. This release allows ribosome recycling and permits the cleavage of TnaC-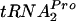 by peptidyl-tRNA hydrolase.
by peptidyl-tRNA hydrolase.
Expression of the tryptophanase (tna) operon of Escherichia coli is regulated by catabolite repression and tryptophan-induced transcription antitermination. Induction by tryptophan requires the translation of a 24-residue peptide coding region, tnaC, located in the 319-bp transcribed leader region. This region precedes tnaA, the structural gene for tryptophanase. Both Trp12 and Pro24 of TnaC have been shown to be essential for induction (5, 8). The key feature of this antitermination mechanism is tryptophan's ability to bind to ribosomes containing TnaC-peptidyl-tRNA; the bound tryptophan inhibits the cleavage of this peptidyl-tRNA. This inhibition results in the stalling of the ribosome translating tnaC mRNA (6). The stalled ribosome blocks the rut binding site in tna leader RNA, preventing Rho factor from binding and terminating transcription (7). Plasmid-mediated overexpression of tnaC results in growth inhibition (5). This inhibition was shown to be primarily a consequence of the reduced availability of 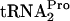 for general protein synthesis (9). This deficiency was caused by the accumulation of uncleaved TnaC-
for general protein synthesis (9). This deficiency was caused by the accumulation of uncleaved TnaC-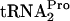 within stalled, translating ribosomes (9). Growth inhibition was relieved by overproducing
within stalled, translating ribosomes (9). Growth inhibition was relieved by overproducing 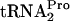 (9).
(9).
During the biosynthesis of polypeptides in bacteria, amino acid assembly is terminated when the mRNA stop codon of a coding region is encountered in the A site of the 70S ribosomal complex. The nascent polypeptide chain is then cleaved from its associated tRNA in a factor-dependent manner. The translating ribosome with the deacylated tRNA initially remains bound to mRNA and is subsequently separated by the action of accessory translation factors (13, 15). Six different factors are known to participate in the translation termination process: (i) release factors RF1 and RF2 (these factors are responsible for the recognition of the three stop codons and for activating the cleavage of each peptidyl-tRNA) (26), (ii) release factor RF3 (this factor increases the rate of dissociation of RF1 and RF2 from the ribosome following polypeptide release) (4, 32, 33), (iii) ribosome recycling factor (RRF) (this factor is responsible for ribosome separation from a transcript, and it releases the mRNA and tRNA from the ribosomal complex) (14), (iv) elongation factor G (EF-G) (this factor and RRF are believed to be jointly responsible for recycling bacterial ribosomes following the termination of polypeptide synthesis) (13), and (v) translation initiation factor IF3 (this factor prevents the reassociation of the dissociated ribosomal subunits by binding to the transiently formed 30S subunit during or following the action of RRF and EF-G) (13, 16, 27).
In addition to the normal mechanism of translation termination, there are two auxiliary mechanisms that bacteria use to hydrolyze a peptidyl-tRNA: the action of peptidyl-tRNA hydrolase (Pth) (19) and the participation of tmRNA (17). An uncleaved peptidyl-tRNA may be released from a stalled ribosome by a phenomenon called peptidyl-tRNA dropoff (21). The peptidyl-tRNA liberated is hydrolyzed by Pth, separating the peptide from its tRNA (19). It has also been reported that nascent peptide chains encoded by truncated mRNAs containing contiguous rare codons, or containing an inefficient stop codon, are targeted and tagged by the action of tmRNA (17, 24, 25). When trans-translation is completed, the unfinished peptide or polypeptide will have an 11-amino-acid-residue tag (AANDENYALAA) added at its carboxy-terminal end. This tag and its associated peptide or polypeptide are subjected to rapid degradation by various cellular proteases (10, 17). This process allows a translating ribosome to be recycled so that it may participate in a new round of protein synthesis (23).
In this study, various plasmids were used to overproduce the different translation factors, and in vivo levels of TnaC-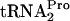 and free
and free 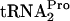 were continuously monitored. The assay procedure used to measure the levels of TnaC-
were continuously monitored. The assay procedure used to measure the levels of TnaC-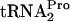 was developed previously (9). Our data indicate that when tnaC is overexpressed in the presence of tryptophan, (i) RF2 action is prevented, (ii) RRF and RF3 cause TnaC-
was developed previously (9). Our data indicate that when tnaC is overexpressed in the presence of tryptophan, (i) RF2 action is prevented, (ii) RRF and RF3 cause TnaC- dropoff, (iii) the released TnaC-
dropoff, (iii) the released TnaC-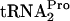 is cleaved by Pth, (iv) RRF and RF3 promote ribosome recycling, and (v) RRF, RF3, and Pth action relieves the growth inhibition caused by
is cleaved by Pth, (iv) RRF and RF3 promote ribosome recycling, and (v) RRF, RF3, and Pth action relieves the growth inhibition caused by 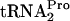 depletion. tmRNA did not act on tryptophan-induced stalled ribosome complexes containing TnaC-
depletion. tmRNA did not act on tryptophan-induced stalled ribosome complexes containing TnaC-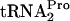 . Under normal induction conditions, in strains lacking a tnaC plasmid, the extent of TnaC-
. Under normal induction conditions, in strains lacking a tnaC plasmid, the extent of TnaC-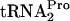 accumulation is insufficient to cause growth inhibition. However, RRF and RF3 appear to play the same role, promoting TnaC-
accumulation is insufficient to cause growth inhibition. However, RRF and RF3 appear to play the same role, promoting TnaC-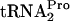 dropoff and ribosome recycling. This allows the released
dropoff and ribosome recycling. This allows the released  and freed ribosome to participate in new rounds of protein synthesis.
and freed ribosome to participate in new rounds of protein synthesis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All the strains and plasmids used in this study are listed in Table 1. The parental E. coli strain SVS1144, bearing a tnaA′-′lacZ translational fusion (30), and strains C600 and a C600 derivative with a temperature-sensitive Pth, C600[pth(Ts)], have been described previously (3). Strain SVS1144[frr(Ts) prfCΔ2::kan] was prepared by P1 transduction.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| SVS1144 | W3110 bglR551 Δ(lac-argF)U169 (λtnaptnaA-lacZ) | 30 |
| SVS1144(prfCΔ2::kan) | prfC gene deleted | 30 |
| SVS1144[frr(Ts)] | frr(Ts) | Lab collection |
| SVS1144[frr(Ts) prfCΔ2::kan] | frr(Ts); prfC gene deleted | This study |
| C600 | Wild type | 3 |
| C600[pth(Ts)] | pth(Ts) | 3 |
| Plasmids | ||
| pUC18 | ColE1 vector; Apr | Lab collection |
| pACYC184 | p15E replicon vector; Tetr Cmr | Lab collection |
| PTnaC | pUC18 derivative; tnaptnaC-rpoBC Apr | 9 |
| PTnaC-RRF | Derived from pTnaC, contains the frr gene with its own promoter and terminator; Apr | This study |
| pUC-IF3 | Contains the infC gene with its promoter and initiation codon AUU; Apr | 27 |
| pKW1 | Derived from pACYC184; Cmr gene deleted, Tetr | 25 |
| pKW24 | Derived from pKW1, contains the tmRNA-His6 mutant gene; Tetr | 25 |
| PHSG299 | Derived from pUC18; Apr gene deleted, Kmr | 14 |
| pRR1 | Derived from pHSG299; contains the frr gene with its promoter and terminator; Kmr | 14 |
| pIQ-RF3 | Derived from pACYC 184, contains lacIq and plac-prfC; Cmr | 30 |
| pEF-G | Derived from pACYC184, contains the fusA gene; Cmr | K. Ito and Y. Nakamura |
| PACD-IF3 | Derived from pACYC184, contains the infC gene with its promoter and initiation codon AUU; Tetr | U. Varshney |
Plasmid pTnaC-RRF was constructed by cloning an EcoRI-HindIII DNA fragment containing the frr gene with its own promoter, obtained from plasmid pRR1, into plasmid pTnaC. Plasmid pRR1 was kindly provided by Akira Kaji (14). Its frr-containing fragment was inserted immediately following the rpoBC terminator sequence (see Fig. 2A).
