Abstract
Proteases play a crucial role in remodeling the bacterial proteome in response to changes in cellular environment. Escherichia coli ZntR, a zinc-responsive transcriptional regulator, was identified by proteomic experiments as a likely ClpXP substrate, suggesting that protein turnover may play a role in regulation of zinc homeostasis. When intracellular zinc levels are high, ZntR activates expression of ZntA, an ATPase essential for zinc export. We find that ZntR is degraded in vivo in a manner dependent on both the ClpXP and Lon proteases. However, ZntR degradation decreases in the presence of high zinc concentrations, the level of ZntR rises, and transcription of the zntA exporter is increased. Mutagenesis experiments reveal that zinc binding does not appear to be solely responsible for the zinc-induced protection from proteolysis. Therefore, we tested whether DNA binding was important in the zinc-induced stabilization of ZntR by mutagenesis of the DNA binding helices. Replacement of a conserved arginine (R19A) in the DNA binding domain both enhances ZntR degradation and abolishes zinc-induced transcriptional activation of zntA. Biochemical and physical analysis of ZntRR19A demonstrates that it is structurally similar to, and binds zinc as well as does, the wild-type protein but is severely defective in binding DNA. Thus, we conclude that two different ligands—zinc and DNA—function together to increase ZntR stability and that ligand-controlled proteolysis of ZntR plays an important role in fine-tuning zinc homeostasis in bacteria.
The intracellular levels of transcription factors are critical for the modulation of gene expression, and therefore their levels must be tightly controlled. Proteolysis is used by all forms of life both for shaping the proteome in response to changes in environmental conditions and for protein quality control by degrading damaged, misfolded, or mislocalized proteins. Regulated proteolysis is an important mechanism for controlling the levels of transcriptional regulators in response to environmental stimuli. Degradation of key regulatory proteins influences multiple stress responses in Escherichia coli (15, 21), progression of the cell cycle in Caulobacter crescentus (12), and competence and sporulation in Bacillus subtilis (22, 44). In this study we investigate the role of regulated proteolysis in zinc homeostasis in E. coli.
Zinc is an essential trace element and a key structural component of a large number of proteins (1, 7). Zinc also serves as an essential cofactor in numerous enzymes and regulatory proteins (8, 9). However, excess zinc is toxic and therefore intracellular zinc levels must be kept tightly in check. In prokaryotes, zinc homeostasis is mainly achieved by regulating the uptake and efflux of zinc (16). The primary zinc import system in E. coli is ZnuABC, an ABC-type transporter (33, 34), and the primary zinc export system is the P-type ATPase ZntA (37). Zinc import by the ZnuABC transporter is regulated by Zur, a zinc-responsive homolog of the iron uptake regulator Fur. In the presence of zinc, Zur binds to the znu operator and acts as a repressor. In the absence of zinc, Zur does not bind DNA and therefore fails to compete with RNA polymerase, and transcription proceeds (30). In contrast, transcription of zntA is activated by ZntR, a member of the MerR family of metal-responsive transcriptional regulators. The binding of zinc to ZntR converts it into a strong transcriptional activator of the zntA gene (31), resulting in increased efflux of zinc. Based on the finding that there is no substantial pool of free zinc in the E. coli cytoplasm under normal growth conditions, the transcription of the zinc efflux or uptake system is considered to be activated by femtomolar concentrations of free zinc (30).
In this study we address factors that control changes in intracellular ZntR levels and thus likely influence zinc homeostasis. ZntR was identified as a substrate captured by an inactive variant of ClpXP (ClpXPtrap) (29), indicating that it is a likely substrate for degradation by the ClpXP protease. ClpXP is one of the five energy-dependent cytoplasmic proteases in E. coli (13). ClpXP is known to recognize, unfold, and degrade a number of important regulatory proteins (17, 24, 39).
We investigate the proteases involved in ZntR proteolysis and the role of two functional ligands in modulating the degradation of ZntR. We demonstrate that ZntR is an in vivo substrate of both ClpXP and Lon proteases and establish that it is directly degraded by Lon in vitro. We also show that ZntR binding to DNA protects ZntR against proteolysis and its stability is further increased in the presence of added zinc. Binding to DNA couples ZntR stabilization against degradation with zinc-induced activation of zntA transcription, which in turn triggers increased efflux of zinc. Therefore, we propose that ligand-controlled turnover of ZntR via ATP-dependent proteolysis contributes to the robust feedback loop that maintains appropriate intracellular zinc levels.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strain MC4100 or MC4100 derivatives were grown at 37°C in Luria-Bertani medium (LB) (27) or EZ rich defined medium (Teknova) supplemented with 0.4% glycerol as a carbon source.
Construction of zntR::cat mutant.
The MC4100 zntR mutant was constructed by one-step inactivation using the λ Red system (10). The chromosomal zntR gene was replaced with the cat gene generated by PCR using pKD3 DNA as a template and the following primers with homology extensions to the noncoding regions located upstream and downstream of the zntR gene: 5′-GACAAGTTGTTGGACAAAATCAACGATAACTAGTGGAGTATGCATATGAATATCCTCCTTAG-3′ and 5′-GGTTATTTAACGGCGCGAGTGTAATCCTGCCAGTGCAAAAGTGTAGGCTGGAGCTGCTTC-3′.
DNA manipulation.
For PCRs, restriction digestions, ligations, transformations, and agarose gel electrophoresis, standard genetic and molecular techniques were followed (38, 42). Plasmid DNA preparations and recovery of DNA from agarose gels were performed with QIAGEN kits. The nucleotide sequence of the insert and adjacent region was confirmed by DNA sequencing (MIT/CCR/HHMI Biopolymers Laboratory) for all constructs generated for this study.
ZntR cloning, purification, and preparation of a polyclonal antibody.
