Abstract
The genome of Streptococcus sanguinis is a circular DNA molecule consisting of 2,388,435 bp and is 177 to 590 kb larger than the other 21 streptococcal genomes that have been sequenced. The G+C content of the S. sanguinis genome is 43.4%, which is considerably higher than the G+C contents of other streptococci. The genome encodes 2,274 predicted proteins, 61 tRNAs, and four rRNA operons. A 70-kb region encoding pathways for vitamin B12 biosynthesis and degradation of ethanolamine and propanediol was apparently acquired by horizontal gene transfer. The gene complement suggests new hypotheses for the pathogenesis and virulence of S. sanguinis and differs from the gene complements of other pathogenic and nonpathogenic streptococci. In particular, S. sanguinis possesses a remarkable abundance of putative surface proteins, which may permit it to be a primary colonizer of the oral cavity and agent of streptococcal endocarditis and infection in neutropenic patients.
Streptococcus sanguinis (formerly known as “S. sanguis,” but renamed for grammatical correctness [91]) is an indigenous gram-positive bacterium that has been recognized for a long time as a key player in colonization of the human oral cavity (81). Like most oral streptococci, this bacterium produces alpha-hemolysis on blood agar, a characteristic linked to the ability of viridans streptococci to oxidize hemoglobin in erythrocytes by secretion of H2O2 (6). S. sanguinis binds directly to saliva-coated teeth, probably by a variety of mechanisms (46). Studies employing saliva-coated hydroxyapatite as a tooth model have revealed both lectin-carbohydrate and nonlectin interactions (27, 38, 42, 64). Some of the salivary components to which S. sanguinis binds have been identified, including salivary immunoglobulin A and α-amylase (27). Once bound, S. sanguinis serves as a tether for the attachment of other oral microorganisms that colonize the tooth surface, form dental plaque, and contribute to development of caries and periodontal disease (46). S. sanguinis may also interfere with colonization of the tooth by Streptococcus mutans, the primary species associated with dental caries (16), and its presence therefore may also be beneficial for oral health.
The viridans streptococci are the most common cause of native-valve infective endocarditis, and S. sanguinis is the viridans streptococcus most commonly implicated in this disease (66). S. sanguinis and other viridans streptococci are also emerging as important bloodstream pathogens in infections that threaten neutropenic patients (1), and these infections may be complicated by an increasing frequency of antibiotic resistance (71). The reasons underlying this previously unrecognized virulence are unknown, and antibiotic resistance is disquieting because viridans streptococci, including S. sanguinis, have been classified historically as penicillin sensitive and for many years were believed to be unable to become resistant to β-lactam antibiotics.
Here, we describe the sequence and an analysis of the genome of S. sanguinis strain SK36, which was originally isolated from human dental plaque (43). Analysis of the predicted proteins yielded new insights into potential pathogenicity and virulence factors in this important bacterium, which allowed comparison with virulence mechanisms in other streptococci. Furthermore, about 28% of the predicted proteins were confirmed with high confidence by mass spectrometry (MS).
MATERIALS AND METHODS
Strain and culture conditions.
S. sanguinis SK36 was isolated from human dental plaque (38, 42). This strain was selected because it (i) has the defining features of S. sanguinis as determined by accepted diagnostic tests, (ii) can aggregate human platelets (35), (iii) is naturally competent (69), (iv) binds to saliva-coated hydroxyapatite (38, 42), (v) coaggregates with other oral bacteria (R. N. Andersen and P. E. Kolenbrander, personal communication), and (vi) is virulent in the rat and rabbit models of infective endocarditis (69). For genomic DNA isolation, cells were grown in an atmosphere containing 10% H2, 10% CO2, and 80% N2 at 37°C in brain heart infusion broth (Difco Inc., Detroit, MI).
Genome sequencing and annotation.
The genome was sequenced using a modified whole-genome shotgun strategy, as previously described (98). In short, two shotgun libraries (1- to 2-kb and 2- to 4-kb inserts) and one BAC library (∼500 clones, 25- to 100-kb inserts) were constructed, and approximately 74,000 sequences were generated (∼15-fold coverage of the genome) by using an ABI 3700 96-lane capillary DNA sequencer (Applied Biosystems). The genomic sequence was assembled as previously described (98). Gaps were closed by genome walking (Clontech), alignment with BAC clones, long-distance PCR, and multiplex PCR (89). All remaining low-quality sequence regions were amplified and resequenced for finishing. About 5,000 sequences were added during gap closing and finishing. Genome annotation was performed automatically essentially as previously described (98). Gene predictions were based on Glimmer (77), database searches, and manual verification in Apollo (50). rRNA boundaries were set based on predicted structural criteria (15).
HGT analyses.
To select candidates for horizontal gene transfer (HGT), the phyletic patterns of gene distribution were analyzed. First, S. sanguinis proteins were compared to the NCBI nonredundant protein database using BLASTP. Significant matches (E < 1e-6) were analyzed to find genes without streptococcal sequences among the top six species matching the S. sanguinis protein. The same analysis was performed with Escherichia coli K-12, considering Salmonella and Yersinia the “same” genus (these genera were chosen as the genera that were closest phylogenetically to E. coli since no other species of Escherichia have been sequenced). This analysis overestimated the number of HGT candidates in E. coli due to the narrow sampling of genetic diversity in the genus compared to the broad sampling available for streptococci.
Proteomic analysis of S. sanguinis.
Total protein was extracted from S. sanguinis grown overnight in brain heart infusion broth. Cells were harvested by centrifugation, washed twice in ice-cold phosphate-buffered saline, and suspended in 20 mM morpholinepropanesulfonic acid (MOPS)-62.5 mM NaCl-0.5 mM MgSO4 (pH 7.8) with a protease inhibitor cocktail (Sigma-Aldrich). The cells were mechanically disrupted with an FP120 FastPrep cell disruptor (Bio 101 Systems, Qbiogen, Inc.) by using three 30-s cycles of homogenization at the maximum speed with 1-min intervals between cycles in ice. The suspension was centrifuged (5,000 × g for 15 min at 4°C) to remove unbroken cells and large cellular debris. The supernatant was suspended in solubilization buffer as previously described (68) and was precipitated with a 2D clean-up kit (GE Healthcare). After reduction with dithiothreitol and iodoacetamide alkylation, proteins (∼75 μg) were digested overnight with trypsin. The resulting tryptic peptides were desalted using C8 cartridges (Michrom BioResources) and were subjected to two-dimensional nano liquid chromatography-MS/MS analyses with a Michrom BioResources Paradigm MS4 multidimensional separation module, a Michrom NanoTrap platform, and an LCQ Deca XP Plus ion trap mass spectrometer. The mass spectrometer was operated in the data-dependent mode, and the four most abundant ions in each MS spectrum were selected and fragmented to produce tandem mass spectra. The MS/MS spectra were recorded in the profile mode. Proteins were identified by searching the MS/MS spectra against our S. sanguinis database using Bioworks v3.2. Peptide and protein hits were scored and ranked using the new probability-based scoring algorithm incorporated in Bioworks v3.2. Only peptides identified as possessing fully tryptic termini with cross-correlation scores greater than 1.9 for singly charged peptides, 2.3 for doubly charged peptides, and 3.75 for triply charged peptides were used for peptide identification. In addition, the delta-correlation scores had to be greater than 0.1, and for increased stringency, a protein was accepted only if its probability score was <0.0001.
Nucleotide sequence accession number.
The S. sanguinis SK36 genome sequence has been deposited in the GenBank database under accession no. CP000387.
RESULTS AND DISCUSSION
General genomic features.