FIG. 2.
tnaC overexpression does not affect the half-life of TnaC- in vivo. Strain SVS1144 (A) or SVS144 harboring plasmid pTnaC (B) was grown at 37°C in minimal medium supplemented with 0.05% acid-hydrolyzed casein and 0.2% glycerol with (+) or without (−) the addition of 100 μg/ml tryptophan (Trp). The OD600 at the time of glucose addition (final concentration of 1.0%) to each culture was 0.6 to 0.8. Samples were taken at the indicated times, harvested by centrifugation, and disrupted by sonication, and equivalent amounts of total protein were electrophoresed on a 10% Tricine-sodium dodecyl sulfate acrylamide gel. Northern blotting was then performed to quantify the level of TnaC-
in vivo. Strain SVS1144 (A) or SVS144 harboring plasmid pTnaC (B) was grown at 37°C in minimal medium supplemented with 0.05% acid-hydrolyzed casein and 0.2% glycerol with (+) or without (−) the addition of 100 μg/ml tryptophan (Trp). The OD600 at the time of glucose addition (final concentration of 1.0%) to each culture was 0.6 to 0.8. Samples were taken at the indicated times, harvested by centrifugation, and disrupted by sonication, and equivalent amounts of total protein were electrophoresed on a 10% Tricine-sodium dodecyl sulfate acrylamide gel. Northern blotting was then performed to quantify the level of TnaC-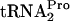 (TC-tRNAP). The TnaC-
(TC-tRNAP). The TnaC-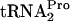 band was detected at 26 kDa. The
band was detected at 26 kDa. The  (tRNAP) molecule was also detected at 14 kDa; however, measurements of
(tRNAP) molecule was also detected at 14 kDa; however, measurements of 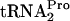 levels were not accurate quantitatively (10). The measured half-life (t1/2) of TC-tRNAP, calculated using the curves in C, is given to the right of each set of data. Little or no TnaC-
levels were not accurate quantitatively (10). The measured half-life (t1/2) of TC-tRNAP, calculated using the curves in C, is given to the right of each set of data. Little or no TnaC-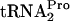 was detected in the samples from uninduced cultures. The percentage of TnaC-
was detected in the samples from uninduced cultures. The percentage of TnaC- relative to the level at 0 min was calculated by dividing the densitometry units obtained from the TnaC-
relative to the level at 0 min was calculated by dividing the densitometry units obtained from the TnaC-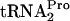 band in each lane by the units obtained for the TnaC-
band in each lane by the units obtained for the TnaC-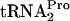 band in the 0-min lane. Note that there is less TnaC-
band in the 0-min lane. Note that there is less TnaC-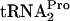 in the 0-min sample from chromosomal tnaC expression (compare +Trp TC-tRNAP values in A with those in B). (C) Plot of TnaC-
in the 0-min sample from chromosomal tnaC expression (compare +Trp TC-tRNAP values in A with those in B). (C) Plot of TnaC-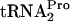 decay curves based on the data shown in A and B. Plots are not shown for the cultures grown without tryptophan. At least three independent experiments were performed, and the TnaC-
decay curves based on the data shown in A and B. Plots are not shown for the cultures grown without tryptophan. At least three independent experiments were performed, and the TnaC-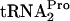 values obtained for any strain and condition varied by less than 20%.
values obtained for any strain and condition varied by less than 20%.
Growth conditions and viability curves.
Cultures were grown aerobically with shaking at the temperatures indicated. Strains C600 and C600[pth(Ts)] were grown in Luria-Bertani (LB) medium. Strain SVS1144 and its derivatives were grown in Vogel-Bonner minimal medium (29) supplemented with 0.2% glycerol and 0.05% acid-hydrolyzed casein with or without the addition of 100 μg/ml l-tryptophan. The following antibiotics were used: ampicillin (100 μg/ml), tetracycline (25 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (30 μg/ml). Growth rates were determined for cultures grown with or without the appropriate antibiotics at 37°C. Cell growth was monitored by measuring the optical density at 600 nm (OD600).
β-Gal assays.
β-Galactosidase (β-Gal) assays were performed on cultures grown with shaking at 37°C in minimal medium with or without 100 μg/ml tryptophan and with or without 10 mM isopropyl-β-d-thiogalactopyranoside (IPTG). β-Gal assays were performed as described previously (22); β-Gal activity is reported in Miller units.
Measurement of levels of TnaC-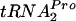 in vivo.
in vivo.
Various E. coli strains, several bearing the plasmids listed in Table 1, were grown in appropriate media at 30°C, 37°C, or 42°C. Cells were harvested at an OD600 of 0.8. The levels of TnaC-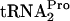 in samples taken from each culture were determined by preparing sonic extracts, centrifugation, and analysis of equal amounts of protein in the supernatants by electrophoresis and Northern blotting. The probe used was unique to
in samples taken from each culture were determined by preparing sonic extracts, centrifugation, and analysis of equal amounts of protein in the supernatants by electrophoresis and Northern blotting. The probe used was unique to 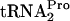 . The procedures used have been described previously (9). TnaC-
. The procedures used have been described previously (9). TnaC- and
and  were detected as bands at 26 kDa and 14 kDa, respectively, under the conditions used (9). For quantification, each gel was exposed to films for different lengths of time, the developed films were scanned, and values were compared to ensure the accuracy of the final values (densitometry imager; Bio-Rad). The band densities (densitometry units) were determined using imaging software (Molecular Analysis; Bio-Rad).
were detected as bands at 26 kDa and 14 kDa, respectively, under the conditions used (9). For quantification, each gel was exposed to films for different lengths of time, the developed films were scanned, and values were compared to ensure the accuracy of the final values (densitometry imager; Bio-Rad). The band densities (densitometry units) were determined using imaging software (Molecular Analysis; Bio-Rad).
RESULTS
Overproduction of both RRF and RF3 in vivo decreases tna operon expression (TnaC-LacZ production).
In E. coli, when a translating ribosome encounters one of the three stop codons, release factor RF1 or RF2 mediates peptidyl-tRNA cleavage. RRF and EF-G as well as other factors, e.g., RF3 and IF3, then participate in the ribosome release/recycling process to provide ribosomal subunits for additional protein synthesis (13). Interestingly, the inactivation of prfC, the structural gene for RF3, has been reported to increase basal-level expression of the tna operon two- to threefold (31). Overexpression of prfC had the opposite effect: it reduced both the basal and induced levels of tna operon expression (31). On the basis of this knowledge, experiments were performed to determine whether the overproduction of any of the translation factors, alone or in combination, would affect tna operon expression. We initially determined β-Gal levels produced by a lacZ deletion strain, SVS1144, containing a tnaA′-′lacZ translational fusion (Table 1). This strain was transformed with combinations of two plasmids that either do or do not overproduce individual translation factors (Table 1). Whenever two plasmids were introduced into the same strain, they were compatible with one another. Initially, we observed that the overproduction of either the RRF or the RF3 protein consistently reduced both the basal and induced levels of β-Gal observed by about 25%, compared to levels of control cultures (Table 2). However, when either IF3 or EF-G was overproduced, no detectable effect on the β-Gal level was observed (Table 2). When RFF and RF3 were overproduced in the same cell, the β-Gal levels were reduced even further (Table 2). When RRF was overproduced together with EFG or IF3 or when RF3 was overproduced with IF3, the reduction in the β-Gal level observed was no greater than the reduction observed upon overproducing either RRF or RF alone (Table 2). These results indicate that these other factors, when tested in combination with RRF or RF3, do not enhance their action. Also, the overproduction of EFG and IF3 in the same cell did not affect the β-Gal levels (Table 2). Thus, the overproduction of both RRF and RF3 has the greatest negative effect on tna operon expression in vivo, reducing both the basal and induced levels. The overproduction of tmRNA had no observable effect on the β-Gal level. Strains containing the empty vector or the plasmid overexpressing tmRNA had the same β-Gal levels (Table 2).