The ZntR coding region was amplified from chromosomal DNA by PCR using the primers 5′-CTAGTGGAGTACATATGTATCGCATTGG-3′ (NdeI) and 5′-GTAATCCTGCGGATCCAAAAAATCAACAACC-3′ (BamHI). After digestion with NdeI and BamHI the PCR fragment was cloned into digested pET-11a (Novagen), resulting in pMP4. For expression of ZntR, pMP4 was transformed into the bacterial strain BL21(DE3) carrying the pLysS plasmid (Novagen). Cells were grown in LB with ampicillin (100 μg ml−1) and chloramphenicol (30 μg ml−1) at 25°C to an optical density at 600 nm (OD600) of 0.6 before IPTG (isopropyl-β-d-thiogalactopyranoside) was added. After a 4-h induction, the cells from a 2-liter culture were harvested by centrifugation and ZntR was purified as described previously (31). Samples with a protein concentration of 77 μM were used for rabbit polyclonal antibody production (Covance Research Products).
pMP32 (carrying the untagged ZntR) was constructed by ligating the NheI/HindIII-treated PCR fragment (containing the ZntR coding region) into pBAD18 vector digested with the same restriction enzymes. The PCR fragment was generating using the primers 5′-CAAAATCAACGCTAGCTAGAGGAGAATGTATG-3′ (NheI), containing the Shine-Dalgarno sequence, and 5′-CGAGTGTAAGCTTGCCAGTGCAAAAAATC-3′ (HindIII).
Expression and purification of tagged ZntR protein.
N- and C-terminally polyhistidine (six-His)-tagged variants of ZntR were constructed using the pBAD/His or pBAD/Myc-His vector (Invitrogen), respectively. For N-terminally six-His-tagged ZntR, a PCR fragment was generated using chromosomal DNA as a template and the primers 5′-GAAGATCTTGATGTATCGCATTGGTGAGCTGG-3′ (BglII) and 5′-GCGAATTCTCAACAACCACTCTTAACGCCAC-3′ (EcoRI). The BglII/EcoRI-cleaved PCR product was ligated into the equally treated pBAD/His vector. The same template and the primers 5′-TCGCCTAGGATGTATCGCATTGGTGAGCTGG-3′ (AvrII) and 5′-GCGAATTCGCACAACCACTCTTAACGCCACTCG-3′ (EcoRI) were used to amplify the PCR product for generation of the C-terminally six-His-tagged ZntR. After digestion with AvrII and EcoRI, the PCR fragment was cloned into the pBAD/Myc-His vector cleaved with the same restriction enzymes.
The pBAD/Myc-His vector carrying the C-terminally six-His-tagged ZntR was transformed into Top10 cells (Invitrogen). Cells were grown in LB with ampicillin (100 μg ml−1) at 30°C to an OD600 of 0.6 before induction with arabinose. After an additional 4-h growth period, the cells from a 2-liter culture were harvested by centrifugation. The pellet was resuspended in lysis buffer (50 mM Tris, pH 8, 0.1% Tween 20, 10 mM β-mercaptoethanol, 300 mM NaCl), and the cells were disrupted with a French press. The whole-cell extract was centrifuged for 30 min at 16,000 rpm (Sorvall SS-34 rotor), and the supernatant was incubated (for 1 h at 4°C) with 1.5 ml nickel-nitrilotriacetic acid resin (QIAGEN) equilibrated in lysis buffer. After mixing, the resin was packed into a column and washed with 200 ml wash buffer (50 mM Tris, pH 8, 0.1% Tween 20, 10 mM β-mercaptoethanol, 300 mM NaCl, 20% glycerol, 20 mM imidazole). The C-terminally six-His-tagged ZntR was eluted with 10 ml elution buffer (50 mM Tris, pH 8, 0.1% Tween 20, 10 mM β-mercaptoethanol, 300 mM NaCl, 10% glycerol, 250 mM imidazole). The eluted protein was loaded onto a Sephacryl S-100 high-resolution gel filtration column equilibrated in storage buffer (50 mM Tris, pH 8, 250 mM NaCl, 5 mM dithiothreitol [DTT], 5% glycerol).
The N-terminally six-His-tagged derivatives of ZntR contain an Xpress epitope, whereas the C-terminally six-His-tagged variants carry a c-Myc epitope.
Generation of ZntR mutants.
For construction of different deletions and mutations in the zntR gene, the QuikChange II site-directed mutagenesis kit (Stratagene) was used (primer sequences are available upon request).
Construction of chromosomal zntA′::lacZ(op) fusion.
Chromosomal lacZ fusion to zntA was isolated using the fusion vector pRS415 (43). To generate the zntA′::lacZ transcriptional fusion, a zntA fragment containing 129 nucleotides of the noncoding region upstream of zntA (including the promoter region of zntA) and 294 nucleotides from the 5′ end of the zntA coding region was amplified by PCR. The primers 5′-GGATAACGCGAATTCTGCGGCCTGCT-3′ (EcoRI) and 5′-GCAGCGCGGATCCAACTTATGCACGAATG-3′ (BamHI) containing a stop codon for zntA′ and the chromosomal MC4100 DNA as a template were used for PCR. The EcoRI/BamHI-treated fragment was cloned into pRS415 digested with the same restriction enzymes, resulting in pMP5. This transcriptional fusion [zntA′::lacZ(op)] was transferred to the att site of MC4100 via λRS45 as described previously (43). Monolysogens were identified by whole-colony PCR (35).
β-Galactosidase assay.
β-Galactosidase activity was assayed using o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate and is reported as micromoles of o-nitrophenol per minute per milligram of cellular protein (27).
Degradation and Western blotting assays.
Protein half-lives (t1/2s) were determined using samples from exponentially growing cells. For proteins expressed from pBAD plasmids, synthesis was induced for 30 min at an OD600 of 0.3 by addition of arabinose to 0.005%. For the degradation of proteins in the presence of added zinc, ZnSO4 (Fluka; ≥99.9% purity) was added to the cultures (0.5 mM final concentration) concomitantly with the addition of arabinose. Protein synthesis was stopped by addition of 100 μg/ml chloramphenicol, and the samples were taken at different time points as described in the respective figure legends. Following the addition of ice-cold trichloroacetic acid to a 10% concentration, the samples were harvested by centrifugation, washed with 100% acetone, and resuspended in Tris-Tricine loading buffer to an OD600 of 10. The protein levels were detected by Western blotting using polyclonal antibodies to ZntR, Xpress, or c-Myc epitope and an ECF substrate (Amersham).
Degradation in vitro.
Lon6 (0.6 μM), ATP (16 mM), and an ATP regeneration system (200 μg/ml creatine kinase and 20 mM creatine phosphate) were mixed in Lon degradation buffer (50 mM Tris, pH 8, 15 mM MgCl2, 5 mM KCl, 1 mM DTT) and incubated for 2 min at 37°C. For all degradation experiments, 5 μM of protein was added, and samples were removed at specific times and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis.