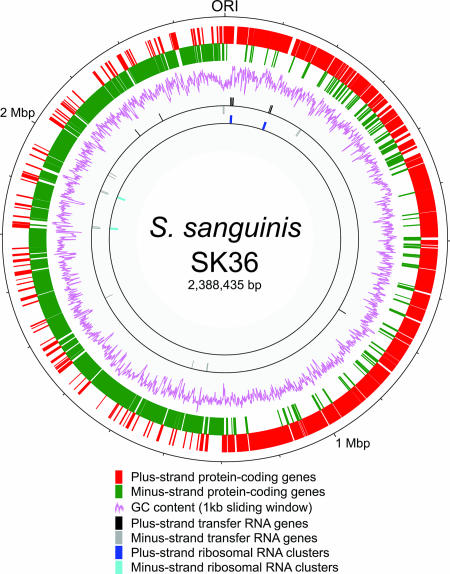
The S. sanguinis genome is comprised of a 2,388,435-bp circular DNA molecule, which is 7 to 24% larger than previously described streptococcal genomes (Table 1). The genome start point was assigned to the putative origin of replication, as determined by GC skew (61), the location of the dnaA gene, and similarity to other genomic sequences (54). The putative replication termination region is ∼1.2 Mbp downstream from the origin of replication (Fig. 1). The G+C content of the genome is 43.40%, which is higher than the G+C content of any of the 21 other completed streptococcal genomes (35.62 to 39.72%) (Table 1). For protein-encoding genes, the G+C contents are 53.55, 35.46, and 44.35% for positions 1, 2, and 3, respectively. Based on the relationship between the G+C contents of whole genomes and the G+C contents of position 3 of coding sequences, which were recently determined for 232 eubacterial genomes (93), the expected value for position 3 in S. sanguinis is 42.5%, in good agreement with the observed value. This observation suggests that unlike the findings for Lactobacillus bulgaricus, the higher overall G+C content of S. sanguinis is not due to an ongoing process of compositional change or to a different relationship of whole-genome and third-position G+C values. There are four rRNA operons containing the 5S, 16S, and 23S rRNA genes, which is less than the number in most other streptococci (Table 1), despite the larger genome size and in contrast to a reported correlation between the numbers of rRNA and tRNA genes and the genome sizes in the Firmicutes (93). The 61 predicted tRNA genes encode all 20 amino acids, but wobble rules are required for several abundant codons (www.sanguinis.mic.vcu.edu/supplemental.htm). Most tRNA genes are clustered near the rRNA operons; i.e., 48 of 61 of these genes were less than 1 kb from an rRNA operon (Fig. 1), as in Streptococcus pneumoniae (88).
TABLE 1.
Comparison of S. sanguinis SK36 genome with other streptococcal genomes
| Strain | Accession no. | Genome size (Mbp) | G+C content of genome (%) | No. of predicted proteins | No. of rRNA operons | No. of tRNA operonsa | Reference |
|---|---|---|---|---|---|---|---|
| S. sanguinis SK36 | CP000387 | 2.39 | 43.40 | 2,274 | 4 | 61 | This study |
| S. agalactiae 2603 V/R | AE009948 | 2.16 | 35.65 | 2,124 | 7 | 80 | 87 |
| S. agalactiae A909 | CP000114 | 2.13 | 35.62 | 1,996 | 7 | 80 | 86 |
| S. agalactiae NEM316 | AL732656 | 2.21 | 35.63 | 2,094 | 7 | 80 | 26 |
| S. mutans UA159 | AE014133 | 2.03 | 36.83 | 1,960 | 5 | 65 | 2 |
| S. pneumoniae D39 | CP000410 | 2.05 | 39.71 | 1,914 | 4 | 58 | 48 |
| S. pneumoniae R6 | AE007317 | 2.04 | 39.72 | 2,043 | 4 | 58 | 37 |
| S. pneumoniae TIGR4 | AE005672 | 2.16 | 39.70 | 2,094 | 4 | 58 | 88 |
| S. pyogenes M1 GAS | AE004092 | 1.85 | 38.51 | 1,697 | 6 | 60 | 22 |
| S. pyogenes MGAS10270 | CP000260 | 1.93 | 38.43 | 1,987 | 6 | 67 | 10 |
| S. pyogenes MGAS10394 | CP000003 | 1.90 | 38.69 | 1,886 | 6 | 67 | 5 |
| S. pyogenes MGAS10750 | CP000262 | 1.94 | 38.32 | 1,979 | 6 | 67 | 10 |
| S. pyogenes MGAS2096 | CP000261 | 1.86 | 38.73 | 1,898 | 6 | 67 | 10 |
| S. pyogenes MGAS315 | AE014074 | 1.90 | 38.59 | 1,865 | 6 | 67 | 11 |
| S. pyogenes MGAS5005 | CP000017 | 1.84 | 38.53 | 1,865 | 6 | 67 | 83 |
| S. pyogenes MGAS6180 | CP000056 | 1.90 | 38.35 | 1,894 | 6 | 67 | 28 |
| S. pyogenes MGAS8232 | AE009949 | 1.90 | 38.55 | 1,845 | 6 | 67 | 80 |
| S. pyogenes MGAS9429 | CP000259 | 1.84 | 38.54 | 1,877 | 6 | 67 | 10 |
| S. pyogenes SSI-1 | BA000034 | 1.89 | 38.55 | 1,861 | 5 | 57 | 67 |
| S. thermophilus CNRZ1066 | CP000024 | 1.80 | 39.08 | 1,915 | 6 | 67 | 13 |
| S. thermophilus LMD-9 | CP000419 | 1.86 | 39.08 | 1,710 | 6 | 67 | 56 |
| S. thermophilus LMG18311 | CP000023 | 1.80 | 39.09 | 1,889 | 6 | 67 | 13 |
Genomes were scanned using the tRNAscan-SE program for comparison.
FIG. 1.
Circular S. sanguinis SK36 genome map. Starting from the outside, the circles show (i) the genome positions (in base pairs) starting from the origin of replication (ORI); (ii and iii) predicted coding regions on the two strands (different colors are used for clarity); (iv) G+C content (in 1-kb windows); (v and vi) rRNA clusters on the two strands; and (vii and viii) tRNA on the two strands.
The genome contains genes encoding 2,274 predicted proteins and accounting for more than 90% of the sequence (www.sanguinis.mic.vcu.edu/supplemental.htm). About 86% (1,965) of these genes are transcribed in the direction of replication, like the genes in other streptococci (2, 87, 88). The average gene contains 935 bp of coding sequence, and the average intergenic region is 115 bp long. The latter value is smaller than the values for other streptococcal genomes that have been sequenced, whose average intergenic region lengths are 130 to 177 bp, or for the E. coli genome, whose average intergenic region length is 139 bp. This observation suggests that S. sanguinis has a more compact genome, although differences in annotation methods may also explain the differences. Of the predicted proteins, 89% exhibit significant similarity to proteins from other organisms. About 22% are conserved hypothetical proteins (present in multiple species but having unknown functions), and approximately 645 of the predicted proteins were confirmed by MS (www.sanguinis.mic.vcu.edu/supplemental.htm).
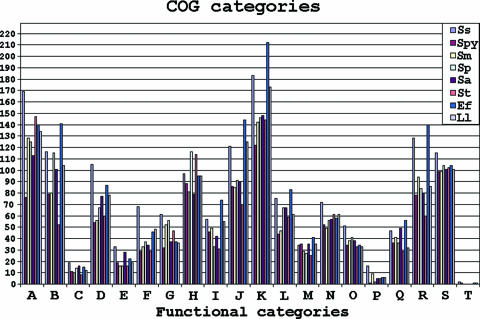
The S. sanguinis SK36 genome was compared with other genomes to identify the proteins that are conserved among streptococci. Figure 2 shows the homologous proteins that are shared by S. sanguinis, S. mutans, and S. pneumoniae. This analysis indicated that S. sanguinis shares 23 more proteins with S. mutans than with S. pneumoniae and that the latter two species share only 19 proteins not present in S. sanguinis. Previous analyses based on rRNA (41) and our more broadly based phylogenetic analysis confirmed that S. sanguinis is more closely related to S. pneumoniae than to S. mutans, suggesting that the similarity with S. mutans reflects the shared oral niche of these two species. The proteins shared by only S. sanguinis and S. mutans include 60 proteins that are hypothetical or have unknown functions and, interestingly, 34 putative transcriptional regulators. All proteins in the S. sanguinis genome were functionally categorized and compared (Fig. 3) essentially as previously described (98).