TABLE 2.
TnaA-LacZ expression by strain SVS1144 containing different combinations of plasmids expressing mutant and nonmutant translational factorsa
| Strain | Gene(s) (protein[s]) overexpressed | ß-Gal (TnaA-LacZ) activity (Miller units)c
|
+Trp/−Trp ratio | |
|---|---|---|---|---|
| −Trp | +Trp | |||
| SVS114 | 499 | 8,310 | 17 | |
| SVS1144/pACYC184/pHSG299 | 430 | 7,430 | 17 | |
| SVS1144/pACYC184/pUC18 | 483 | 7,830 | 16 | |
| SVS1144/pACYC184/pRR1 | frr (RRF) | 272 | 5,220 | 19 |
| SVS1144/pIQ-RF3/pHSG299 | prfC (RF3) | 457 | 7,670 | 17 |
| 224 (IPTG)b | 4,480 (IPTG) | 20 | ||
| SVS1144/pACYC184/pUC-IF3 | infC (IF3) | 436 | 8,170 | 19 |
| SVS1144/pEF-G/pHSG299 | fusA (EF-G) | 456 | 7,900 | 17 |
| SVS1144/pIQ-RF3/pUC-IF3 | prfC and infC (RF3 and IF3) | 466 | 7,860 | 17 |
| 238 (IPTG) | 4,600 (IPTG) | 19 | ||
| SVS1144/pEF-G/pRR1 | fusA and frr (EF-G and RRF) | 202 | 5,300 | 26 |
| SVS1144/pEF-G/pUC-IF3 | fusA and infC (EF-G and IF3) | 402 | 7,300 | 18 |
| SVS1144/pACD-IF3/pRR1 | infC and frr (IF3 and RRF) | 302 | 5,090 | 17 |
| SVS1144/pIQ-RF3/pRR1 | prfC and frr (RF3 and RRF) | 184 | 5,430 | 30 |
| 74 (IPTG) | 840 (IPTG) | 11 | ||
| SVS1144/pKW1 | 525 | 7,900 | 15 | |
| SVS1144/pKW24 | ssrA (tmRNA) | 560 | 8,180 | 14 |
Cultures were grown in minimal medium plus 0.2% glycerol and 0.05% acid-hydrolyzed casein with or without additions. Added compounds were 100 μg/ml tryptophan, 30 μg/ml chloramphenicol (pIQ-RF3 and pEF-G), 25 μg/ml tetracycline (pACD-IF3, pKW1, and pKW24), 100 μg/ml ampicillin (pUC-IF3), or 50 μg/ml kanamycin (pHSG299 and pRR1).
A total of 10 mM IPTG was added for the production of RF3 from pIQ-RF3.
β-Gal assays and the averaged results are presented. Repeat experiments gave values that varied approximately 10%. Values reflecting significant changes are underlined.
Growth inhibition caused by plasmid-mediated overexpression of tnaC is relieved by overproduction of both RRF and RF3.
On the basis of the decrease in TnaA-LacZ synthesis observed upon the overproduction of RRF and/or RF3 in a strain with chromosomal tnaC (Table 2), we next determined whether the overproduction of both of these factors could rescue ribosomes stalled as a consequence of tnaC overexpression. It was shown previously that when tnaC was overexpressed from a multicopy plasmid, pTnaC, the availability of free  for continued protein synthesis was reduced (9). This was due to the accumulation of uncleaved TnaC-
for continued protein synthesis was reduced (9). This was due to the accumulation of uncleaved TnaC-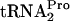 within the ribosomes translating tnaC mRNA (9). This inhibition was relieved upon
within the ribosomes translating tnaC mRNA (9). This inhibition was relieved upon 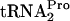 overproduction (9). If the overproduction of RRF and RF3 could promote TnaC-
overproduction (9). If the overproduction of RRF and RF3 could promote TnaC-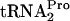 cleavage, their overproduction would be expected to relieve the growth inhibition resulting from TnaC-
cleavage, their overproduction would be expected to relieve the growth inhibition resulting from TnaC-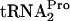 accumulation (9). To examine this possibility, it was necessary to overproduce TnaC, RRF, and RF3 in the same bacterium. To achieve this objective, the frr gene (encoding RRF), driven by its own promoter, pfrr, was inserted into plasmid pTnaC immediately following the rpoBC terminator sequence (Fig. 1A). After confirming the location and sequence of the DNA insert, the overproduction of RRF in vivo was verified by Western blotting using anti-RRF antibodies. The amount of RRF produced was estimated to be three- to fourfold greater than that produced by the control strain (data not shown). Reverse transcription-PCR analyses also indicated that there was a 2.5-fold increase in tnaC mRNA in strains with the tnaC plasmid and that the insertion of the frr gene and its promoter had no effect on the level of tnaC mRNA (data not shown). SVS1144 strains transformed with plasmids (i) pTnaC, (ii) pTnaC and pIQ-RF3, (iii) pTnaC-RRF, and (iv) pTnaC-RRF and pIQ-RF3 were grown in supplemented minimal medium with or without 100 μg/ml tryptophan and with or without 10 mM IPTG (Fig. 1B). The production of RF3 by plasmid pIQ-RF3 is dependent upon the presence of the inducer, IPTG (the RF3 level has been reported to increase 10-fold when 10 mM IPTG is present in the growth medium) (20). Our results indicate that the growth inhibition caused by the presence of plasmid pTnaC in strains grown in the presence of tryptophan was relieved only when both RRF and RF3 were overproduced in transformants containing both plasmids pTnaC-RRF and pIQ-RF3 grown with 10 mM IPTG (Fig. 1B). Transformants overproducing RRF or RF3 alone did not exhibit relief from this growth inhibition (Fig. 1B). These findings demonstrate that an excess of both factors RRF and RF3 is necessary in order to reverse the growth inhibition caused by plasmid-mediated tnaC overexpression in strains grown in the presence of tryptophan.
accumulation (9). To examine this possibility, it was necessary to overproduce TnaC, RRF, and RF3 in the same bacterium. To achieve this objective, the frr gene (encoding RRF), driven by its own promoter, pfrr, was inserted into plasmid pTnaC immediately following the rpoBC terminator sequence (Fig. 1A). After confirming the location and sequence of the DNA insert, the overproduction of RRF in vivo was verified by Western blotting using anti-RRF antibodies. The amount of RRF produced was estimated to be three- to fourfold greater than that produced by the control strain (data not shown). Reverse transcription-PCR analyses also indicated that there was a 2.5-fold increase in tnaC mRNA in strains with the tnaC plasmid and that the insertion of the frr gene and its promoter had no effect on the level of tnaC mRNA (data not shown). SVS1144 strains transformed with plasmids (i) pTnaC, (ii) pTnaC and pIQ-RF3, (iii) pTnaC-RRF, and (iv) pTnaC-RRF and pIQ-RF3 were grown in supplemented minimal medium with or without 100 μg/ml tryptophan and with or without 10 mM IPTG (Fig. 1B). The production of RF3 by plasmid pIQ-RF3 is dependent upon the presence of the inducer, IPTG (the RF3 level has been reported to increase 10-fold when 10 mM IPTG is present in the growth medium) (20). Our results indicate that the growth inhibition caused by the presence of plasmid pTnaC in strains grown in the presence of tryptophan was relieved only when both RRF and RF3 were overproduced in transformants containing both plasmids pTnaC-RRF and pIQ-RF3 grown with 10 mM IPTG (Fig. 1B). Transformants overproducing RRF or RF3 alone did not exhibit relief from this growth inhibition (Fig. 1B). These findings demonstrate that an excess of both factors RRF and RF3 is necessary in order to reverse the growth inhibition caused by plasmid-mediated tnaC overexpression in strains grown in the presence of tryptophan.