Zinc content determination.
The metallochromic indicator 4-(2-pyridylazo)resorcinol (PAR) was used for determination of zinc content of the purified C-terminally six-His-tagged ZntR proteins (19). The zinc-bound form of the C-terminally six-His-tagged wild-type ZntR and ZntRR19A were prepared under aerobic conditions using 2 to 4 equivalents of ZnSO4. To calculate the fraction of zinc released, the absorbance of the zinc-PAR complex was read at 500 nm and normalized against an identical sample containing 1 mM methyl methanethiosulfonate. Methyl methanethiosulfonate alkylates cysteine residues and releases the zinc bound to the protein.
The growth media were also spectrophotometrically tested for metal contamination by the addition of PAR and EDTA (19).
Electrophoretic mobility shift assay.
For the gel mobility shift assays a DNA fragment encoding the zntA promoter region, which contains the ZntR binding site, was generated by PCR. The primers 5′-GTCCGCTCGCTGTATCTCTG-3′ and 5′-CATCCTCCGGTTAAGTTTTTTC-3′, and pMP5 as a template, were used for PCR. The binding of ZntR to the defined PCR fragment including the zntA regulatory region was carried out in a 20-μl reaction mixture containing increasing amounts of the purified C-terminally six-His-tagged wild-type ZntR or ZntRR19A protein (from 0.65 μM to 26 μM) and 50 nM of DNA, as previously described (32).
CD and GdnHCl denaturation.
Circular dichroism (CD) spectra of purified proteins (36 μM) were recorded at 25°C using an Aviv Associates (Lakewood, NJ) model 62DS instrument. Far-UV CD spectra from 200 nm to 250 nm (step size, 1 nm; averaging time, 5 s) were corrected by subtracting a corresponding buffer blank. The CD buffer contained 10 mM potassium phosphate, pH 8.0, 2 mM DTT, and 5% glycerol. CD signals were recorded as residue ellipticity (θmrw).
Protein samples (12 μM) containing different GdnHCl concentrations (from 0 to 7 M) were prepared using appropriate dilutions of the stock solution (8 mM). Ellipticity at 220 nm was monitored at 25°C for each sample as an average of 60 data points taken at 1-s intervals after 4 min of sample mixing. The molar fraction of folded peptide was calculated as previously described (28).
RESULTS
clpX and lon mutants increase ZntR stability.
ZntR was identified by a proteomic study (29) as an in vivo ClpXPtrap-associated protein. To understand why this protein was captured and to identify proteases involved in ZntR degradation, we measured the rate of in vivo degradation of ZntR protein in wild-type and several protease mutant cells. Because endogenous ZntR protein could not be detected by Western blotting using a polyclonal antibody to ZntR (data not shown), N-terminally six-His-tagged or untagged ZntR was expressed from a plasmid containing an arabinose-inducible promoter (see Materials and Methods). After a 30-min induction, protein synthesis was stopped by addition of chloramphenicol and the rates of ZntR proteolysis were determined by Western blotting. In wild-type cells, untagged ZntR was degraded with a t1/2 of about 30 min (Fig. 1A and B). The N-terminally six-His-tagged variant was more rapidly degraded, with a t1/2 of about 9 min (see below). Enhanced degradation by cooperative interactions between intrinsic degradation signals and added regions of flexible polypeptide sequence appears to be common (18, 36); this is likely the explanation for the increased degradation rate observed with the tagged protein. The disruption of clpX and lon genes increased the stability of both the six-His-tagged (data not shown) and untagged forms of ZntR (Fig. 1A and B), indicating that ZntR is likely degraded by both ClpXP and Lon proteases in vivo. As the protein was substantially stabilized in each of these mutant strains, ClpXP and Lon are likely the major proteases involved in ZntR degradation. Based on our observations that six-His-tagged ZntR behaves similarly to the untagged protein, we decided to use the tagged form for many of the following experiments. However, critical conclusions were confirmed using the untagged ZntR (see below).
FIG. 1.
ZntR is a ClpXP and Lon substrate and is stabilized in the presence of added zinc in vivo. Untagged ZntR was expressed from a pBAD promoter in MC4100 (circles in panels B and D) and its clpX::kan (squares in panel B) or lon::kan (diamonds in panel B) derivative. The strains were grown in rich defined medium, and at an OD600 of 0.3, arabinose and ZnSO4 (when specified; closed symbols in panel D) were added. After a 30-min induction, protein synthesis was inhibited by the addition of chloramphenicol, and samples for Western blots (shown in panels A and C) were removed at specific time points. Data obtained from quantitative Western blot analysis are shown in panels B and D. wt, wild type.
To determine if ZntR is directly recognized and degraded by Lon and ClpXP, we purified the C-terminally six-His-tagged ZntR protein for in vitro studies. ZntR was efficiently degraded by Lon in vitro, in a reaction requiring ATP (Fig. 2A and B). However, we failed to observe robust degradation of the purified ZntR by ClpXP in vitro (data not shown). These results suggest that an adaptor protein may be needed to deliver ZntR to ClpXP protease, as is the case for several well-characterized ClpXP substrates (39).
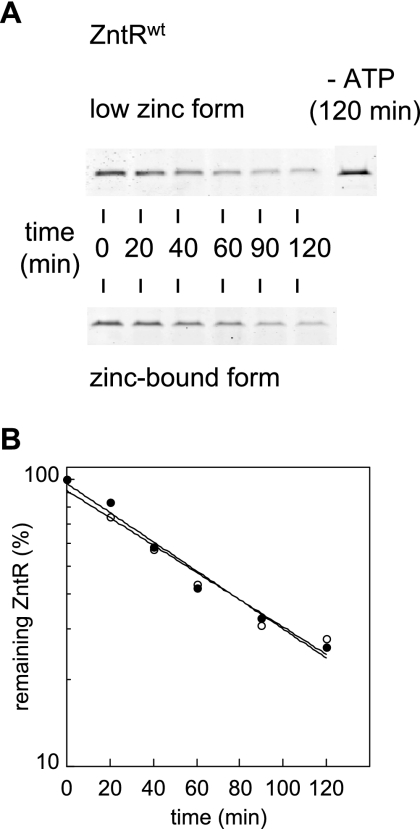
FIG. 2.