FIG. 2.
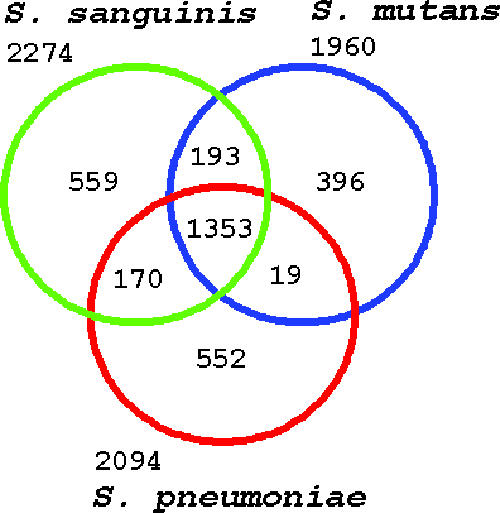
In silico comparisons of streptococci. The protein sets of S. sanguinis SK36, S. mutans UA159, and S. pneumoniae TIGR4 were compared. The numbers under and above the species names indicate the total numbers of genes; the numbers in the intersections indicate the numbers of genes shared by two or three species.
FIG. 3.
COG classification of the S. sanguinis SK36 genome and comparison with other microbial genomes. The numbers of genes of eight species were compared based on the functional classification in the COG database. Ss, S. sanguinis SK36; Spy, S. pyogenes M1GAS; Sm, S. mutans UA159; Sp, S. pneumoniae R6; Sa, S. agalactiae NEM316; St, S. thermophilus CNZR1066; Ef, Enterococcus faecalis V583; Ll, Lactococcus lactis IL-1403. The functional categories are indicated as follows: A, amino acid transport and metabolism; B, carbohydrate transport and metabolism; C, cell division and chromosome partitioning; D, cell envelope biogenesis, outer membrane; E, cell motility and secretion; F, coenzyme metabolism; G, defense mechanisms; H, DNA replication, recombination, and repair; I, energy production and conversion; J, unknown function; K, general function prediction; L, inorganic ion transport and metabolism; M, lipid metabolism; N, nucleotide transport and metabolism; O, posttranslational modification, protein turnover, chaperones; P, secondary metabolite biosynthesis, transport, and catabolism; Q, signal transduction mechanisms; R, transcription; S, translation, ribosomal structure, and biogenesis; T, other.
Energy and metabolism.
Consistent with previous observations (43), S. sanguinis can apparently use a broad range of carbohydrate sources for survival. We identified more than 50 putative carbohydrate transporters, including phosphotransferase system enzymes specific for transport of glucose, fructose, mannose, cellobiose, glucosides, fructose, lactose, trehalose, mannose, galactitol, and maltose (see Table S1 in the supplemental material). Thus, this bacterium seems to possess a robust system for energy generation by fermentation of sugars and other carbohydrates.
Similar to S. mutans (2) and other streptococci, S. sanguinis has an incomplete citrate cycle and contains only the enzymes to convert oxaloacetate into 2-oxoglutarate. Although clearly incapable of direct ATP production, this pathway fragment likely generates intermediates in the synthesis of aspartate and glutamate.
Our analysis suggests that S. sanguinis has a robust biosynthetic capacity. All key enzymes for gluconeogenesis are present. This bacterium has both pyruvate-phosphate dikinase (EC 2.7.9.1) (encoded by SSA_1053) found in other streptococci and phosphoenolpyruvate synthase (EC 2.7.9.2) (encoded by SSA_1012 and SSA_1016) that is absent in other streptococci. There is also a Firmicutes-specific fructose-1,6-bisphosphatase (EC 3.1.3.11) (encoded by SSA_1056) that is present in Streptococcus agalactiae but not in S. pneumoniae, S. mutans, Streptococcus pyogenes, or Streptococcus thermophilus. Phyletic pattern analyses suggested that the genes for these enzymes were acquired by HGT (www.sanguinis.mic.vcu.edu/supplemental.htm). Similarly, enzymes in the pentose phosphate pathway and enzymes in the purine and pyrimidine pathways, which are required for de novo synthesis of nucleotides with the possible exception of dTTP, seem to be available. Enzymes necessary for converting glutamate and glutamine to intermediates in purine and pyrimidine synthesis are also present. However, as in S. mutans (2), the gene for nucleoside diphosphate kinase (EC 2.7.4.6), which phosphorylates dTDP to dTTP, could not be identified. Since these enzymes are highly conserved in other streptococci, it is unlikely that we missed identifying their genes, assuming that they are derived from common progenitors.
S. sanguinis seems to have the ability to synthesize de novo all essential amino acids except the branched amino acids (leucine, isoleucine, and valine), lysine, and tryptophan (www.sanguinis.mic.vcu.edu/supplemental.htm). This conclusion is in agreement with our finding that S. sanguinis cannot grow in a semidefined biofilm medium (52) if supplemental amino acids are not included (data not shown). Synthesis of asparagine likely relies on a two-step process in which aspartate is bound to tRNAAsn by a nondiscriminating Asp-tRNA synthetase, followed by conversion of the aspartate to asparagine via a three-subunit aspartyl/glutamyl-tRNA amidotransferase, as has been shown for Deinococcus radiodurans (62). The latter enzyme is probably also responsible for conversion of Glu-tRNAGln to Gln-tRNAGln, thus explaining the lack of a gene encoding glutaminyl-tRNA synthetase in the genome (72). As noted above, enzymes for gluconeogenesis are present and could permit the bacterium to convert some amino acids (e.g., serine) into fructose-6-phosphate, an entry point of the pentose phosphate pathway. In this way, amino acids can be converted into the precursors of nucleotide biosynthesis. Marri et al. (58) recently reported that among the streptococci, S. mutans is unique in possessing the genes responsible for biosynthesis of histidine and that S. pyogenes is unique in its apparent ability to convert histidine to glutamate. S. sanguinis possesses the genes for both of these processes.
Lipid biosynthesis apparently follows the classical bacterial type II fatty acid synthase complex pathway (34). As shown previously for S. pneumoniae (33, 57), S. sanguinis encodes the enoyl-(acyl-carrier protein) reductase (EC 1.3.1.9) FabK instead of the widespread and conserved FabI type enzyme of other bacteria and plants. The FabK enzyme of S. pneumoniae is less sensitive to inhibition by the antimicrobial triclosan than FabI is (33, 57). Therefore, S. sanguinis is probably more resistant than FabI-containing bacteria to inhibition of lipid biosynthesis by the triclosan used in some toothpastes. Fatty acids can be generated from amino acids since enzymes needed for the conversion of some amino acids (e.g., serine) into acetyl coenzyme A are present (www.sanguinis.mic.vcu.edu/supplemental.htm).
As expected, the S. sanguinis genome contains the genes required for cell wall sugar, peptidoglycan, and teichoic acid biosynthesis and degradation (www.sanguinis.mic.vcu.edu/supplemental.htm). Single copies of the genes encoding homologs of the S. mutans signal recognition particle components Ffh, FtsY, and small cytoplasmic RNA are present in S. sanguinis, as are single copies of the genes encoding the secretion components YidC1, YidC2, YajC, SecA, and SecYEG (31).
HGT.