FIG. 1.
Overproduction of RRF and RF3 relieves the growth inhibition caused by tnaC overexpression. (A) Schematic representation of the organization of plasmids pTnaC and pTnaC-RRF. Both plasmids are pUC18 derivatives and contain the tna operon promoter, the tnaC open reading frame, the noncoding region located between tnaC and tnaA genes, and an added rpoBC terminator. In preparing pTnaC-RRF, a fragment containing the RRF coding region and its own promoter from plasmid pRR1 was subcloned into plasmid pTnaC immediately following the rpoBC terminator. (B) Overproduction of both RRF and RF3 relieves the growth inhibition caused by tnaC overexpression by pTnaC in the presence of tryptophan. E. coli cells bearing the different plasmids listed in the figure (Table 1) were diluted from cultures grown overnight and were grown with shaking at 37°C in minimal medium supplemented with 0.05% acid-hydrolyzed casein and 0.2% glycerol with (+) or without (−) 100 μg/ml tryptophan (Trp). Cell growth was monitored at an OD600. A total of 10 mM IPTG (+) was added at time zero for RF3 overproduction. Four independent growth experiments were performed. The growth curves for the different strains grown with or without added tryptophan were all similar to those shown in this figure.
In the parental strain and a strain overexpressing tnaC, the half-life of TnaC-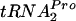 is 8 min under induction conditions in vivo.
is 8 min under induction conditions in vivo.
Cultures of strain SVS1144 with or without pTnaC (Table 1), a plasmid overexpressing tnaC, were grown to an OD600 of 0.6 to 0.8 in supplemented minimal medium in the presence or absence of 100 μg/ml tryptophan. Glucose was then added to each culture (final level, 1%) to inhibit the initiation of transcription of the tna operon, and portions of each culture were harvested at the times indicated (Fig. 2). The levels of TnaC- were then measured, and its half-life was determined (Fig. 2). The parental SVS1144 culture, expressing tnaC chromosomally, when grown in the absence of tryptophan, had no detectable TnaC-
were then measured, and its half-life was determined (Fig. 2). The parental SVS1144 culture, expressing tnaC chromosomally, when grown in the absence of tryptophan, had no detectable TnaC-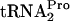 (Fig. 2A). When grown in the presence of tryptophan, its TnaC-
(Fig. 2A). When grown in the presence of tryptophan, its TnaC- level in the sample at time zero was low, and its half-life was approximately 8 min (Fig. 2A and C). In previous in vitro-coupled transcription-translation assays performed with RF2-deficient cell extracts, it was shown that the transcription of tnaC and translation of tnaC mRNA were not affected by the absence of tryptophan (8). Thus, the absence of detectable TnaC-
level in the sample at time zero was low, and its half-life was approximately 8 min (Fig. 2A and C). In previous in vitro-coupled transcription-translation assays performed with RF2-deficient cell extracts, it was shown that the transcription of tnaC and translation of tnaC mRNA were not affected by the absence of tryptophan (8). Thus, the absence of detectable TnaC-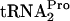 in cultures grown without tryptophan is probably due to its rapid cleavage; its half-life under these conditions is presumably <8 min. When strain SVS1144 containing plasmid pTnaC was grown in the presence of tryptophan, the TnaC-
in cultures grown without tryptophan is probably due to its rapid cleavage; its half-life under these conditions is presumably <8 min. When strain SVS1144 containing plasmid pTnaC was grown in the presence of tryptophan, the TnaC- half-life was also approximately 8 min (Fig. 2B and C). In the absence of tryptophan, no TnaC-
half-life was also approximately 8 min (Fig. 2B and C). In the absence of tryptophan, no TnaC- was detected; therefore, its half-life is presumably <8 min. These findings demonstrate that under induction conditions, with or without tnaC overexpression, TnaC-
was detected; therefore, its half-life is presumably <8 min. These findings demonstrate that under induction conditions, with or without tnaC overexpression, TnaC-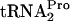 accumulates. However, it is eventually hydrolyzed, yielding TnaC and
accumulates. However, it is eventually hydrolyzed, yielding TnaC and  . The freed
. The freed 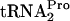 and ribosomes thus would become available for additional protein synthesis.
and ribosomes thus would become available for additional protein synthesis.
Overproduction of RRF and RF3 in a strain overexpressing tnaC reduces TnaC-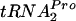 accumulation and shortens its half-life.
accumulation and shortens its half-life.