Lon degradation of ZntR in vitro. (A) Degradation of the purified low-zinc form and the prepared zinc-bound form of the C-terminally six-His-tagged wild-type ZntR by the Lon protease in vitro. (B) Quantification of the ZntR levels from the SDS-polyacrylamide gels in panel A (open circles represent the low-zinc form and closed circles represent the zinc-bound form of the wild-type ZntR).
ZntR is more stable in the presence of added zinc.
Transcription of the zntA gene is strongly activated by the zinc-bound form of ZntR (31). To investigate the impact of metal binding on ZntR recognition by proteases, we determined the in vivo degradation rate of untagged ZntR in the presence and absence of added zinc. Zinc (ZnSO4), at a concentration of 0.5 mM that is known not to affect cell growth (3, 46), was added to the culture simultaneously with arabinose to induce ZntR protein expression. ZntR levels were then monitored following arrest of protein synthesis 30 min after zinc addition.
In the presence of added zinc, the t1/2 of untagged ZntR was increased (t1/2 of >60 min) more than twofold compared to its stability (t1/2 of ∼30 min) in the culture grown in the absence of added zinc (Fig. 1C and D). Also, the stability of the N-terminally six-His-tagged variant was enhanced in the presence of added zinc, from 9 min to 15 min (see Fig. 3A). Thus, the zinc-bound protein appears more stable to proteolysis than the apo or “low-zinc” form of the protein. We prefer to use the term “low zinc” as, based on our measurements, the medium contains an estimated concentration of 80 to 200 nM heavy metal (see Materials and Methods), and at least some of this trace metal is likely to be zinc; thus, some fraction of the ZntR is very likely zinc bound, even in the condition of no added zinc.
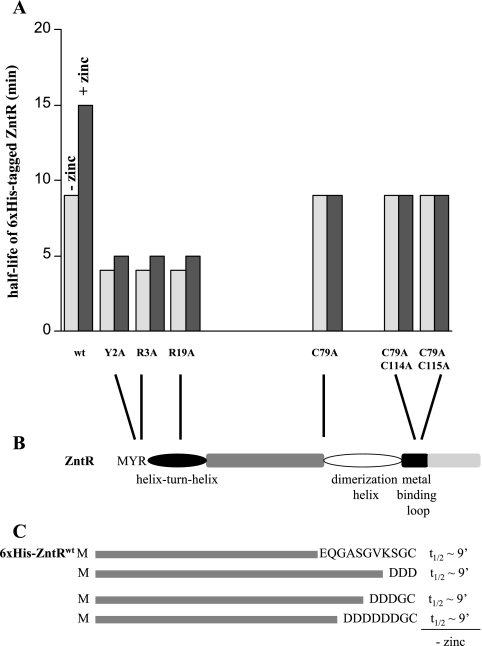
FIG. 3.
Substitutions at the N terminus decrease ZntR stability. (A) The t1/2s of several N-terminally six-His-tagged ZntR variants with amino acid substitutions in the DNA binding motif or in the residues known to coordinate zinc. wt, wild type. (B) Schematic representation of ZntR domains (based on UniProtKB/Swiss-Prot prediction and reference 5). (C) Illustration of the N-terminally six-His-tagged variants of ZntR carrying substitutions at the C terminus and their t1/2s (in minutes).
Previous experiments established that zntA transcription is induced in wild-type E. coli 5 min after zinc addition and that after 30 min the level of zntA transcript decreased to the level prior to addition of zinc (46). We were interested in whether ZntR stability would change on a shorter timescale after the addition of zinc. Therefore, we also tested ZntR degradation 5 and 10 min after zinc addition. Under these experimental conditions, ZntR was found to have a t1/2 similar to that measured 30 min after zinc addition (data not shown). In our experiments ZntR was overexpressed; therefore, in the experiments performed by Yamamoto and Ishihama (46), the levels of endogenous zinc-bound ZntR may have fluctuated more rapidly than we observed.
To start to address the mechanism of increased stability of ZntR in the presence of added zinc, we first replaced the residues known to coordinate zinc. The crystal structure of ZntR missing the N-terminal 43 or 45 amino acid residues from each monomer was recently solved (5). ZntR binds two zinc ions per monomer, with one atom coordinated by C114 and C124 of a metal-binding loop and C79 from the other monomer. The other zinc ion is coordinated with C115 and H119 of the metal-binding loop and C79 from the other monomer. Thus, C79 connects the metal-binding domain of one monomer and the dimerization domain of the second monomer. ZntR derivatives with single amino acid substitutions (C79A/S, C114A/S, C115A/S, or H119A) were previously assayed for their response to added zinc by measuring activation of zntA transcription (23). The replacement of the cysteines abolished transcriptional induction of zntA in the presence of added zinc.
Our N-terminally six-His-tagged ZntR mutant proteins, containing single or double amino acid substitutions (C79A, C79A/C114A, or C79A/C115A), were not stabilized by zinc in vivo (Fig. 3A). These data suggest that zinc binding mutants lose the zinc responsiveness of the stability against proteolysis. However, the six-His-tagged mutant proteins were all degraded with a rate similar to that of the six-His-tagged wild-type ZntR in the absence of added zinc (Fig. 3A); thus, the proteins are not hyperunstable, suggesting that their structures were not grossly altered.
Zinc binding to ZntR protein is not sufficient to protect ZntR against degradation.
To test whether increased ZntR stability was due solely to zinc binding to the protein, we performed in vitro degradation experiments using the low-zinc or zinc-bound form of the C-terminally six-His-tagged ZntR. First, we determined the zinc content of the purified C-terminally six-His-tagged wild-type ZntR using an assay employing the metallochromic indicator PAR and found that it contains 0.075 ± 0.01 zinc/monomer. In contrast, the zinc-bound form of this protein (prepared under aerobic conditions) contained 0.75 ± 0.075 zinc/monomer. These data are in concert with the previously published results for the purified untagged ZntR, which contained less than 0.05 zinc/monomer for the low-zinc and 0.9 ± 0.3 zinc/monomer for the zinc-bound forms, respectively (31).
Lon in vitro degradation of the low-zinc or zinc-bound form of the C-terminally six-His-tagged ZntR showed that the two proteins were degraded with similar t1/2s of about 50 min (Fig. 2A and B). Therefore, zinc binding to ZntR is not sufficient to protect ZntR against degradation by the Lon protease, although our in vivo experiments showed an increase of ZntR stability in the presence of added zinc. These data suggest that another ligand acts together with zinc binding to stabilize ZntR in vivo.