In contrast to S. pneumoniae, in which ∼5% of the genome is composed of insertion sequences (IS) (88), we found only two apparently functional IS elements (SSA_0265 and SSA_0266; SSA_1361 and SSA_1362) in S. sanguinis. These elements are flanked by 4-bp direct repeats and are ∼80% identical at the nucleotide level to IS3 elements flanked by 3-bp repeats in S. mutans (55). Neither IS interrupts a known gene or open reading frame (ORF). Other evidence of transposable elements includes remnants of IS elements (SSA_1477 to SSA_1479 and SSA_0732) and a truncated transposase (SSA_2029). No intact prophages were found, although some apparent remnants (SSA_0235, SSA_2032, and SSA_2295 encoding an integrase/recombinase; SSA_2383 encoding a prophage maintenance system killer protein; and SSA_2282 encoding a phage infection protein) are present (www.sanguinis.mic.vcu.edu/supplemental.htm). No evidence of the presence of integrons was found. Homologs of the dpnM, dpnA, and dpnB genes of S. pneumoniae encoding the DpnII restriction-modification system are present in the S. sanguinis genome (SSA_1716 to SSA_1718). This system reduces the efficiency of HGT by phage infection, conjugative transfer, and transformation by plasmid (but not chromosomal) DNA (47). We did not find genes for the R.StsI and M.StsI components previously found in S. sanguinis 54 (44).
In spite of the relative paucity of transposon- and phage-related genes, at least 270 S. sanguinis genes (12% of the genes) were identified as candidates for HGT by observing the phyletic pattern of gene distribution (www.sanguinis.mic.vcu.edu/supplemental.htm) (see Materials and Methods). The apparent lack of phage genes and conjugative transposable elements suggests that transformation is the predominant method by which HGT occurs in S. sanguinis. Like certain other streptococci, S. sanguinis is naturally competent for transformation (25). In S. pneumoniae, 22 proteins necessary for chromosomal transformation have been identified (70). We found that 20 of these proteins have apparent orthologs in S. sanguinis (www.sanguinis.mic.vcu.edu/supplemental.htm). Neither ComW, an 80-amino-acid protein which stabilizes and activates the alternative sigma factor ComX (84) and for which there are no database matches in any other bacterium in the GenBank database, nor ComB, which functions with ComA to cleave and export competence-stimulating peptide (CSP), was identified. The SSA_1100 product exhibits similarity to ComA. However, the best matches for SSA_1100 in the GenBank database were matches to genes encoding transporters for RTX-type toxins from gram-negative bacteria (94). Since the adjacent gene encodes a putative RTX toxin, it appears that this protein transports the toxin rather than CSP. Therefore, it appears that ComA and ComB are not present in S. sanguinis. This absence may be related to the previous observation that ComC, the CSP precursor in S. sanguinis, is unique among all 125 ComC sequences from 13 streptococcal species in the GenBank database in that it lacks a double-glycine cleavage site (32). This unique cleavage site could be paired with unique proteins for processing and export.
One 70-kb cluster of 68 HGT candidates (SSA_0463 to SSA_0541) encodes an anaerobic cobalamin (vitamin B12) biosynthetic (cob) pathway, as well as propanediol utilization (pdu) and ethanolamine utilization (eut) pathways (Fig. 4; see Table S2 in the supplemental material). Many of the proteins in this cluster were identified by MS, proving that these genes are expressed.
FIG. 4.
Schematic map of the 70-kb HGT region for vitamin B12 biosynthesis and related pathways. The colors indicate genes in different pathways based on homology with Salmonella, as follows: red, cob; blue, pdu; black, eut; gray, not predicted to be part of any of these three pathways; white, genes flanking the transferred region.
Vitamin B12 is an important nutrient for human health; a deficiency leads to pernicious anemia. However, synthesis of this compound in prokaryotes (40) occurs only by two alternative routes: an aerobic pathway that incorporates molecular oxygen during biosynthesis and an anaerobic pathway that incorporates chelated cobalt ions in the absence of oxygen (78). All genes required for anaerobic cobalamin biosynthesis are present in S. sanguinis. It appears that the complete vitamin B12 biosynthesis pathway is available. If so, this is the first time that the complete B12 biosynthesis pathway has been identified in streptococci, although three proteins involved in cobalamin biosynthesis and cobalt transport (cbiMQO) have been found in Streptococcus salivarius 57.I and S. thermophilus (18).
Cobalamin-dependent utilization of 1,2-propanediol via the pdu pathway plays an important role in Salmonella enterica serovar Typhimurium infection (20), and the pdu genes are correlated with cobalamin biosynthetic genes in terms of both location and coregulation. The S. enterica serovar Typhimurium pdu pathway contains 23 genes for the coenzyme B12-dependent catabolism of 1,2-propanediol (12). S. sanguinis has all of these genes except pduM and pduS, which encode proteins with unknown functions, and pduN, which encodes polyhedral bodies that may not be directly related to the catabolism of 1,2-propanediol (12) (see Table S2 in the supplemental material).
The eut pathway in S. enterica serovar Typhimurium is required for utilization of ethanolamine as a carbon and nitrogen source (75). Only 4 (eutB, eutC, eutD, and eutE) of the 17 genes in the S. enterica serovar Typhimurium eut operon have been correlated directly with an enzymatic activity known to be required for ethanolamine utilization (79). Three of these four genes, eutB (SSA_0519), eutC (SSA_0520), and eutE (SSA_0523), have homologs in S. sanguinis. eutD encodes a protein with phosphotransacetylase activity (14) and exhibits 40% identity with the S. sanguinis SSA_1207 ORF, which is annotated as phosphate acetyltransferase ORF. A two-component system (SSA_0516 and SSA_0517) that may regulate ethanolamine utilization in response to environmental factors is upstream of eutA. Since ethanolamine and propanediol sources in the environment seem largely man-made (e.g., toothpaste, mouthwash, and antifreeze) and their utilization is dependent on vitamin B12, it is interesting to speculate that this large ∼70-kb gene cluster may have been selected in S. sanguinis by exposure to these man-made products.
Although very few of these cobalamin-related genes are present in previously published streptococcal genomes, many are present in other oral pathogens, including Porphyromonas gingivalis, Treponema denticola, and Fusobacterium nucleatum (see Table S2 in the supplemental material). Our analyses suggest that the 70-kb cluster of HGT genes has an origin similar to the origin of orthologs in Listeria (www.sanguinis.mic.vcu.edu/supplemental.htm), but a more in-depth phylogenetic analysis involving more prokaryotic genomes is necessary to confirm its origin.
Two small discrete blocks of HGT candidate genes (SSA_1012 to SSA_1017 and SSA_1053 to SSA_1056) contain three genes involved in gluconeogenesis. The two genes in the second block (SSA_1053 and SSA_1056), encoding EC 2.7.9.1 and EC 3.1.3.11, are sufficient, in combination with other apparently native genes, to enable gluconeogenesis. These two genes are also found in S. agalactiae, theoretically enabling gluconeogenesis in this organism, while all other streptococcal genomes that have been sequenced seem to lack the complete set of genes required for gluconeogenesis. The results of our analysis (see Materials and Methods) are consistent with the hypothesis that these genes were transferred by HGT to these streptococci from other bacteria belonging to the phylum Firmicutes (www.sanguinis.mic.vcu.edu/supplemental.htm).
Putative virulence factors and adhesins.
Several proteins potentially relevant to adhesion in the oral cavity or to virulence in invasive disease were identified in the S. sanguinis genome (see Table S3 in the supplemental material). Perhaps the most surprising is the protein encoded by SSA_1099 (Stx), which exhibits homology to RTX-type toxins in gram-negative bacteria (94). To our knowledge, this is the first occurrence of this class of toxin gene in a gram-positive bacterium. Consistent with this unique setting, orthologs of the HylB ATPase and HlyD “membrane fusion protein” components of an RTX toxin export system are encoded by adjacent ORFs (SSA_1100 and SSA_1101, respectively), but no homolog of the TolC outer membrane component (36) was found. Both Stx and the putative ATPase transporter component, encoded by SSA_1100, were detected in the proteomic analysis (www.sanguinis.mic.vcu.edu/supplemental.htm). Although the leukotoxin from the oral bacterium Actinobacillus actinomycetemcomitans is a well-known ortholog of the Stx protein, the products of SSA_1099 to SSA_1101 are, as a whole, most similar to proteins in plant-pathogenic pseudomonads. Thus, the origin of these S. sanguinis genes and their functions are unclear.