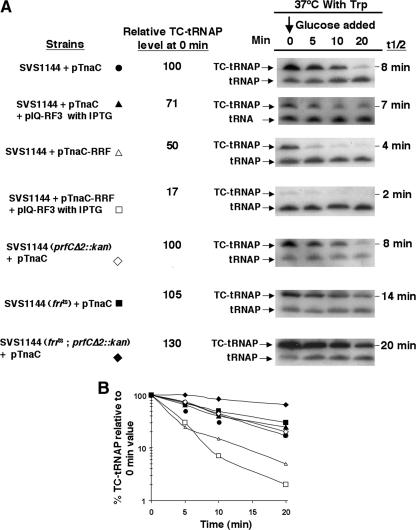
Next, experiments to determine if the stability of TnaC-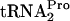 was affected in strains containing plasmid pTnaC when RF3 and/or RRF was limiting or overproduced were performed (Fig. 3). Cultures of wild-type and mutant strains with plasmid pTnaC were grown at 37°C in supplemented minimal medium in the presence of 100 μg/ml tryptophan to an OD600 of 0.6 to 0.8. At this temperature, it was observed by Western blot analysis that the RRF level is reduced in strains containing frr(Ts) (data not shown). Glucose was then added to each culture (final level, 1%) to shut off tna operon transcription initiation. Portions of each culture were harvested at the times indicated, and the level and half-life of TnaC-
was affected in strains containing plasmid pTnaC when RF3 and/or RRF was limiting or overproduced were performed (Fig. 3). Cultures of wild-type and mutant strains with plasmid pTnaC were grown at 37°C in supplemented minimal medium in the presence of 100 μg/ml tryptophan to an OD600 of 0.6 to 0.8. At this temperature, it was observed by Western blot analysis that the RRF level is reduced in strains containing frr(Ts) (data not shown). Glucose was then added to each culture (final level, 1%) to shut off tna operon transcription initiation. Portions of each culture were harvested at the times indicated, and the level and half-life of TnaC-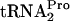 were measured (Fig. 3). In the control culture containing plasmid pTnaC (SVS1144 plus pTnaC), the TnaC-
were measured (Fig. 3). In the control culture containing plasmid pTnaC (SVS1144 plus pTnaC), the TnaC-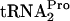 half-life was approximately 8 min. When either RF3 (pIQ-RF3) or RRF (pTnaC-RRF) was overproduced alone in strains with pTnaC, the TnaC-
half-life was approximately 8 min. When either RF3 (pIQ-RF3) or RRF (pTnaC-RRF) was overproduced alone in strains with pTnaC, the TnaC-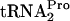 half-life was reduced from 8 min to 7 min and from 8 min to 4 min, respectively (Fig. 3A and B). When both RRF and RF3 were overproduced in the same cell, the half-life of TnaC-
half-life was reduced from 8 min to 7 min and from 8 min to 4 min, respectively (Fig. 3A and B). When both RRF and RF3 were overproduced in the same cell, the half-life of TnaC- was reduced further, to 2 min (Fig. 3A and B). In pTnaC strains producing a temperature-sensitive RRF protein [frr(Ts)] incubated at 37°C for 1 h after shifting from 30°C, the half-life of TnaC-
was reduced further, to 2 min (Fig. 3A and B). In pTnaC strains producing a temperature-sensitive RRF protein [frr(Ts)] incubated at 37°C for 1 h after shifting from 30°C, the half-life of TnaC-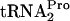 was increased only slightly (Fig. 3A and B). A similar result was observed with a strain lacking the RF3 protein (prfCΔ2::kan) (Fig. 3A and B). However, when both RRF and RF3 were limiting {strain SVS1144[frr(Ts) prfCΔ2::kan] plus pTnaC}, the half-life of TnaC-
was increased only slightly (Fig. 3A and B). A similar result was observed with a strain lacking the RF3 protein (prfCΔ2::kan) (Fig. 3A and B). However, when both RRF and RF3 were limiting {strain SVS1144[frr(Ts) prfCΔ2::kan] plus pTnaC}, the half-life of TnaC- was increased appreciably, from 8 min to 20 min (Fig. 3A and B). These findings demonstrate that changing the levels of these two factors, RRF and RF3, has the most profound effect on the half-life of TnaC-
was increased appreciably, from 8 min to 20 min (Fig. 3A and B). These findings demonstrate that changing the levels of these two factors, RRF and RF3, has the most profound effect on the half-life of TnaC-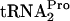 . Consistent with this conclusion and the calculated half-lives are the levels of TnaC-
. Consistent with this conclusion and the calculated half-lives are the levels of TnaC-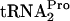 detected in the 0-min samples that were analyzed (Fig. 3A, second column). These 0-min levels reflect the sensitivity of TnaC-
detected in the 0-min samples that were analyzed (Fig. 3A, second column). These 0-min levels reflect the sensitivity of TnaC-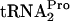 to cleavage in the respective strains. The half-life of TnaC-
to cleavage in the respective strains. The half-life of TnaC-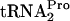 was also examined in strains overproducing other translational factors such as EF-G and IF3; these had no observable effects (data not shown).
was also examined in strains overproducing other translational factors such as EF-G and IF3; these had no observable effects (data not shown).
FIG. 3.
Overproduction of RRF and RF3 reduces the half-life of TnaC-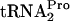 in vivo. (A) Strains bearing the indicated plasmid or plasmids were grown at 30°C or 37°C in minimal medium supplemented with 0.05% acid-hydrolyzed casein and 0.2% glycerol with the addition of 100 μg/ml tryptophan (Trp). In strain SVS1144(prfCΔ2::kan), the prfC gene was deleted. Strain SVS1144[frr(Ts)] produces an RRF protein that is temperature sensitive. The frr(Ts) allele was also present in SVS1144[frr(Ts) prfCΔ2::kan]. Strains with plasmid pIQ-RF3 were grown with 10 mM IPTG, which induces RF3 production. Plasmid pTnaC-RRF was used to overexpress tnaC and overproduce RRF. Strain SVS1144[frr(Ts)] pTnaC was grown initially at 30°C and then shifted to 37°C for 1 h. Presumably, most of its RRF protein would be inactivated during growth at 37°C. The OD600 at the time of glucose addition (final concentration of 1.0%) to each culture was 0.6 to 0.8. Samples were taken and sonicated, and Northern blot assays performed to detect TnaC-
in vivo. (A) Strains bearing the indicated plasmid or plasmids were grown at 30°C or 37°C in minimal medium supplemented with 0.05% acid-hydrolyzed casein and 0.2% glycerol with the addition of 100 μg/ml tryptophan (Trp). In strain SVS1144(prfCΔ2::kan), the prfC gene was deleted. Strain SVS1144[frr(Ts)] produces an RRF protein that is temperature sensitive. The frr(Ts) allele was also present in SVS1144[frr(Ts) prfCΔ2::kan]. Strains with plasmid pIQ-RF3 were grown with 10 mM IPTG, which induces RF3 production. Plasmid pTnaC-RRF was used to overexpress tnaC and overproduce RRF. Strain SVS1144[frr(Ts)] pTnaC was grown initially at 30°C and then shifted to 37°C for 1 h. Presumably, most of its RRF protein would be inactivated during growth at 37°C. The OD600 at the time of glucose addition (final concentration of 1.0%) to each culture was 0.6 to 0.8. Samples were taken and sonicated, and Northern blot assays performed to detect TnaC-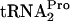 (TC-tRNAP) and
(TC-tRNAP) and 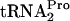 (tRNAP), as indicated in the legend of Fig. 2. The half-life (t1/2) of TnaC-
(tRNAP), as indicated in the legend of Fig. 2. The half-life (t1/2) of TnaC-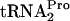 , calculated using the curve in C, is shown to the right of each set of data. The relative TnaC-
, calculated using the curve in C, is shown to the right of each set of data. The relative TnaC- level at 0 min is shown to the left of each panel. These values were calculated by dividing the densitometry units obtained for the TnaC-
level at 0 min is shown to the left of each panel. These values were calculated by dividing the densitometry units obtained for the TnaC- band in each 0-min lane by the densitometry units obtained for the TnaC-
band in each 0-min lane by the densitometry units obtained for the TnaC-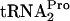 band in the 0-min lane for the control SVS1144(pTnaC) culture. (B) The TnaC-
band in the 0-min lane for the control SVS1144(pTnaC) culture. (B) The TnaC- decay curves are based on the percentage of TnaC-
decay curves are based on the percentage of TnaC- detected relative to the 0-min value, as indicated in Fig. 2. At least three independent experiments were performed, and the TnaC-
detected relative to the 0-min value, as indicated in Fig. 2. At least three independent experiments were performed, and the TnaC-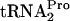 values obtained for any strain and condition varied by less than 20%.
values obtained for any strain and condition varied by less than 20%.
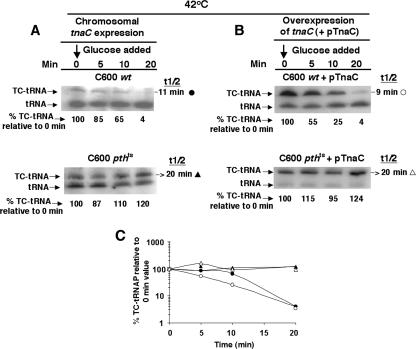
Limiting Pth availability affects the half-life of TnaC-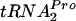 .
.