Dissection of the protease recognition signals within ZntR.
A recent proteomic analysis of ClpXP-trapped proteins revealed five classes of ClpX recognition motifs that are bound by the enzyme as unstructured peptides and are usually located near either the N or the C terminus of the substrate protein (11). The Lon protease has also been reported to recognize terminal signals (20, 41).
ZntR does not appear to carry any of these characterized signals. To identify potential recognition sequences, we constructed and expressed several variants of ZntR carrying N- or C-terminal epitope tags and deletions or substitutions at the N or C terminus and measured the rate of in vivo proteolysis for each of these protein variants. The N-terminally six-His-tagged variants with deletion of 5 or 8 amino acids from the C terminus or carrying variable residues at the C terminus (DDD, DDDGC, or DDDDDDGC) were each degraded with a t1/2 (of about 9 min) similar to that of the N-terminally six-His-tagged wild-type ZntR (Fig. 3C). These results suggested that the C terminus is not important for ZntR recognition. In contrast, substitutions at the N terminus (Y2A or R3A) of the N- or C-terminally six-His-tagged ZntR resulted in more rapid degradation of ZntR (Fig. 3A). These six-His-tagged ZntR mutant proteins were degraded with a t1/2 of about 4 min. A potential explanation for these results is that the amino acid replacements may compromise the ability of ZntR to bind DNA, as the helix-turn-helix motif involved in site-specific DNA binding is predicted to start at residue 4 of the protein (Fig. 3B). These data suggest that potential recognition signals may be located at or near the N terminus. Perhaps these determinants overlap with the helix-turn-helix motif, making a mutational analysis of the protease recognition signal(s) difficult.
Next, we wanted to know whether the N-terminally six-His-tagged ZntR variants carrying replacements at the N terminus (Y2A or R3A) were responsive to zinc. These mutants, probably compromised in their ability to bind DNA, were less stable than the N-terminally six-His-tagged wild-type ZntR protein in both the presence and the absence of added zinc (Fig. 3A). These data lead us to hypothesize that the interactions of ZntR with both zinc and DNA play roles in protecting the protein from destruction by ATP-dependent proteases. How these protein-ligand interactions function together to stabilize the protein is probed further below.
zntA transcription is not activated by ZntRR19A.
Previous studies showed that both the apo and zinc-bound forms of ZntR bind to DNA (31). However, ZntR dramatically activates the transcription of the zntA gene only in the presence of added zinc (2).
ZntR is a member of the MerR family transcriptional regulators, which activate transcription through distortion of the DNA at the center of the operator (4). Alignment of the N-terminal helix-turn-helix DNA binding regions of the MerR family members revealed several conserved positively charged amino acids (23). To isolate a mutant variant defective in operator binding, we replaced in ZntR one of these conserved residues, arginine 19, with alanine and tested ZntRR19A activation of a chromosomal zntA::lacZ transcriptional fusion.
ZntRR19A abolished zinc-induced activation of zntA in vivo. The expression of the zntA::lacZ operon fusion was measured in a zntR mutant strain, with N-terminally six-His-tagged wild-type ZntR or ZntRR19A expressed from a pBAD vector in the presence or absence of added zinc. In the presence of added zinc, wild-type ZntR activated the expression of the zntA fusion more than 10-fold over the activity measured in the absence of added zinc (Fig. 4). When ZntRR19A was expressed from the plasmid, no significant increase in the expression of zntA fusion was observed in the presence of added zinc (Fig. 4). The R19A substitution did not affect the ability of ZntR protein to bind zinc. The purified C-terminally six-His-tagged ZntRR19A contained 0.075 ± 0.01 zinc/monomer, as did the C-terminally six-His-tagged wild-type protein. Likewise, the zinc-bound form, prepared under aerobic conditions as was the six-His-tagged wild-type ZntR, has the same content of 0.74 ± 0.06 zinc/monomer.
FIG. 4.
ZntRR19A does not activate zntA transcription. N-terminally six-His-tagged wild-type ZntR and ZntRR19A were expressed from a pBAD promoter in the MC4100 zntR::cat derivative carrying the transcriptional zntA::lacZ fusion. Specific β-galactosidase activities were determined 30 min after induction of ZntR with arabinose (0.005% final concentration) in the presence (hatched bars) or absence (open bars) of ZnSO4 (0.5 mM final concentration). The data are the averages of three independent cultures with a standard deviation of 0.001 in the absence of added zinc and 0.01 or 0.002 in the presence of added zinc expressing wild-type ZntR or ZntRR19A, respectively.
The lack of zntA activation by ZntRR19A in the presence of added zinc suggests that this mutant protein is defective in binding to operator DNA due to disruption in the helix-turn-helix motif. Thus, this mutant was attractive for further probing the link between DNA binding and zinc-controlled proteolysis.
ZntRR19A does not bind DNA.
ZntR dimer interacts with the palindromic sequence located in the suboptimal spacer, between the −35 and −10 region, of the zntA promoter region, to make the zntA promoter a better substrate for the RNA polymerase (2). To determine whether ZntRR19A can bind specifically to DNA, gel mobility shift assays were performed (Fig. 5A and B). Increasing concentrations of purified C-terminally six-His-tagged wild-type ZntR or ZntRR19A proteins (from 0.65 μM to 26 μM) were incubated with a defined 92-nucleotide DNA fragment containing the zntA regulatory region. A protein-DNA complex was clearly detected when the wild-type protein was incubated with the DNA fragment (Fig. 5A), whereas no complex band was observed when the same DNA fragment was incubated with the purified ZntRR19A (Fig. 5B). There is no published information on the Kd of ZntR for DNA, but the protein concentrations used in our experiments are in excess compared to that (75 nM) used by Outten et al. (31) for their footprinting experiments, indicating that ZntRR19A is severely defective in DNA binding. Our data confirm that the lack of activation of zntA transcription by ZntRR19A (Fig. 4) can be explained by compromised binding to zntA operator DNA.
FIG. 5.