The genes associated with pathogenicity in S. sanguinis also include genes encoding orthologs of the major known adhesins in other viridans species. SspC and SspD are orthologs of the SspA and SspB adhesins of Streptococcus gordonii (39, 53). Whereas the latter proteins are encoded by adjacent genes in S. gordonii, this is not true in S. sanguinis. Conversely, the cshA and cshB adhesin genes are not contiguous in S. gordonii (60), whereas the S. sanguinis crpABC orthologs are contiguous. The ligand specificity of SspA orthologs in viridans streptococci is determined by their sequences (39, 53). Neither SspC nor SspD is closely related to any SspA homolog that has been characterized previously. As determined by BLASTP analysis (3), SspC has only 55% identity with its closest relative (SspA), and SspD has 33% identity with its closest relative (PaaA of Streptococcus criceti). Therefore, it is not clear what ligand(s), if any, SspC and SspD bind. However, the 27-amino-acid region of SspB that has been shown to mediate binding of S. gordonii to P. gingivalis is conserved in SspC (18 identical residues and five similar residues), including perfect identity of the critical NITVK subsequence (21). This observation suggests that SspC may also adhere to P. gingivalis.
Lipoproteins (LP) and cell-wall anchored proteins (CWA), two classes of proteins that are surface exposed and prevalent among reported virulence factors, were predicted (www.sanguinis.mic.vcu.edu/supplemental.htm). The lgt and lspA genes expected for LP processing are present (SSA_1546 and SSA_1069, respectively), as are genes encoding three sortases (SSA_0022, SSA_1219, and SSA_1631) for CWA processing. Interestingly, the numbers of these surface proteins (60 LPs and 33 CWAs) are striking compared to the numbers in related species. As determined by the same search criteria used for S. sanguinis, S. mutans has only 29 LPs and six CWAs. S. pneumoniae TIGR4 possesses 40 LPs and 12 CWAs, while R6 has 39 LPs and 13 CWAs. However, many of the additional ORFs in S. sanguinis appear to be redundant. Thus, S. sanguinis contains nine paralogous CWAs in three families and seven paralogous LPs in three families. In addition, functional redundancy may occur in the absence of overall sequence similarity; five CWAs possess the collagen-binding domain, Pfam05737 (23). This vast array of surface proteins may contribute to the ability of S. sanguinis to colonize the tooth and interact with a diverse group of oral bacteria (46) and may account for its predominance as a cause of streptococcal endocarditis (66).
Fibrils or pili are involved in streptococcal adherence and virulence (7, 59, 82). S. sanguinis strains possess both short fibrils and long fibrils (30). Fap1 of Streptococcus parasanguinis, an ortholog of the CWA encoded by SSA_0829 or SrpA, is thought to be the structural component of long fibrils (82), and its orthologs are important for adhesion to platelets (9), saliva-coated hydroxyapatite (96), and salivary agglutinin (39). The products of SSA_0830 to SSA_0841 exhibit homology to the proteins shown to be required for the glycosylation and export of SrpA orthologs in S. parasanguinis and S. gordonii (9, 17, 85). In fact, the 11 genes downstream from srpA are most similar in terms of sequence to, and are in the same order as, the 11 genes that form the export locus of the SrpA ortholog, GspB, in S. gordonii (85). Shorter fibrils in S. gordonii are comprised of CshA and possibly also CshB (59), which are orthologs of CWAs encoded by SSA_0904 to SSA_0906. The fact that S. sanguinis has both classes of proteins, as well as the locus dedicated to SrpA export, could account for the apparent presence of both short and long fibrils. In addition, in recent studies workers have identified long pili in S. agalactiae (49), S. pyogenes (63), and S. pneumoniae (7). In these bacteria, a single locus contains three putative pilin subunit genes encoding CWA motifs and one to three sortase genes that are required for assembly of the pili (7, 49, 63). S. sanguinis also contains an apparent pilus locus, with SSA_1632 to SSA_1635 encoding LPXTG proteins and SSA_1631 encoding a sortase. SSA_1632 to SSA_1634 also each contain a conserved “E box” domain found in many pilin genes (90).
The SSA_2302 to SSA_2318 sequences exhibit homology to ORFs required for production of type IV pili. Such pili were originally believed to exist only in gram-negative bacteria, although the gram-positive bacterium Ruminococcus albus appears to possess a type IV pilus that serves as an adhesin (73). Our analysis suggests that the S. sanguinis ORFs were acquired by HGT, perhaps from a clostridial species, and are distinct from the ORFs in S. sanguinis that apparently encode the pseudopilus involved in genetic competence (data not shown).
Cell wall polysaccharides (CWP) serve as important receptors for agglutination and coaggregation in oral streptococci (19, 45, 46). S. sanguinis SK36 is similar to type strain ATCC 10556 in that it coaggregates with numerous species of Streptococcus, Actinomyces, and Fusobacterium (38, 45) (Kolenbrander and Andersen, personal communication). These interactions are inhibited by addition of 60 mM N-acetyl-d-galactosamine, confirming the polysaccharide composition of the receptor (45). Six structures have been defined for CWP in oral streptococci (19), and the loci responsible for synthesis of one of these structures have been characterized in S. gordonii (97). Orthologs of these genes are located mostly in two genomic segments in S. sanguinis, SSA_1509 to SSA_1519 and SSA_2211 to SSA_2225. However, these segments also contain apparent CWP synthesis genes that have close orthologs in S. thermophilus, Streptococcus suis, S. pneumoniae, or Streptococcus iniae but no orthologs in S. gordonii. These CWP loci, therefore, appear to be unlike any loci characterized previously, and it is not clear whether they direct the synthesis of a type 1 N-acetylgalactosamine-β1→3-galactose CWP like that found in previously characterized S. sanguinis strains (19).
Other interesting features.
The S. sanguinis genome contains only two homologs of the twin-arginine translocation (Tat) system, which exports folded proteins with the characteristic N-terminal twin-arginine motif across the cytoplasmic membrane (65). SSA_1132 and SSA_1133 apparently encode the TatC Sec-independent protein translocase and the TatA Sec-independent protein secretion pathway component, respectively. Of the streptococcus genomes examined to date, this system has been found only in S. thermophilus. Our analysis showed that three genes, encoding a periplasmic lipoprotein involved in iron transport (SSA_1129), an iron-dependent peroxidase (SSA_1130), and a high-affinity Fe2+/Pb2+ permease (SSA_1131) associated with the Tat genes in S. sanguinis, are similarly associated in other genomes, including the genomes of S. thermophilus, Staphylococcus aureus MRSA252, and Staphylococcus haemolyticus. Using the TatP server (8) to search for Tat secretion substrates, we found that the iron-dependent peroxidase gene SSA_1130 was the only ORF in the genome that encoded both a consensus Tat motif and a Tat signal peptide.
Two glucosyltransferases (GTF) were found in S. sanguinis. The SSA_0613 product is a homolog of GtfR of Streptococcus oralis ATCC 10557, which synthesizes water-soluble glucans with no primer dependence (24). The SSA_1006 product is a homolog of GtfA, an enzyme that, in the presence of inorganic phosphate, converts sucrose to fructose and glucose-1-phosphate (4). Furthermore, the products of several ORFs exhibit homology to S. mutans non-GTF glucan-binding proteins (GBP), including the products of SSA_0019, SSA_0303, and SSA_0956. Non-GTF GBPs are cell surface receptors for glucan or secreted proteins that can become cell associated when glucan coats the bacterial cells. Although all GBPs have glucan-binding properties, they are a heterogeneous group of proteins with variations in size, glucan-binding domains, glucan-binding affinity, and function (4).