When the synthesis of a peptidyl-tRNA is completed, either it is hydrolyzed in response to the action of one of the release factors or, if peptidyl-tRNA dropoff occurs, it is cleaved by Pth (21). Since the presence of tryptophan inhibits RF2-initiated hydrolysis of TnaC-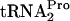 , we wished to determine if the slow cleavage of TnaC-
, we wished to determine if the slow cleavage of TnaC- observed under tryptophan induction conditions was due to peptidyl-tRNA dropoff and Pth cleavage. The stability and half-life of TnaC-
observed under tryptophan induction conditions was due to peptidyl-tRNA dropoff and Pth cleavage. The stability and half-life of TnaC-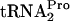 were determined in strains C600 and C600[pth(Ts)] as well as in strains C600 pTnaC and C600[pth(Ts)] pTnaC grown at 42°C (Fig. 4). These strains were grown under conditions chosen to maximally reduce the level of the Pth enzyme. Cultures were initially grown at 30°C in LB broth, a medium rich in tryptophan, to an OD600 of 0.8 (mid-log phase). The cultures were then shifted to 42°C for an hour of additional growth, with the objective of reducing the cellular level of the Pth(Ts) protein (3). Glucose was then added, and cell samples were harvested at the times indicated. The TnaC-
were determined in strains C600 and C600[pth(Ts)] as well as in strains C600 pTnaC and C600[pth(Ts)] pTnaC grown at 42°C (Fig. 4). These strains were grown under conditions chosen to maximally reduce the level of the Pth enzyme. Cultures were initially grown at 30°C in LB broth, a medium rich in tryptophan, to an OD600 of 0.8 (mid-log phase). The cultures were then shifted to 42°C for an hour of additional growth, with the objective of reducing the cellular level of the Pth(Ts) protein (3). Glucose was then added, and cell samples were harvested at the times indicated. The TnaC-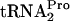 level was then determined. In wild-type strain C600 grown at 42°C, the half-life of TnaC-
level was then determined. In wild-type strain C600 grown at 42°C, the half-life of TnaC-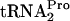 was 11 min. A similar value was obtained for strain C600 pTnaC; the half-life of its TnaC-
was 11 min. A similar value was obtained for strain C600 pTnaC; the half-life of its TnaC- was 9 min. (Fig. 4, compare A and B; also see Fig. 4C). When the TnaC-
was 9 min. (Fig. 4, compare A and B; also see Fig. 4C). When the TnaC-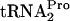 half-life was determined in mutant strains C600[pth(Ts)] and C600[pth(Ts)] pTnaC grown at 42°C, half-life values greater than 20 min were calculated for both strain (Fig. 4, compare A and B; also see C). These findings indicate that TnaC-
half-life was determined in mutant strains C600[pth(Ts)] and C600[pth(Ts)] pTnaC grown at 42°C, half-life values greater than 20 min were calculated for both strain (Fig. 4, compare A and B; also see C). These findings indicate that TnaC-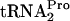 was essentially uncleaved in these strains during the 20-min test period. This important result indicates that peptidyl-tRNA dropoff followed by Pth cleavage is primarily responsible for the processing of this peptidyl-tRNA under these conditions.
was essentially uncleaved in these strains during the 20-min test period. This important result indicates that peptidyl-tRNA dropoff followed by Pth cleavage is primarily responsible for the processing of this peptidyl-tRNA under these conditions.
FIG. 4.
Pth (peptidyl-tRNA hydrolase) is responsible for the cleavage of TnaC- in vivo. Cultures of strains C600 (wild type) and C600[pth(Ts)] (temperature-sensitive Pth) either without (A) or with (B) plasmid pTnaC (+pTnaC) were grown in LB medium {C600[pth(Ts)]} at 30°C to an OD600 of 0.8. They were then shifted to 42°C and incubated for 1 h. Glucose was then added to 1%, and samples were harvested at the indicated times. Cultures of wild-type (wt) strain C600 grown at 42°C were used as controls. Northern blot assays were performed to detect TnaC-
in vivo. Cultures of strains C600 (wild type) and C600[pth(Ts)] (temperature-sensitive Pth) either without (A) or with (B) plasmid pTnaC (+pTnaC) were grown in LB medium {C600[pth(Ts)]} at 30°C to an OD600 of 0.8. They were then shifted to 42°C and incubated for 1 h. Glucose was then added to 1%, and samples were harvested at the indicated times. Cultures of wild-type (wt) strain C600 grown at 42°C were used as controls. Northern blot assays were performed to detect TnaC- (TC-tRNAP) and
(TC-tRNAP) and 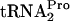 (tRNAP), and the percentage of TnaC-
(tRNAP), and the percentage of TnaC- detected relative to the 0-min value was calculated as described in the legend of Fig. 2. The half-life (t1/2) of TnaC-
detected relative to the 0-min value was calculated as described in the legend of Fig. 2. The half-life (t1/2) of TnaC-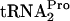 , calculated using the curve in C, is shown to the right of each set of data in A and B. (C) TnaC-
, calculated using the curve in C, is shown to the right of each set of data in A and B. (C) TnaC- decay curves based on the data in A and B. At least two independent experiments were performed, and the TnaC-
decay curves based on the data in A and B. At least two independent experiments were performed, and the TnaC-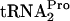 values obtained for any strain and condition varied by less than 15%.
values obtained for any strain and condition varied by less than 15%.
DISCUSSION
Previous studies on the mechanism of tryptophan induction of tna operon expression have established that ribosome recognition of features of the newly synthesized TnaC peptidyl-tRNA, TnaC- , creates a tryptophan binding site in the ribosome at which bound tryptophan inhibits TnaC-
, creates a tryptophan binding site in the ribosome at which bound tryptophan inhibits TnaC- cleavage (1, 2, 6). This uncleaved TnaC-
cleavage (1, 2, 6). This uncleaved TnaC-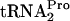 is temporarily retained within the translating ribosome, stalling its movement (7, 9). This stalling at the tnaC stop codon blocks Rho factor binding to tna operon leader RNA, thereby permitting a paused RNA polymerase to continue transcription into the structural genes of the tna operon (7). It has also been shown that tryptophan induction in strains containing a plasmid overexpressing tnaC results in the retention of sufficient uncleaved TnaC-
is temporarily retained within the translating ribosome, stalling its movement (7, 9). This stalling at the tnaC stop codon blocks Rho factor binding to tna operon leader RNA, thereby permitting a paused RNA polymerase to continue transcription into the structural genes of the tna operon (7). It has also been shown that tryptophan induction in strains containing a plasmid overexpressing tnaC results in the retention of sufficient uncleaved TnaC-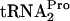 within the translating ribosome to create a
within the translating ribosome to create a 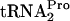 deficiency (9). This deficiency reduces the cell growth rate. Providing additional uncharged
deficiency (9). This deficiency reduces the cell growth rate. Providing additional uncharged  was shown to restore normal growth (9).
was shown to restore normal growth (9).