ZntRR19A does not bind DNA and is properly folded. (A and B) Gel shift assays using purified ZntR protein. The zntA promoter fragment was incubated without protein and with increasing amounts of purified C-terminally six-His-tagged wild-type ZntR (A) or ZntRR19A (B) protein (0.65, 1.3, 2.6, 4, 8, 13, and 26 μM) or with bovine serum albumin (BSA). The DNA-protein complexes (formed using wild-type ZntR) are shown by arrows. (C) Far-UV CD spectra of C-terminally six-His-tagged wild-type ZntR (open circles) and ZntRR19A (closed circles). (D) Thermodynamic stability of ZntR protein probed by GdnHCl denaturation (open circles represent C-terminally six-His-tagged wild-type ZntR and closed circles represent C-terminally six-His-tagged ZntRR19A).
ZntRR19A is properly folded.
CD spectroscopy was used to confirm that the inability of C-terminally six-His-tagged ZntRR19A to bind DNA is due to a change in affinity and not to unfolding of the protein. Far-UV CD spectra were collected to detect potential changes in the secondary structure introduced by replacing R19 from the DNA binding domain. The presence of two minima at 208 nm and 222 nm demonstrates that both proteins have substantial alpha-helix content (Fig. 5C). The spectrum of the mutant protein is nearly identical to that of the wild type, indicating that ZntR secondary structure was not affected by the R19A substitution.
As a further control, the thermodynamic stability of the proteins was assessed through chemically induced unfolding of C-terminally six-His-tagged wild-type ZntR and ZntRR19A, which was monitored by CD at 220 nm. The denaturation profiles of the two proteins were virtually identical and showed the same denaturation midpoint of 2.6 M GdnHCl (Fig. 5D). These data demonstrate that ZntRR19A is properly folded.
The R19A substitution accelerates degradation and destroys the zinc responsiveness of ZntR proteolysis in vivo.
We established that ZntRR19A is defective in activation of zntA transcription and DNA binding but remains properly folded. Therefore, we used this protein to test the hypothesis that DNA binding is important for zinc-regulated stability of ZntR in vivo.
ZntRR19A was more rapidly degraded in vivo (Fig. 6A and B). This untagged mutant protein was degraded with a t1/2 of about 15 min, approximately twofold faster than the wild-type untagged ZntR (t1/2 of ∼30 min). Whereas wild-type ZntR was more stable upon addition of zinc (t1/2 of >60 min), no significant changes in the rate of proteolysis could be observed for ZntRR19A. These data indicate that the binding to DNA is needed for zinc-enhanced ZntR stability. In vitro degradation by Lon protease of the low-zinc or zinc-bound form of the C-terminally six-His-tagged ZntRR19A showed that the two forms are degraded with a t1/2, of about 50 min, similar to that of the C-terminally six-His-tagged wild-type ZntR (Fig. 6C and D). These data argue against the explanation that ZntRR19A is more rapidly degraded in vivo because it is intrinsically a better protease substrate and therefore support our conclusion that enhanced degradation observed in vivo is due to the defect in DNA binding. The influence of DNA on Lon degradation of the mutant and wild-type ZntR could not be investigated in vitro, as Lon protease binds nonspecifically to nucleic acid and its enzymatic activity is generally inhibited (reference 6 and our unpublished observations), an effect that confounds the interpretation of these experiments.
FIG. 6.
Effect of R19A replacement on ZntR stability. (A) R19A substitution reduces ZntR stability in vivo. Strain MC4100 carrying untagged ZntRR19A under pBAD control was grown in rich defined medium. At an OD600 of 0.3, arabinose and ZnSO4 (when specified; closed symbols in panel B) were added. The cells were grown for an additional 30 min followed by the addition of chloramphenicol to stop protein synthesis. Samples for Western blots were taken at specified time points. (B) Quantification of the levels of ZntR in panel A (diamonds represent ZntRR19A in comparison with the data for the wild-type untagged ZntR, represented by circles). (C) Degradation of the C-terminally six-His-tagged ZntRR19A by Lon in vitro. (D) Quantification of the ZntR levels from the SDS-polyacrylamide gels shown in panel C (open diamonds represent low-zinc form and closed diamonds represent zinc-bound form of ZntRR19A).
DISCUSSION
The intracellular levels of transcriptional regulators are critical for cellular physiology and homeostasis. Regulated proteolysis of transcriptional factors involved in the influx or efflux of metal ions contributes to metal homeostasis.
In this study we have shown that ZntR is degraded in vivo by the ClpXP and Lon proteases (Fig. 1). The ATP-dependent proteases typically recognize their substrate proteins by binding to short unstructured peptides located usually at either the N or the C terminus (11, 14, 20, 40, 41). Our deletion and mutational analyses demonstrate that the C terminus is not important for ZntR recognition by those proteases. The identification of potential recognition signals at or near the N terminus was difficult because any important determinants located in this region appear likely to overlap with the DNA binding helices. In fact, we find that the protein is preferentially degraded when defective in DNA binding, consistent with the idea that the protease recognition signals and the DNA binding determinants may overlap. In the absence of added zinc, untagged ZntR is degraded with a t1/2 of about 30 min, whereas the addition of high zinc concentrations to the culture results in a more-than-twofold increase in ZntR t1/2 (Fig. 1C and D). ZntR has a femtomolar sensitivity to free zinc (30); therefore, the ZntR t1/2 of about 30 min in the absence of added zinc is likely an underestimate of the turnover rate of the apo form, as there is certainly some zinc present in the medium and in the environment under these experimental conditions.
The in vivo trapping and degradation experiments strongly indicate that the ClpXP and Lon proteases are responsible for turnover of ZntR. We were able to confirm that ZntR is a Lon substrate by in vitro degradation experiments (Fig. 2). We were not able to find robust in vitro degradation by ClpXP. Therefore, the in vivo degradation defect seen in a clpX mutant could be indirect. However, we favor the idea that ZntR is a direct substrate, as it is trapped by ClpXPtrap in vivo and stabilized in a clpX mutant. Perhaps it needs an adaptor protein to be efficiently recognized, explaining its inefficient degradation by purified ClpXP.
The mechanistic basis for the increase of ZntR stability in the presence of added zinc was dissected by replacing the amino acids in ZntR known to coordinate zinc. These substitutions do not affect ZntR recognition and proteolysis in the absence of added zinc, but they cause the loss of zinc-enhanced stability observed with the N-terminally tagged wild-type protein upon zinc addition (Fig. 3A). In contrast, replacement of the conserved arginine 19 from the predicted helix-turn-helix motif of the DNA binding domain results in a failure to bind DNA in vitro (Fig. 5B) and a decrease of ZntR t1/2 in vivo (Fig. 6A and B). A t1/2 of about 15 min was observed for the untagged ZntRR19A regardless of the presence or absence of added zinc; these data strongly suggest that binding to DNA stabilizes ZntR against proteolytic attack.