More than 100 putative transcriptional regulators were identified in the S. sanguinis genome (www.sanguinis.mic.vcu.edu/supplemental.htm). Like the genomes of some other streptococci, the S. sanguinis genome contains genes encoding a major sigma factor 70 (SSA_0825, rpoD) and an ortholog of the competence-specific sigma factor, ComX (SSA_0016). Genes encoding NusA (SSA_1900), NusB (SSA_0452), and NusG (SSA_2205) were found, although no obvious Rho protein was identified. This was also true for the other streptococcal genomes examined. Two genes, SSA_1187 and SSA_1695, code for additional putative antitermination proteins. Two-component regulatory systems, composed of a sensor histidine kinase and a transcriptional response regulator, provide a mechanism for bacteria to sense and respond to environmental signals. We found 29 genes that apparently comprise 14 two-component regulatory systems (www.sanguinis.mic.vcu.edu/supplemental.htm). This number is comparable to the numbers found in other streptococci (2, 26, 37, 80, 87). The “orphan” two-component response regulator encoded by SSA_1810 is an ortholog of the tissue-specific virulence factor RitR that represses the hemin-iron transport system in S. pneumoniae (92) and of the virulence factor CsrR in S. pyogenes (29), suggesting that this regulator may have a similar role in virulence in S. sanguinis.
S. sanguinis is one of the pioneer colonizers of the oral cavity and may initiate biofilm formation on tooth surfaces. Several putative biofilm-related genes are found in S. sanguinis and most other streptococci. For example, SSA_0135 to SSA_0137 are clustered in an arrangement similar to that observed for their orthologs in the adc operon, which is involved in biofilm formation in S. gordonii (52). Genes of the inducible fructose phosphotransferase operon, which is also related to biofilm formation in S. gordonii (51), are similarly clustered in S. sanguinis (SSA_1080 to SSA_1082). The SSA_1909 product is more than 60% identical to biofilm regulatory protein A (BrpA) in S. mutans. BrpA codes for a predicted surface-associated protein with functions not only in biofilm formation, autolysis, and cell division but also in the regulation of acid and oxidative stress tolerance in S. mutans (95).
SSA_1853 is an ortholog of the LuxS gene in S. oralis 34, which is responsible for the catabolism of S-ribosylhomocysteine, producing autoinducer 2, a universal signal molecule mediating cell-cell and interspecies communication (quorum sensing) among bacteria, biofilm formation, and virulence (74).
Conclusion.
S. sanguinis is one of the most frequently recognized pioneering inhabitants of human oral plaque (76). Completion of its genome sequence provided unique insight into the biology, virulence, and pathogenesis of this important bacterium. The greater size and G+C content of the S. sanguinis genome reflect the differences between this organism and other streptococci. The genome has clearly been molded by HGT, and the mechanisms by which the large cluster of genes in the cob, pdu, and eut pathways were transferred and confer a selective advantage to S. sanguinis are rich subjects for future investigations. Our analysis of the genome also provided fundamental genetic data for investigating the etiology of caries by comparison with cariogenic S. mutans. The biology and metabolism of this important bacterium have been described so that new prophylactic and therapeutic strategies can now be explored. Finally, in previous studies workers have used many different strains of S. sanguinis, several of which would now be classified as S. gordonii, S. parasanguinis, or other species. The availability of the SK36 sequence, as well as the bacterium, which has been deposited in the American Type Culture Collection (catalog no. BAA-1455), should facilitate future studies with this species.
Supplementary Material
Acknowledgments
This work was supported by PHS grants DE12882 from the National Institute of Dental and Craniofacial Research (to F.L.M. and G.A.B.) and AI47841 and AI054908 from the National Institute of Allergy and Infectious Disease (to T.K.) and by grant J743 from the Jeffress Trust (to P.X.).
Sequence analysis was performed in the Nucleic Acids Research Facilities at Virginia Commonwealth University.
Footnotes
Published ahead of print on 2 February 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahmed, R., T. Hassall, B. Morland, and J. Gray. 2003. Viridans streptococcus bacteremia in children on chemotherapy for cancer: an underestimated problem. Pediatr. Hematol. Oncol. 20:439-444. [PubMed] [Google Scholar]
- 2.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Banas, J. A., and M. M. Vickerman. 2003. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral Biol. Med. 14:89-99. [DOI] [PubMed] [Google Scholar]
- 5.Banks, D. J., S. F. Porcella, K. D. Barbian, S. B. Beres, L. E. Philips, J. M. Voyich, F. R. DeLeo, J. M. Martin, G. A. Somerville, and J. M. Musser. 2004. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 190:727-738. [DOI] [PubMed] [Google Scholar]
- 6.Barnard, J. P., and M. W. Stinson. 1996. The alpha-hemolysin of Streptococcus gordonii is hydrogen peroxide. Infect. Immun. 64:3853-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barocchi, M. A., J. Ries, X. Zogaj, C. Hemsley, B. Albiger, A. Kanth, S. Dahlberg, J. Fernebro, M. Moschioni, V. Masignani, K. Hultenby, A. R. Taddei, K. Beiter, F. Wartha, A. von Euler, A. Covacci, D. W. Holden, S. Normark, R. Rappuoli, and B. Henriques-Normark. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. USA 103:2857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bendtsen, J. D., H. Nielsen, D. Widdick, T. Palmer, and S. Brunak. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bensing, B. A., and P. M. Sullam. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 44:1081-1094. [DOI] [PubMed] [Google Scholar]
- 10.Beres, S. B., E. W. Richter, M. J. Nagiec, P. Sumby, S. F. Porcella, F. R. DeLeo, and J. M. Musser. 2006. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc. Natl. Acad. Sci. USA 103:7059-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beres, S. B., G. L. Sylva, K. D. Barbian, B. F. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. M. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brinsmade, S. R., and J. C. Escalante-Semerena. 2004. The eutD gene of Salmonella enterica encodes a protein with phosphotransacetylase enzyme activity. J. Bacteriol. 186:1890-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caufield, P. W., A. P. Dasanayake, Y. Li, Y. Pan, J. Hsu, and J. M. Hardin. 2000. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect. Immun. 68:4018-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, Q., H. Wu, and P. M. Fives-Taylor. 2004. Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguis FW213. Mol. Microbiol. 53:843-856. [DOI] [PubMed] [Google Scholar]
- 18.Chen, Y. Y., and R. A. Burne. 2003. Identification and characterization of the nickel uptake system for urease biogenesis in Streptococcus salivarius 57.I. J. Bacteriol. 185:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cisar, J. O., A. L. Sandberg, G. P. Reddy, C. Abeygunawardana, and C. A. Bush. 1997. Structural and antigenic types of cell wall polysaccharides from viridans group streptococci with receptors for oral actinomyces and streptococcal lectins. Infect. Immun. 65:5035-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conner, C. P., D. M. Heithoff, S. M. Julio, R. L. Sinsheimer, and M. J. Mahan. 1998. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl. Acad. Sci. USA 95:4641-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daep, C. A., D. M. James, R. J. Lamont, and D. R. Demuth. 2006. Structural characterization of peptide-mediated inhibition of Porphyromonas gingivalis biofilm formation. Infect. Immun. 74:5756-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujiwara, T., T. Hoshino, T. Ooshima, S. Sobue, and S. Hamada. 2000. Purification, characterization, and molecular analysis of the gene encoding glucosyltransferase from Streptococcus oralis. Infect. Immun. 68:2475-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaustad, P. 1979. Genetic transformation in Streptococcus sanguis. Distribution of competence and competence factors in a collection of strains. Acta Pathol. Microbiol. Scand. Sect. B 87B:123-128. [PubMed] [Google Scholar]
