Abstract
Burkholderia vietnamiensis has both the cepIR quorum-sensing system that is widely distributed among the Burkholderia cepacia complex (BCC) and the bviIR system. Comparison of the expression of cepI, cepR, bviI, and bviR-luxCDABE fusions in B. vietnamiensis G4 and the G4 cepR and bviR mutants determined that the expression of bviI requires both a functional cognate regulator, BviR, and functional CepR. The cepIR system, however, is not regulated by BviR. Unlike the cepIR genes in other BCC species, the cepIR genes are not autoregulated in G4. N-Acyl-homoserine lactone (AHL) production profiles in G4 cepI, cepR, bviI, and bviR mutants confirmed the regulatory organization of the G4 quorum-sensing systems. The regulatory network in strain PC259 is similar to that in G4, except that CepR positively regulates cepI and negatively regulates cepR. AHL production and the bviI expression levels in seven B. vietnamiensis isolates were compared. All strains produced N-octanoyl-homoserine lactone and N-hexanoyl-homoserine lactone; however, only one of four clinical strains but all three environmental strains produced the BviI synthase product, N-decanoyl-homoserine lactone (DHL). The three strains that did not produce DHL expressed bviR but not bviI. Heterologous expression of bviR restored DHL production in these strains. The bviIR loci of the non-DHL-producing strains were sequenced to confirm that bviR encodes a functional transcriptional regulator. Lack of expression of G4 bviI in these three strains indicated that an additional regulatory element may be involved in the regulation of bviIR expression in certain strains of B. vietnamiensis.
Burkholderia vietnamiensis is a part of the B. cepacia complex (BCC), a group of nine closely related bacterial species with extreme metabolic versatility. The beneficial metabolic properties of B. vietnamiensis, including bioremediation and plant growth promotion, have been well characterized (38, 45). B. vietnamiensis G4 produces ortho-monooxygenase that degrades the groundwater contaminant trichloroethylene. B. vietnamiensis strains are also able to fix atmospheric nitrogen and enhance crop yields, presumably by increasing the available nitrogen in the rhizosphere (24). Inoculation of rice with B. vietnamiensis significantly increases grain yield and is potentially an economic alternative to nitrogen-based fertilizers (60).
In addition to having biocontrol, bioremediation, and plant growth-enhancing applications (38, 45), BCC species are also opportunistic pathogens of particular importance for people with cystic fibrosis (CF) and chronic granulomatous disease (41). Each BCC species has been isolated from CF patients; however, the species vary in frequency of colonization, transmissibility, and geographic distribution. Burkholderia cenocepacia is the most commonly reported BCC species isolated from CF patients in North America, where 80 percent of BCC isolates recovered from CF patients in Canada and 50 percent of BCC isolates recovered from patients in the United States are B. cenocepacia (35, 56). There is a low incidence of B. vietnamiensis infections in North American CF patients; such infections comprise 1.6 percent of Canadian and 5.1 percent of U.S. CF patient BCC isolates (47, 57). An epidemiological study conducted in Brazil with 11 patients, however, reported equal incidences of B. cenocepacia and B. vietnamiensis isolates from CF patients, illustrating the potential of B. vietnamiensis as a CF pathogen (16). The pathogenic potential of B. vietnamiensis has delayed further research on its biotechnological applications (9).
BCC species utilize N-acyl-homoserine lactone (AHL)-based quorum-sensing systems for the regulation of diverse physiological processes, including those involved in virulence. Quorum sensing is a form of genetic regulation typically mediated by the accumulation of self-produced signal compounds in the environment. AHL-mediated quorum-sensing systems are comprised of a luxI homologue, which encodes an AHL synthase that catalyzes the synthesis of an AHL signal molecule(s), and a luxR homologue that encodes a transcriptional regulator that mediates gene expression in its active, AHL-bound form (22).
The cepIR quorum-sensing system is widely distributed among BCC strains (26, 36). CepI directs the synthesis of N-octanoyl-homoserine lactone (OHL) and N-hexanoyl-homoserine lactone (HHL) (32, 33). In B. cepacia, the cepIR quorum-sensing system is autoregulated in that CepR positively regulates the expression of cepI (2). Transcriptional analysis of a B. cepacia promoter library identified 28 genes that are positively regulated by CepR (2). The B. cepacia cepIR system also negatively regulates the stationary-phase sigma factor rpoS, positively regulates protease production, and contributes to onion maceration by positively regulating polygalacturonase production (1).