We propose a model (Fig. 7A) in which two different ligands, zinc and DNA, act together to increase ZntR stability in vivo: (i) the apoprotein is unstable, although the MerR-like regulators in their apo form do bind DNA (4); (ii) ZntR t1/2 increases upon addition of zinc; and (iii) DNA binding is needed for zinc-enhanced ZntR stability. We conclude that it is the zinc-bound, operator-bound form of ZntR that is most stable against proteolysis. The structural basis of the increased stability of ZntR within this complex is unknown; however, studies with several MerR family members demonstrate differences in the conformation of the protein-DNA complex upon metal binding. For example, the apo form of the transcription factor binds to the operator DNA and induces bending of the DNA. Metal binding then induces an additional conformational change, detected by further distortion of the operator DNA (4). Thus, there is substantial evidence that the zinc-ZntR-operator ternary complex is likely to have a unique structure that could explain its enhanced protease resistance.
FIG. 7.
Stabilization of ZntR and its effect on zinc homeostasis. (A) Model of cooperative ZntR stabilization by zinc and DNA. Apo-ZntR is unstable, and addition of zinc enhances ZntR t1/2 in a DNA-dependent manner. (B) Regulated degradation of ZntR contributes to zinc homeostasis in E. coli through a feedback loop. At low intracellular zinc levels, apo-ZntR is unstable and leads to lack of zinc export due to poor activation of zntA transcription. Lack of zinc export and activation of zinc import result in high intracellular zinc concentrations. ZntR binds zinc, and the zinc-bound form of ZntR is more stable and strongly activates zntA transcription. This change leads to high efflux of zinc and inactivation of zinc uptake, which closes the feedback loop with a low zinc concentration in the cell.
This study provides the first example of a combination of two ligands, metal and DNA, that contribute to increase the stability of a protein, here the transcriptional regulator ZntR. Each of these cofactors has been shown previously to contribute singly to protein stability. It has been shown that the degradation of SoxS by Lon is inhibited by protein-DNA or protein-RNA polymerase interactions (40). Another ClpXP substrate, the λO initiator protein, is rapidly degraded but is protected against proteolysis when it is a member of the replication complex (45). Metal involvement in recognition of a substrate by ClpXP protease has also been shown for the global regulatory protein FNR (26). Apo-FNR, which lacks the Fe-S cluster, is degraded more quickly than the active [4Fe-4S]2+ form of FNR. In contrast, at high copper concentrations the turnover of CopZ metallochaperone in Enterococcus hirae is enhanced (25). The same effect was observed for the copper transporter Ctr1 of Saccharomyces cerevisiae and the yeast transcription factor Mac1 (47), whose degradation is critical for copper resistance. Thus, it appears that metal ion control of protein stability will be a recurring theme in metal response pathways.
It is very attractive to consider that ligand-controlled proteolysis of ZntR contributes to the homeostatic control of zinc export in E. coli through a feedback loop (Fig. 7B). Our study revealed that apo-ZntR is less stable in vivo than zinc-ZntR (Fig. 1D). It has been previously shown that the ZntR apo form only slightly activates zntA transcription (31). Therefore, at low intracellular zinc levels there is poor activation of zntA expression or zinc export, but the zinc uptake is activated and zinc accumulates in the cell. At high intracellular zinc concentrations, ZntR binds zinc and the protein's stability against degradation increases. This complex formation and stabilization lead to strong activation of zntA transcription and high zinc export. High zinc efflux and inactivation of zinc import result in a low intracellular zinc concentration, closing the feedback loop. Increased ZntR stability against proteolysis, in the presence of toxic zinc concentrations, thus may be necessary to ensure a high zinc export.
Acknowledgments
We thank Aliaa Abdelhakim, Peter Chien, Anne Meyer, and Jade Wang for comments on the manuscript and members of the Baker lab for helpful discussion. We also thank Ian Buchanan for construction of several ZntR variants, Elizabeth Oakes for kindly providing the Lon protease, and Jim Butler for his help with zinc content determination. We gratefully acknowledge the receipt of plasmid pRS415 and the phage λRS45 provided by T. Silhavy and R. Simons.
This work was supported by NIH grant GM49224. T.A.B. and M.P. are employees of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Blencowe, D. K., and A. P. Morby. 2003. Zn(II) metabolism in prokaryotes. FEMS Microbiol. Rev. 27:291-311. [DOI] [PubMed] [Google Scholar]
- 2.Brocklehurst, K. R., J. L. Hobman, B. Lawley, L. Blank, S. J. Marshall, N. L. Brown, and A. P. Morby. 1999. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 31:893-902. [DOI] [PubMed] [Google Scholar]
- 3.Brocklehurst, K. R., and A. P. Morby. 2000. Metal-ion tolerance in Escherichia coli: analysis of transcriptional profiles by gene-array technology. Microbiology 146:2277-2282. [DOI] [PubMed] [Google Scholar]
- 4.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 5.Changela, A., K. Chen, Y. Xue, J. Holschen, C. E. Outten, T. V. O'Halloran, and A. Mondragon. 2003. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301:1383-1387. [DOI] [PubMed] [Google Scholar]
- 6.Charette, M. F., G. W. Henderson, L. L. Doane, and A. Markovitz. 1984. DNA-stimulated ATPase activity on the Lon (CapR) protein. J. Bacteriol. 158:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudhury, R., and S. Srivastava. 2001. Zinc resistance in bacteria. Curr. Sci. 81:768-775. [Google Scholar]
- 8.Coleman, J. E. 1998. Zinc enzymes. Curr. Opin. Chem. Biol. 2:222-234. [DOI] [PubMed] [Google Scholar]
- 9.Coleman, J. E. 1992. Zinc proteins: enzymes, storage proteins, transcription factors, and replication proteins. Annu. Rev. Biochem. 61:897-946. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flynn, J. M., S. B. Neher, Y. I. Kim, R. T. Sauer, and T. A. Baker. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671-683. [DOI] [PubMed] [Google Scholar]