- 26.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 27.Gong, K., L. Mailloux, and M. C. Herzberg. 2000. Salivary film expresses a complex, macromolecular binding site for Streptococcus sanguis. J. Biol. Chem. 275:8970-8974. [DOI] [PubMed] [Google Scholar]
- 28.Green, N. M., S. Zhang, S. F. Porcella, M. J. Nagiec, K. D. Barbian, S. B. Beres, R. B. LeFebvre, and J. M. Musser. 2005. Genome sequence of a serotype M28 strain of group A streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect. Dis. 192:760-770. [DOI] [PubMed] [Google Scholar]
- 29.Gryllos, I., J. C. Levin, and M. R. Wessels. 2003. The CsrR/CsrS two-component system of group A Streptococcus responds to environmental Mg2+. Proc. Natl. Acad. Sci. USA 100:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Handley, P. S., P. L. Carter, J. E. Wyatt, and L. M. Hesketh. 1985. Surface structures (peritrichous fibrils and tufts of fibrils) found on Streptococcus sanguis strains may be related to their ability to coaggregate with other oral genera. Infect. Immun. 47:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hasona, A., P. J. Crowley, C. M. Levesque, R. W. Mair, D. G. Cvitkovitch, A. S. Bleiweis, and L. J. Brady. 2005. Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc. Natl. Acad. Sci. USA 102:17466-17471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Håvarstein, L. S., R. Hakenbeck, and P. Gaustad. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heath, R. J., and C. O. Rock. 2000. A triclosan-resistant bacterial enzyme. Nature 406:145-146. [DOI] [PubMed] [Google Scholar]
- 34.Heath, R. J., S. W. White, and C. O. Rock. 2001. Lipid biosynthesis as a target for antibacterial agents. Prog. Lipid Res. 40:467-497. [DOI] [PubMed] [Google Scholar]
- 35.Herzberg, M. C., A. Nobbs, L. Tao, A. Kilic, E. Beckman, A. Khammanivong, and Y. Zhang. 2005. Oral streptococci and cardiovascular disease: searching for the platelet aggregation-associated protein gene and mechanisms of Streptococcus sanguis-induced thrombosis. J. Periodontol. 76:2101-2105. [DOI] [PubMed] [Google Scholar]
- 36.Holland, I. B., L. Schmitt, and J. Young. 2005. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway. Mol. Membr. Biol. 22:29-39. [DOI] [PubMed] [Google Scholar]
- 37.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu, S. D., J. O. Cisar, A. L. Sandberg, and M. Kilian. 1994. Adhesive properties of viridans group streptococcal species. Microb. Ecol. Health Dis. 7:125-137. [Google Scholar]
- 39.Jakubovics, N. S., N. Stromberg, C. J. van Dolleweerd, C. G. Kelly, and H. F. Jenkinson. 2005. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol. Microbiol. 55:1591-1605. [DOI] [PubMed] [Google Scholar]
- 40.Kapadia, C. R. 1995. Vitamin B12 in health and disease: part I—inherited disorders of function, absorption, and transport. Gastroenterologist 3:329-344. [PubMed] [Google Scholar]
- 41.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 42.Kilian, M., and K. Holmgren. 1981. Ecology and nature of immunoglobulin A1 protease-producing streptococci in the human oral cavity and pharynx. Infect. Immun. 31:868-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kilian, M., L. Mikkelsen, and J. Henrichsen. 1989. Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven 1946), Streptococcus oralis (Bridge and Sneath 1982), and Streptococcus mitis (Andrewes and Horder 1906). Int. J. Syst. Bacteriol. 39:471-484. [Google Scholar]
- 44.Kita, K., H. Kotani, H. Ohta, H. Yanase, and N. Kato. 1992. StsI, a new FokI isoschizomer from Streptococcus sanguis 54, cleaves 5′ GGATG(N)10/14 3′. Nucleic Acids Res. 20:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolenbrander, P. E., R. N. Andersen, and L. V. Moore. 1990. Intrageneric coaggregation among strains of human oral bacteria: potential role in primary colonization of the tooth surface. Appl. Environ Microbiol. 56:3890-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolenbrander, P. E., and J. London. 1993. Adhere today, here tomorrow: oral bacterial adherence. J. Bacteriol. 175:3247-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lacks, S. A., and S. S. Springhorn. 1984. Transfer of recombinant plasmids containing the gene for DpnII DNA methylase into strains of Streptococcus pneumoniae that produce DpnI or DpnII restriction endonucleases. J. Bacteriol. 158:905-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lanie, J. A., W. L. Ng, K. M. Kazmierczak, T. M. Andrzejewski, T. M. Davidsen, K. J. Wayne, H. Tettelin, J. I. Glass, and M. E. Winkler. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lauer, P., C. D. Rinaudo, M. Soriani, I. Margarit, D. Maione, R. Rosini, A. R. Taddei, M. Mora, R. Rappuoli, G. Grandi, and J. L. Telford. 2005. Genome analysis reveals pili in group B Streptococcus. Science 309:105. [DOI] [PubMed] [Google Scholar]
- 50.Lewis, S. E., S. M. Searle, N. Harris, M. Gibson, V. Lyer, J. Richter, C. Wiel, L. Bayraktaroglir, E. Birney, M. A. Crosby, J. S. Kaminker, B. B. Matthews, S. E. Prochnik, C. D. Smithy, J. L. Tupy, G. M. Rubin, S. Misra, C. J. Mungall, and M. E. Clamp. 2002. Apollo: a sequence annotation editor. Genome Biol. 3:RESEARCH0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loo, C. Y., K. Mitrakul, I. B. Voss, C. V. Hughes, and N. Ganeshkumar. 2003. Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J. Bacteriol. 185:6241-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loo, C. Y., K. Mitrakul, I. B. Voss, C. V. Hughes, and N. Ganeshkumar. 2003. Involvement of the adc operon and manganese homeostasis in Streptococcus gordonii biofilm formation. J. Bacteriol. 185:2887-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love, R. M., M. D. McMillan, Y. Park, and H. F. Jenkinson. 2000. Coinvasion of dentinal tubules by Porphyromonas gingivalis and Streptococcus gordonii depends upon binding specificity of streptococcal antigen I/II adhesin. Infect. Immun. 68:1359-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mackiewicz, P., J. Zakrzewska-Czerwinska, A. Zawilak, M. R. Dudek, and S. Cebrat. 2004. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 32:3781-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Macrina, F. L., K. R. Jones, and P. Laloi. 1996. Characterization of IS199 from Streptococcus mutans V403. Plasmid 36:9-18. [DOI] [PubMed] [Google Scholar]
- 56.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marrakchi, H., W. E. Dewolf, Jr., C. Quinn, J. West, B. J. Polizzi, C. Y. So, D. J. Holmes, S. L. Reed, R. J. Heath, D. J. Payne, C. O. Rock, and N. G. Wallis. 2003. Characterization of Streptococcus pneumoniae enoyl-(acyl-carrier protein) reductase (FabK). Biochem. J. 370:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marri, P. R., W. Hao, and G. B. Golding. 2006. Gene gain and gene loss in Streptococcus: is it driven by habitat? Mol. Biol. Evol. 23:2379-2391. [DOI] [PubMed] [Google Scholar]