If tryptophan induction of tna operon expression led to permanent ribosome stalling, induced cells would suffer from the accumulation of inactive ribosomes and a deficiency of 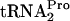 . In previous studies, the half-life of TnaC-
. In previous studies, the half-life of TnaC- in cultures grown under inducing conditions was observed to be 10 to 15 min, implying that E. coli employs some natural mechanism to cleave the accumulated TnaC-
in cultures grown under inducing conditions was observed to be 10 to 15 min, implying that E. coli employs some natural mechanism to cleave the accumulated TnaC-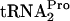 (9). In the present study, the mechanisms used by E. coli to release and cleave ribosome-bound TnaC-
(9). In the present study, the mechanisms used by E. coli to release and cleave ribosome-bound TnaC-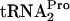 were investigated. It was found that when tnaC was overexpressed in vivo, increased production of RRF plus RF3 reduced the half-life of TnaC-
were investigated. It was found that when tnaC was overexpressed in vivo, increased production of RRF plus RF3 reduced the half-life of TnaC- from 8 min to 2 min (Fig. 3). It was also observed that the inactivation of RRF in a temperature-sensitive frr mutant and the deletion of prfC, the structural gene for RF3, stabilized TnaC-
from 8 min to 2 min (Fig. 3). It was also observed that the inactivation of RRF in a temperature-sensitive frr mutant and the deletion of prfC, the structural gene for RF3, stabilized TnaC-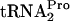 appreciably; its half-life increased from 8 min to >20 min (Fig. 3). It was further established that the principal means of cleavage of TnaC-
appreciably; its half-life increased from 8 min to >20 min (Fig. 3). It was further established that the principal means of cleavage of TnaC- was by the action of the enzyme Pth following TnaC-
was by the action of the enzyme Pth following TnaC- dropoff from the translating ribosome. In a mutant producing a temperature-sensitive Pth grown at an elevated, nonpermissive temperature, the TnaC-
dropoff from the translating ribosome. In a mutant producing a temperature-sensitive Pth grown at an elevated, nonpermissive temperature, the TnaC-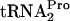 produced was essentially stable (Fig. 4). This stability was observed regardless of whether tnaC was expressed chromosomally or was overexpressed from a plasmid (Fig. 4). These findings suggest that the peptidyl-tRNA dropoff mediated by RRF and RF3 action is the normal mechanism employed by E. coli to relieve tryptophan-induced ribosome stalling at the tnaC stop codon. Dropoff presumably makes the TnaC-
produced was essentially stable (Fig. 4). This stability was observed regardless of whether tnaC was expressed chromosomally or was overexpressed from a plasmid (Fig. 4). These findings suggest that the peptidyl-tRNA dropoff mediated by RRF and RF3 action is the normal mechanism employed by E. coli to relieve tryptophan-induced ribosome stalling at the tnaC stop codon. Dropoff presumably makes the TnaC-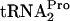 available for Pth cleavage, and this cleavage provides additional
available for Pth cleavage, and this cleavage provides additional 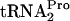 for protein synthesis. RF2 does not appear to play a major role in activating (or inhibiting) TnaC-
for protein synthesis. RF2 does not appear to play a major role in activating (or inhibiting) TnaC-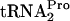 cleavage under tryptophan induction conditions (data not shown). Rather, RRF and RF3 appear to bind to TnaC-
cleavage under tryptophan induction conditions (data not shown). Rather, RRF and RF3 appear to bind to TnaC- -stalled ribosomes and release the TnaC-
-stalled ribosomes and release the TnaC-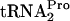 . This would free the previously stalled ribosomes and permit Pth to cleave the TnaC-
. This would free the previously stalled ribosomes and permit Pth to cleave the TnaC-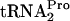 .
.
E. coli is known to use at least three procedures to promote the release of a ribosome stalled at a sense or stop codon: (i) the action of the release factor RF1 or RF2 (26), (ii) RRF/RF3-mediated peptidyl-tRNA dropoff (28), and (iii) trans-translation by tmRNA (17). The in vivo and in vitro experiments described previously suggest that RF2 is ineffective in releasing ribosomes stalled at the tnaC UGA stop codon following the synthesis of TnaC-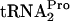 . This is due to bound tryptophan preventing RF2 action (6). Rather, as shown in this study, RRF and RF3 appear to promote TnaC-
. This is due to bound tryptophan preventing RF2 action (6). Rather, as shown in this study, RRF and RF3 appear to promote TnaC- dropoff from the ribosome, and this peptidyl-tRNA is then cleaved by Pth.
dropoff from the ribosome, and this peptidyl-tRNA is then cleaved by Pth.
The possible action of tmRNA on ribosome-bound TnaC-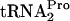 was also studied in vivo and in vitro (data not shown). It was found that tmRNA was incapable of attacking ribosomes containing TnaC-
was also studied in vivo and in vitro (data not shown). It was found that tmRNA was incapable of attacking ribosomes containing TnaC-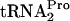 in vivo (overexpression of tmRNA had no observable effect on the β-Gal [TnaA-LacZ] level in induced cells) (Table 2). tmRNA action was also examined in vitro using two truncated inducible and noninducible (Trp12 replaced by Arg12) tnaC mRNAs (data not shown). In both transcripts, the UGA stop codon of tnaC was replaced by UU, with these nucleotides at the 3′ end of the transcript. In reactions performed in the absence of added tryptophan, tmRNA was able to attack the TnaC peptidyl-tRNA produced from either transcript. However, when tryptophan was added, it blocked tmRNA action on the inducible transcript but not on the noninducible transcript. These data provide additional support for the conclusion that Trp12 of TnaC is essential for tryptophan binding and inhibition of TnaC-
in vivo (overexpression of tmRNA had no observable effect on the β-Gal [TnaA-LacZ] level in induced cells) (Table 2). tmRNA action was also examined in vitro using two truncated inducible and noninducible (Trp12 replaced by Arg12) tnaC mRNAs (data not shown). In both transcripts, the UGA stop codon of tnaC was replaced by UU, with these nucleotides at the 3′ end of the transcript. In reactions performed in the absence of added tryptophan, tmRNA was able to attack the TnaC peptidyl-tRNA produced from either transcript. However, when tryptophan was added, it blocked tmRNA action on the inducible transcript but not on the noninducible transcript. These data provide additional support for the conclusion that Trp12 of TnaC is essential for tryptophan binding and inhibition of TnaC-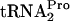 cleavage; they also indicate that tmRNA is not involved in TnaC-
cleavage; they also indicate that tmRNA is not involved in TnaC-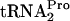 metabolism.
metabolism.
In other unpublished studies, Feng Gong, then in our laboratory, and Akira Kaji showed that RRF plays a role in determining the level of expression of a noninducible tna operon. They found that the expression of a noninducible tnaA′-′lacZ fusion increased when RRF was limiting. They performed these studies using a plasmid with a mutant tnaC gene with the Trp12-to-Arg12 codon replacement. This result suggests that in the absence of RRF, a ribosome translating the tnaC Arg12 sequence is not liberated normally from the tnaC stop codon. It was also shown previously that the overproduction of RF3 influences the expression of tnaA of E. coli; however, the explanation for this observation was not known at that time (31). Previous studies also showed that RRF and RF3 were involved in peptidyl-tRNA dropoff from a translating ribosome (12, 28). In vitro analyses have also suggested that a disruption of prfC, or a down-regulation of RRF expression, results in a decreased rate of peptidyl-tRNA dropoff (12). It was also shown that RF3 can substitute for EF-G in RRF-dependent ribosome recycling reactions in vitro, during the dropoff of a short peptidyl-tRNA (11). The cryoelectron microscopy structure of the translating ribosome suggests that RF3 may induce a conformational change in the ribosome that facilitates the binding of RRF (18). In our studies, other factors such as EF-G or IF3 had no observable effects on either basal or induced tna operon expression (Table 2).
On the basis of studies performed previously by others and the results described in this paper, we propose the following explanation for the events that occur during tryptophan induction of tna operon expression. During induction, features of the TnaC-leader peptidyl-tRNA, most notably, Trp12 and Pro24 (1, 8), create a tryptophan binding site in the A site of the translating ribosome (1). Tryptophan binds at this site, interfering with RF2 action and preventing the cleavage of the newly synthesized TnaC-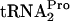 . RF3 and RRF then slowly bind to the ribosome and promote TnaC-
. RF3 and RRF then slowly bind to the ribosome and promote TnaC- dropoff from the ribosome. Pth then hydrolyzes the released TnaC-
dropoff from the ribosome. Pth then hydrolyzes the released TnaC- , freeing
, freeing 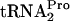 for additional rounds of protein synthesis. The release of TnaC-
for additional rounds of protein synthesis. The release of TnaC-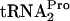 also allows ribosome dissociation from the transcript. Our results raise several additional questions. How is tmRNA prevented from acting on TnaC-
also allows ribosome dissociation from the transcript. Our results raise several additional questions. How is tmRNA prevented from acting on TnaC-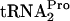 ? What roles do RF3 and RRF play during the liberation of TnaC-
? What roles do RF3 and RRF play during the liberation of TnaC-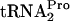 ? Additional studies will be required to answer these questions and identify the exact site(s) and mechanism(s) of action of the various translation termination factors.