In B. cenocepacia, CepR positively regulates cepI expression and negatively controls its own expression (33). The cepIR quorum-sensing system in B. cenocepacia is involved in the regulation of swarming motility, mature biofilm development, chitinase production, extracellular protease production, and the biosynthesis of the siderophore ornibactin (29, 32-34). Proteomic analysis of B. cenocepacia cepI mutant revealed that 55 of 985 examined proteins are differentially expressed in the quorum-sensing mutant and the wild type (48). Animal and nematode infection models have demonstrated a role for the cepIR quorum-sensing system in B. cenocepacia virulence (30, 55).
In epidemic strains of B. cenocepacia that possess the B. cenocepacia pathogenicity island (cci), CepR is required for the expression of an additional quorum-sensing system, cciIR (5, 39). The predominant AHL produced by the AHL synthase, CciI, is HHL with minor amounts of OHL (39). The cciI and cciR genes are cotranscribed, and CciR negatively regulates the expression of the cciIR operon. CciR is involved in the negative regulation of cepI as well as in the regulation of extracellular protease production and swarming motility (39).
B. vietnamiensis strains possess an additional quorum-sensing system, bviIR. PCR amplification and Southern hybridization analysis revealed that B. vietnamiensis is the only BCC species to contain the bviIR system (10, 36). In addition to OHL and HHL, B. vietnamiensis strains produce N-decanoyl-homoserine lactone (DHL), N-dodecanoyl-homoserine lactone, and N-(3oxodecanoyl)-homoserine lactone (10, 11, 44). Mutations constructed in bviI and bviR of B. vietnamiensis strain G4 (bviIG4 and bviRG4, respectively) indicated that BviI is responsible for the synthesis of all AHLs produced by G4 in that the bviI mutant did not produce detectable levels of any AHL except minor amounts of OHL (10). Although B. vietnamiensis possesses the genes for two quorum-sensing systems, its AHL production has been shown to be strain dependent with regard to type and quantity of AHL produced (11, 26, 72).
Few phenotypes are known to be regulated by the bviIR system. One study suggested that a B. vietnamiensis G4 transposon mutant with decreased AHL production showed reduced antibiotic production, although this antibiotic has yet to be characterized (44). In the same study, it was determined that the degradation of toluene and thus the expression of toluene ortho-monooxygenase is not regulated by quorum sensing (44). There is also evidence that the bviIR system is not involved in siderophore production (10).
The objectives of this study were to further characterize the bviIR and cepIR quorum-sensing systems of B. vietnamiensis G4, as well as the regulatory relationship between the two quorum-sensing systems. In this study we also investigate the basis for the variations in AHL production in clinical and environmental B. vietnamiensis strains at a molecular level.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. For genetic manipulations, cultures were routinely grown at 37°C in Luria-Bertani broth (Invitrogen, Burlington, Ontario, Canada) with shaking (200 rpm) or on 1.5% LB agar plates. When appropriate, the following concentrations of antibiotics were used: 100 μg/ml of trimethoprim (Tp), 30 μg/ml of tetracycline (Tc), 20 μg/ml of gentamicin (Gm), and 80 μg/ml of chloramphenicol (Cm) for B. vietnamiensis G4; 100 μg/ml of Tp, 250 μg/ml of Tc, and 400 μg/ml of kanamycin (Km) for B. vietnamiensis PC259; and 1.5 mg/ml Tp, 15 μg/ml of Tc, 25 μg/ml Gm, 35 μg/ml of Cm, and 50 μg/ml of Km for Escherichia coli. Antibiotics were purchased from Sigma-Aldrich Canada, Ltd. (Oakville, Ontario, Canada). For RNA isolation, luminescence assays, biofilm assays, the alfalfa model of infection, and AHL extractions, cultures were grown in Trypticase soy broth (Difco, Franklin Lakes, NJ) at 30°C. For chrome azurol S assays, cultures were grown at 32°C in succinate medium supplemented with ornithine (10 mM) (40). For examination of swarming motility, cultures were grown in nutrient broth (Difco) supplemented with 0.5% glucose.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain (alternate name) or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF) recA1 endA gyrA96 thi-1 hsdR17 supE44 relAl deoR U169 | Invitrogen |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Str) endA1 nupG | Invitrogen |