- 59.McNab, R., H. Forbes, P. S. Handley, D. M. Loach, G. W. Tannock, and H. F. Jenkinson. 1999. Cell wall-anchored CshA polypeptide (259 kilodaltons) in Streptococcus gordonii forms surface fibrils that confer hydrophobic and adhesive properties. J. Bacteriol. 181:3087-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McNab, R., H. F. Jenkinson, D. M. Loach, and G. W. Tannock. 1994. Cell-surface-associated polypeptides CshA and CshB of high molecular mass are colonization determinants in the oral bacterium Streptococcus gordonii. Mol. Microbiol. 14:743-754. [DOI] [PubMed] [Google Scholar]
- 61.Mewes, H. W., D. Frishman, K. F. Mayer, M. Munsterkotter, O. Noubibou, P. Pagel, T. Rattei, M. Oesterheld, A. Ruepp, and V. Stumpflen. 2006. MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res. 34:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Min, B., J. T. Pelaschier, D. E. Graham, D. Tumbula-Hansen, and D. Soll. 2002. Transfer RNA-dependent amino acid biosynthesis: an essential route to asparagine formation. Proc. Natl. Acad. Sci. USA 99:2678-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mora, M., G. Bensi, S. Capo, F. Falugi, C. Zingaretti, A. G. O. Manetti, T. Maggi, A. R. Taddei, G. Grandi, and J. L. Telford. 2005. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. USA 102:15641-15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morris, E. J., and B. C. McBride. 1984. Adherence of Streptococcus sanguis to saliva-coated hydroxyapatite: evidence for two binding sites. Infect. Immun. 43:656-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muller, M., and R. B. Klosgen. 2005. The Tat pathway in bacteria and chloroplasts. Mol. Membr. Biol. 22:113-121. [DOI] [PubMed] [Google Scholar]
- 66.Mylonakis, E., and S. B. Calderwood. 2001. Infective endocarditis in adults. N. Engl. J. Med. 345:1318-1330. [DOI] [PubMed] [Google Scholar]
- 67.Nakagawa, I., K. Kurokawa, A. Yamashita, M. Nakata, Y. Tomiyasu, N. Okahashi, S. Kawabata, K. Yamazaki, T. Shiba, T. Yasunaga, H. Hayashi, M. Hattori, and S. Hamada. 2003. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 13:1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paba, J., C. A. Ricart, W. Fontes, J. M. Santana, A. R. Teixeira, J. Marchese, B. Williamson, T. Hunt, B. L. Karger, and M. V. Sousa. 2004. Proteomic analysis of Trypanosoma cruzi developmental stages using isotope-coded affinity tag reagents. J. Proteome Res. 3:517-524. [DOI] [PubMed] [Google Scholar]
- 69.Paik, S., L. Senty, S. Das, J. C. Noe, C. L. Munro, and T. Kitten. 2005. Identification of virulence determinants for endocarditis in Streptococcus sanguinis by signature-tagged mutagenesis. Infect. Immun. 73:6064-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051-1070. [DOI] [PubMed] [Google Scholar]
- 71.Prabhu, R. M., K. E. Piper, M. R. Litzow, J. M. Steckelberg, and R. Patel. 2005. Emergence of quinolone resistance among viridans group streptococci isolated from the oropharynx of neutropenic peripheral blood stem cell transplant patients receiving quinolone antimicrobial prophylaxis. Eur. J. Clin. Microbiol. Infect. Dis. 24:832-838. [DOI] [PubMed] [Google Scholar]
- 72.Raczniak, G., H. D. Becker, B. Min, and D. Soll. 2001. A single amidotransferase forms asparaginyl-tRNA and glutaminyl-tRNA in Chlamydia trachomatis. J. Biol. Chem. 276:45862-45867. [DOI] [PubMed] [Google Scholar]
- 73.Rakotoarivonina, H., G. Jubelin, M. Hebraud, B. Gaillard-Martinie, E. Forano, and P. Mosoni. 2002. Adhesion to cellulose of the gram-positive bacterium Ruminococcus albus involves type IV pili. Microbiology 148:1871-1880. [DOI] [PubMed] [Google Scholar]
- 74.Rickard, A. H., R. J. Palmer, Jr., D. S. Blehert, S. R. Campagna, M. F. Semmelhack, P. G. Egland, B. L. Bassler, and P. E. Kolenbrander. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60:1446-1456. [DOI] [PubMed] [Google Scholar]
- 75.Roof, D. M., and J. R. Roth. 1988. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170:3855-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosan, B., and R. J. Lamont. 2000. Dental plaque formation. Microbes Infect. 2:1599-1607. [DOI] [PubMed] [Google Scholar]
- 77.Salzberg, S. L., M. Pertea, A. L. Delcher, M. J. Gardner, and H. Tettelin. 1999. Interpolated Markov models for eukaryotic gene finding. Genomics 59:24-31. [DOI] [PubMed] [Google Scholar]
- 78.Scott, A. I., and C. A. Roessner. 2002. Biosynthesis of cobalamin (vitamin B12). Biochem. Soc. Trans. 30:613-620. [DOI] [PubMed] [Google Scholar]
- 79.Sheppard, D. E., J. T. Penrod, T. Bobik, E. Kofoid, and J. R. Roth. 2004. Evidence that a B12-adenosyl transferase is encoded within the ethanolamine operon of Salmonella enterica. J. Bacteriol. 186:7635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Socransky, S. S., A. D. Manganiello, D. Propas, V. Oram, and J. van Houte. 1977. Bacteriological studies of developing supragingival dental plaque. J. Periodontal Res. 12:90-106. [DOI] [PubMed] [Google Scholar]
- 82.Stephenson, A. E., H. Wu, J. Novak, M. Tomana, K. Mintz, and P. Fives-Taylor. 2002. The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol. Microbiol. 43:147-157. [DOI] [PubMed] [Google Scholar]
- 83.Sumby, P., S. F. Porcella, A. G. Madrigal, K. D. Barbian, K. Virtaneva, S. M. Ricklefs, D. E. Sturdevant, M. R. Graham, J. Vuopio-Varkila, N. P. Hoe, and J. M. Musser. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192:771-782. [DOI] [PubMed] [Google Scholar]
- 84.Sung, C. K., and D. A. Morrison. 2005. Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae. J. Bacteriol. 187:3052-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takamatsu, D., B. A. Bensing, and P. M. Sullam. 2005. Two additional components of the accessory sec system mediating export of the Streptococcus gordonii platelet-binding protein GspB. J. Bacteriol. 187:3878-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. DeBoy, T. M. Davidsen, M. Mora, M. Scarselli, I. Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 89.Tettelin, H., D. Radune, S. Kasif, H. Khouri, and S. L. Salzberg. 1999. Optimized multiplex PCR: efficiently closing a whole-genome shotgun sequencing project. Genomics 62:500-507. [DOI] [PubMed] [Google Scholar]
- 90.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 53:251-261. [DOI] [PubMed] [Google Scholar]
- 91.Truper, H. and. L. D. Clari. 1997. Taxonomic note: necessary corrections of specific epithets formed as substantives (nouns) “in apposition”. Int. J. Syst. Bacteriol. 47:908-909. [Google Scholar]
- 92.Ulijasz, A. T., D. R. Andes, J. D. Glasner, and B. Weisblum. 2004. Regulation of iron transport in Streptococcus pneumoniae by RitR, an orphan response regulator. J. Bacteriol. 186:8123-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van de Guchte, M., S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J. M. Batto, T. Walunas, J. F. Gibrat, P. Bessieres, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA 103:9274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Welch, R. A. 1991. Pore-forming cytolysins of gram-negative bacteria. Mol. Microbiol. 5:521-528. [DOI] [PubMed] [Google Scholar]
- 95.Wen, Z. T., H. V. Baker, and R. A. Burne. 2006. Influence of BrpA on critical virulence attributes of Streptococcus mutans. J. Bacteriol. 188:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu, H., K. P. Mintz, M. Ladha, and P. M. Fives-Taylor. 1998. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol. Microbiol. 28:487-500. [DOI] [PubMed] [Google Scholar]
- 97.Xu, D. Q., J. Thompson, and J. O. Cisar. 2003. Genetic loci for coaggregation receptor polysaccharide biosynthesis in Streptococcus gordonii 38. J. Bacteriol. 185:5419-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu, P., G. Widmer, Y. Wang, L. S. Ozaki, J. M. Alves, M. G. Serrano, D. Puiu, P. Manque, D. Akiyoshi, A. J. Mackey, W. R. Pearson, P. H. Dear, A. T. Bankier, D. L. Peterson, M. S. Abrahamsen, V. Kapur, S. Tzipori, and G. A. Buck. 2004. The genome of Cryptosporidium hominis. Nature 431:1107-1112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.