? Additional studies will be required to answer these questions and identify the exact site(s) and mechanism(s) of action of the various translation termination factors.
Acknowledgments
We are greatly indebted to Koichi Ito, Yoshikazu Nakamura, Akira Kaji, and Umesh Varshney for providing materials needed for these investigations. We thank Robert Sauer for generous gifts of tmRNA plasmids. We are also grateful to Catherine Squires, Yoshikazu Nakamura, and Akira Kaji for their helpful advice and for their comments on the manuscript. We thank Feng Gong and Akira Kaji for permission to cite their unpublished results.
These studies were supported by National Science Foundation grants MCB-0093023 and MCB-0615390 to C.Y.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Cruz-Vera, L. R., M. Gong, and C. Yanofsky. 2006. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc. Natl. Acad. Sci. USA 103:3598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cruz-Vera, L. R., S. Rajagopal, C. Squires, and C. Yanofsky. 2005. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell 19:333-343. [DOI] [PubMed] [Google Scholar]
- 3.Cruz-Vera, L. R., I. Toledo, J. Hernandez-Sanchez, and G. Guarneros. 2000. Molecular basis for the temperature sensitivity of Escherichia coli pth(Ts). J. Bacteriol. 182:1523-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freistroffer, D. V., M. Y. Pavlov, J. MacDougall, R. H. Buckingham, and M. Ehrenberg. 1997. Release factor RF3 in E. coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 16:4126-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gish, K., and C. Yanofsky. 1993. Inhibition of expression of the tryptophanase operon in Escherichia coli by extrachromosomal copies of the tna leader region. J. Bacteriol. 175:3380-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong, F., K. Ito, Y. Nakamura, and C. Yanofsky. 2001. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNA(Pro). Proc. Natl. Acad. Sci. USA 98:8997-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong, F., and C. Yanofsky. 2002. Analysis of tryptophanase operon expression in vitro: accumulation of TnaC-peptidyl-tRNA in a release factor 2-depleted S-30 extract prevents Rho factor action, simulating induction. J. Biol. Chem. 277:17095-17100. [DOI] [PubMed] [Google Scholar]
- 8.Gong, F., and C. Yanofsky. 2002. Instruction of translating ribosome by nascent peptide. Science 297:1864-1867. [DOI] [PubMed] [Google Scholar]
-
9.Gong, M., F. Gong, and C. Yanofsky. 2006. Overexpression of tnaC of Escherichia coli inhibits growth by depleting
 availability. J. Bacteriol. 188:1892-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
availability. J. Bacteriol. 188:1892-1898. [DOI] [PMC free article] [PubMed] [Google Scholar] - 10.Gottesman, S., E. Roche, Y. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grentzmann, G., P. J. Kelly, S. Laalami, M. Shuda, M. A. Firpo, Y. Cenatiempo, and A. Kaji. 1998. Release factor RF-3 GTPase activity acts in disassembly of the ribosome termination complex. RNA 4:973-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heurgue-Hamard, V., R. Karimi, L. Mora, J. MacDougall, C. Leboeuf, G. Grentzmann, M. Ehrenberg, and R. H. Buckingham. 1998. Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO J. 17:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirokawa, G., R. M. Nijman, V. S. Raj, H. Kaji, K. Igarashi, and A. Kaji. 2005. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA 11:1317-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ichikawa, S., and A. Kaji. 1989. Molecular cloning and expression of ribosome releasing factor. J. Biol. Chem. 264:20054-20059. [PubMed] [Google Scholar]
- 15.Kaji, A., M. C. Kiel, G. Hirokawa, A. R. Muto, Y. Inokuchi, and H. Kaji. 2001. The fourth step of protein synthesis: disassembly of the posttermination complex is catalyzed by elongation factor G and ribosome recycling factor, a near-perfect mimic of tRNA. Cold Spring Harb. Symp. Quant. Biol. 66:515-529. [DOI] [PubMed] [Google Scholar]
- 16.Karimi, R., M. Y. Pavlov, R. H. Buckingham, and M. Ehrenberg. 1999. Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell 3:601-609. [DOI] [PubMed] [Google Scholar]
- 17.Keiler, K. C., P. R. Waller, and R. T. Sauer. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990-993. [DOI] [PubMed] [Google Scholar]
- 18.Klaholz, B. P., A. G. Myasnikov, and M. van Heel. 2004. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature 427:862-865. [DOI] [PubMed] [Google Scholar]
- 19.Kossel, H. 1970. Purification and properties of peptidyl-tRNA hydrolase from Escherichia coli. Biochim. Biophys. Acta 204:191-202. [DOI] [PubMed] [Google Scholar]
- 20.Matsumura, K., K. Ito, Y. Kawazu, O. Mikuni, and Y. Nakamura. 1996. Suppression of temperature-sensitive defects of polypeptide release factors RF-1 and RF-2 by mutations or by an excess of RF-3 in Escherichia coli. J. Mol. Biol. 258:588-599. [DOI] [PubMed] [Google Scholar]
- 21.Menninger, J. R. 1976. Peptidyl transfer RNA dissociates during protein synthesis from ribosomes of Escherichia coli. J. Biol. Chem. 251:3392-3398. [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 23.Moore, S. D., and R. T. Sauer. 2005. Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol. Microbiol. 58:456-466. [DOI] [PubMed] [Google Scholar]
- 24.Roche, E. D., and R. T. Sauer. 2001. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J. Biol. Chem. 276:28509-28515. [DOI] [PubMed] [Google Scholar]
- 25.Roche, E. D., and R. T. Sauer. 1999. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 18:4579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scolnick, E., R. Tompkins, T. Caskey, and M. Nirenberg. 1968. Release factors differing in specificity for terminator codons. Proc. Natl. Acad. Sci. USA 61:768-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seshadri, A., and U. Varshney. 2006. Mechanism of recycling of post-termination ribosomal complexes in eubacteria: a new role of initiation factor 3. J. Biosci. 31:281-289. [DOI] [PubMed] [Google Scholar]
- 28.Singh, N. S., G. Das, A. Seshadri, R. Sangeetha, and U. Varshney. 2005. Evidence for a role of initiation factor 3 in recycling of ribosomal complexes stalled on mRNAs in Escherichia coli. Nucleic Acids Res. 33:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 30.Yanofsky, C., V. Horn, and P. Gollnick. 1991. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J. Bacteriol. 173:6009-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanofsky, C., V. Horn, and Y. Nakamura. 1996. Loss of overproduction of polypeptide release factor 3 influences expression of the tryptophanase operon of Escherichia coli. J. Bacteriol. 178:3755-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zavialov, A. V., R. H. Buckingham, and M. Ehrenberg. 2001. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107:115-124. [DOI] [PubMed] [Google Scholar]
- 33.Zavialov, A. V., L. Mora, R. H. Buckingham, and M. Ehrenberg. 2002. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell 10:789-798. [DOI] [PubMed] [Google Scholar]