| HB101 | supE44 hsdS20(rB mB) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 49 |
| A. tumefaciens | ||
| A136 | Ti plasmidless host | C. Fuqua |
| B. vietnamiensis | ||
| PC259 (LMG18835) | Isolated from a patient with CF in Washington State | 31 |
| FC466 (LMG18836) | Isolated from a patient with CF in Sweden | 64 |
| FC441 (LMG16232) | Isolated from a patient with septic granulomatous disease in British Columbia, Canada | 37 |
| C2822 | Isolated from a patient with CF in British Columbia, Canada | E. Mahenthiralingam |
| FC369T (LMG10929T) | Isolated from a rice rhizosphere in Vietnam | 24 |
| DBO1 (LMG17830) | Environmental isolate | 67 |
| G4 (R1808) | Isolated from a water treatment facility in Florida | 43 |
| IMT-61 | bviI::aacCI derivative of G4, Gmr | 10 |
| RMT-14 | bviR::aacCI derivative of G4, Gmr | 10 |
| G4cepI | cepI::cat derivative of G4, Cmr | This study |
| G4cepR | cepR::cat derivative of G4, Cmr | This study |
| G4cepIGSV | cepI::pRMSVI derivative of G4, Gmr | This study |
| G4cepRGSV | cepR::pRMSVR derivative of G4, Gmr | This study |
| PC259cepR | cepR::dhfRII derivative of PC259, Tpr | This study |
| M. luteus | ||
| ATCC 9341 | Soil isolate | T. Louie |
| Plasmids | ||
| pEX18Tc | Suicide vector, sacB Tcr | 27 |
| pUCP26 | Broad-host-range vector, Tcr | 70 |
| pUCP28T | Broad-host-range vector, Tpr | 51 |
| pCR2.1Topo | Cloning vector for PCR products, Apr Kmr | Invitrogen |
| pRK2013 | ColE1 Tra (RK2)+ Kmr | 21 |
| pCF218 | IncP plasmid expressing TraR, Tcr | 73 |
| pCF372 | pUCD2 with a traI-lacZ fusion, Spr | 23 |
| p34S-Cm | Source of the Cm resistance cassette | 14 |
| pRM6.8 | pUCP26 with a 6.8-kb BamHI fragment containing the cepIR locus from PC259 | This study |
| pRM282 | pUCP28T with a 1.5-kb PCR-amplified cepR fragment from G4, Tpr | This study |
| pRM284 | pUCP28T with a 1.1-kb PCR-amplified bviR fragment from G4, Tpr | This study |
| pRM285 | pUCP28T with a 997-bp PCR-amplified bviI fragment from G4, Tpr | This study |
| pRM292 | pUCP28T with a 1,426-bp PCR-amplified 7350 fragment from G4, Tpr | This study |
| pRM2X1 | pEX18Tc with a 2.3-kb PCR-amplified cepR fragment from G4, Tcr | This study |
| pRM2X1-Cm | PRM2X2 with the cepR gene disrupted by a Cm resistance cassette at the PstI site, Tcr Cmr | This study |
| pRM2X2-Cm | pEX18Tc with the disrupted cepI fragment from pRM2T2-Cm, Tcr Cmr | This study |
| pEXCEPI | pEX18Tc containing the cepI gene disrupted by the Tp resistance cassette, Tpr Tcr | 32 |
| pEXCEPR | pEX18Tc containing the cepR gene disrupted by the Tp resistance cassette, Tpr Tcr | 33 |
| pMS402 | luxCDABE-based promoter reporter plasmid, Kmr Tpr | 18 |
| pRM452 | pMS402 containing the 1.8-kb PC259 cepR promoter fragment, Kmr Tpr | This study |
| pRM453 | pMS402 containing the 769-bp PC259 cepI promoter fragment, Kmr Tpr | This study |
| pRM455 | pMS402 containing the 983-bp PC259 bviI promoter fragment, Kmr Tpr | This study |
| pRM462 | pMS402 containing the 266-bp G4 cepR promoter fragment, Kmr Tpr | This study |
| pRM463 | pMS402 containing the 266-bp G4 cepI promoter fragment, Kmr Tpr | This study |
| pRM464 | pMS402 containing the 695-bp G4 bviR promoter fragment, Kmr Tpr | This study |
| pRM465 | pMS402 containing the 983-bp G4 bviI promoter fragment, Kmr Tpr | This study |
| pRM475 | pMS402 containing the 983-bp FC466 bviI promoter fragment | This study |
| pRM485 | pMS402 containing the 983-bp FC441 bviI promoter fragment | This study |
| pRM495 | pMS402 containing the 983-bp FC369TbviI promoter fragment | This study |
| pRM415 | pMS402 containing the 983-bp DBO1 bviI promoter fragment | This study |
| pGSV3 | Mobilizable suicide vector, Gmr | 15 |
| pRMSVI | pGSV3 containing the EcoRI fragment from pRM2T5 from G4, Gmr | This study |
| pRMSVR | pGSV3 containing the EcoRI fragment from pRM2T4, Gmr | This study |
Ap, ampicillin; St, streptomycin; Sp, spectinomycin.
DNA manipulations.