- 12.Gorbatyuk, B., and G. T. Marczynski. 2005. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol. Microbiol. 55:1233-1245. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman, S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19:565-587. [DOI] [PubMed] [Google Scholar]
- 14.Griffith, K. L., I. M. Shah, and R. E. Wolf, Jr. 2004. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol. Microbiol. 51:1801-1816. [DOI] [PubMed] [Google Scholar]
- 15.Grigorova, I. L., R. Chaba, H. J. Zhong, B. M. Alba, V. Rhodius, C. Herman, and C. A. Gross. 2004. Fine-tuning of the Escherichia coli sigmaE envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev. 18:2686-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hantke, K. 2001. Bacterial zinc transporters and regulators. Biometals 14:239-249. [DOI] [PubMed] [Google Scholar]
- 17.Hengge, R., and B. Bukau. 2003. Proteolysis in prokaryotes: protein quality control and regulatory principles. Mol. Microbiol. 49:1451-1462. [DOI] [PubMed] [Google Scholar]
- 18.Hoskins, J. R., and S. Wickner. 2006. Two peptide sequences can function cooperatively to facilitate binding and unfolding by ClpA and degradation by ClpAP. Proc. Natl. Acad. Sci. USA 103:909-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt, J. B., S. H. Neece, and A. Ginsburg. 1985. The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal. Biochem. 146:150-157. [DOI] [PubMed] [Google Scholar]
- 20.Ishii, Y., and F. Amano. 2001. Regulation of SulA cleavage by Lon protease by the C-terminal amino acid of SulA, histidine. Biochem. J. 358:473-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenal, U., and R. Hengge-Aronis. 2003. Regulation by proteolysis in bacterial cells. Curr. Opin. Microbiol. 6:163-172. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, X., A. Rubio, S. Chiba, and K. Pogliano. 2005. Engulfment-regulated proteolysis of SpoIIQ: evidence that dual checkpoints control sigma activity. Mol. Microbiol. 58:102-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan, S., K. R. Brocklehurst, G. W. Jones, and A. P. Morby. 2002. The functional analysis of directed amino-acid alterations in ZntR from Escherichia coli. Biochem. Biophys. Res. Commun. 299:438-445. [DOI] [PubMed] [Google Scholar]
- 24.Kim, Y. I., R. E. Burton, B. M. Burton, R. T. Sauer, and T. A. Baker. 2000. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell 5:639-648. [DOI] [PubMed] [Google Scholar]
- 25.Lu, Z. H., and M. Solioz. 2001. Copper-induced proteolysis of the CopZ copper chaperone of Enterococcus hirae. J. Biol. Chem. 276:47822-47827. [DOI] [PubMed] [Google Scholar]
- 26.Mettert, E. L., and P. J. Kiley. 2005. ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe-4S] cluster. J. Mol. Biol. 354:220-232. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 28.Monera, O. D., C. M. Kay, and R. S. Hodges. 1994. Protein denaturation with guanidine hydrochloride or urea provides a different estimate of stability depending on the contributions of electrostatic interactions. Protein Sci. 3:1984-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neher, S. B., J. Villen, E. C. Oakes, C. E. Bakalarski, R. T. Sauer, S. P. Gygi, and T. A. Baker. 2006. Proteomic profiling of ClpXP substrates after DNA damage reveals extensive instability within SOS regulon. Mol. Cell 22:193-204. [DOI] [PubMed] [Google Scholar]
- 30.Outten, C. E., and T. V. O'Halloran. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488-2492. [DOI] [PubMed] [Google Scholar]
- 31.Outten, C. E., F. W. Outten, and T. V. O'Halloran. 1999. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J. Biol. Chem. 274:37517-37524. [DOI] [PubMed] [Google Scholar]
- 32.Parkhill, J., A. Z. Ansari, J. G. Wright, N. L. Brown, and T. V. O'Halloran. 1993. Construction and characterization of a mercury-independent MerR activator (MerRAC): transcriptional activation in the absence of Hg(II) is accompanied by DNA distortion. EMBO J. 12:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patzer, S. I., and K. Hantke. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275:24321-24332. [DOI] [PubMed] [Google Scholar]
- 34.Patzer, S. I., and K. Hantke. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28:1199-1210. [DOI] [PubMed] [Google Scholar]
- 35.Powell, B. S., M. P. Rivas, D. L. Court, Y. Nakamura, and C. L. Turnbough, Jr. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prakash, S., L. Tian, K. S. Ratliff, R. E. Lehotzky, and A. Matouschek. 2004. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 11:830-837. [DOI] [PubMed] [Google Scholar]
- 37.Rensing, C., B. Mitra, and B. P. Rosen. 1997. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc. Natl. Acad. Sci. USA 94:14326-14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 39.Sauer, R. T., D. N. Bolon, B. M. Burton, R. E. Burton, J. M. Flynn, R. A. Grant, G. L. Hersch, S. A. Joshi, J. A. Kenniston, I. Levchenko, S. B. Neher, E. S. Oakes, S. M. Siddiqui, D. A. Wah, and T. A. Baker. 2004. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 119:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah, I. M., and R. E. Wolf, Jr. 2006. Inhibition of Lon-dependent degradation of the Escherichia coli transcription activator SoxS by interaction with ‘soxbox’ DNA or RNA polymerase. Mol. Microbiol. 60:199-208. [DOI] [PubMed] [Google Scholar]
- 41.Shah, I. M., and R. E. Wolf, Jr. 2006. Sequence requirements for Lon-dependent degradation of the Escherichia coli transcription activator SoxS: identification of the SoxS residues critical to proteolysis and specific inhibition of in vitro degradation by a peptide comprised of the N-terminal 21 amino acid residues. J. Mol. Biol. 357:718-731. [DOI] [PubMed] [Google Scholar]
- 42.Silhavy, T., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 44.Turgay, K., J. Hahn, J. Burghoorn, and D. Dubnau. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17:6730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wegrzyn, A., G. Wegrzyn, and K. Taylor. 1995. Protection of coliphage lambda O initiator protein from proteolysis in the assembly of the replication complex in vivo. Virology 207:179-184. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto, K., and A. Ishihama. 2005. Transcriptional response of Escherichia coli to external zinc. J. Bacteriol. 187:6333-6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, Z., S. Labbe, M. M. Pena, and D. J. Thiele. 1998. Copper differentially regulates the activity and degradation of yeast Mac1 transcription factor. J. Biol. Chem. 273:1277-1280. [DOI] [PubMed] [Google Scholar]