DNA manipulations were performed using standard techniques as described by Sambrook et al. (49). Genomic DNA was isolated as described by Ausubel et al. (4). Genomic DNA for PCR was isolated as described by Walsh et al. (66). Restriction endonucleases and T4 DNA polymerase were purchased from Invitrogen. T4 DNA ligase was purchased from New England Biolabs (Mississauga, Ontario, Canada) or Invitrogen. Shrimp alkaline phosphatase was purchased from Roche (Mannheim, Germany). Oligonucleotide primers (Table 2) were synthesized at Invitrogen or at the University of Calgary Core DNA and Protein Services (Calgary, Alberta, Canada). PCR was performed with either Platinum Taq polymerase (Invitrogen) or Phusion Taq polymerase (Finnzymes, Espoo, Finland) according to the manufacturer-recommended protocol. DNA fragments used in cloning procedures were purified with a QIAquick gel extraction kit (QIAGEN, Mississauga, Ontario, Canada). Plasmids were introduced into B. vietnamiensis G4 by electroporation using a Gene Pulser (Bio-Rad, Richmond, CA) as previously described (13). Plasmids were transferred into all other B. vietnamiensis strains by conjugation employing pRK2013 (21) as the mobilization vector. Nucleotide sequencing was performed by University of Calgary Core DNA and Protein Services. Sequence analysis was performed with DNAMAN sequence analysis software (Lynnon Biosoft, Vandreuil, Quebec, Canada). The sequence data for B. vietnamiensis G4 was provided by the U.S. Department of Energy Joint Genome Institute (JGI) (http://www.jgi.doe.gov/).
TABLE 2.
Oligonucleotide primers used in this study
| Name | Sequence (5′-3′)f | Restriction site | Reference |
|---|---|---|---|
| 7350F | GAGCTGCGCAAGGAACTCAa | This study | |
| 7350R | TGGGTAAAGGACGCGGATAACa | This study | |
| bviIKOF | GTCCGAGGATCCAGAGCGb | BamHI | This study |
| bviIKOR | GCACGCGAAGCTTCACGGb | HindIII | This study |
| bviIproB | CGAATTGGATCCATTATCGGb | BamHI | This study |
| bviIproX | AATAGCCCTCGAGTGGCCb | XhoI | This study |
| bviRKOF | CACGGTCGTCTAGACGAGGb | XbaI | This study |
| bviRKOR | GGCCGAAGCTTGATGAATCGb | HindIII | This study |
| bviRUF | GACCAGCTCGAGGTAGCCG3b | XhoI | This study |
| bviRUR | CAGTGGTCGGATCCGAGCGb | BamHI | This study |
| cepIF | CAGGCGGCGATAGCTTGc | This study | |
| cepIR | CACAGATCCGAGGACATCCAc | This study | |
| cepRF | GAGAAAGAATGGAACTGCGCc | This study | |
| cepRProF | GGCCGCTCGCGACATGGTc | This study | |
| cepRProR | CCGCGGCGCTGAATTGTTGGc | This study | |
| cepRR | TTGTTCACGTGGAAGTTGACc | This study | |
| ExcepI | GCCTGCAGGGCACAACGACGCCTATCATGCc | 33 | |
| G4cepIF | TCAATCCCGCCGATCAAGe | This study | |
| G4cepIR | GCGCGAAAGACCTGAGACTGe | This study | |
| G4cepRHind | CATTTCAAGCTTGAGCTGGACCa | HindIII | This study |
| G4cepRKOF | CGGATCGGTACCTTGGGATGa | KpnI | This study |
| G4cepRKOR | CCGCAAGCTTCCCGTTTCACa | HindIII | This study |
| IN7349F | TCAAGACCCAGCATCTCAATGa | This study | |
| IN7349R | ACCCGACAGGTTGATGAGCa | This study | |
| IN7350F | TCCAACGGACCGGTATGTGa | This study | |
| IN7350R | AGGCCACGAACGGAGAGGTAa | This study | |
| INcepI | GCGGATCCACCAGACGCCCATCTACCTGCTTCGc | 33 | |
| R2Cla3RE | GAACGAAGGTCTGCATGGATGc | This study | |
| RTbviIF | CACGGAGAACGCAATGAGGb | This study | |
| RTbviIR | CACGCGGATACCCTTTACGTCb | This study | |
| RTbviRF | CAGACGTGGGTCGAACGCTAb | This study | |
| RTbviRR | ATAGTTGGCCGTGTTGGCGb | This study | |
| RTsigAF | AATGACCGAGGCGAACCTGa | This study | |
| RTsigAR | TCTTGTCTTCCGGCATCTCCa | This study | |
| UnicepIF | GACCTTCGTTCACGAGGAAGd | This study | |
| UnicepIR | CGTCACGCCGATCAGCTGCd | This study |
Primer designed based on the sequence from the incomplete B. vietnamiensis G4 sequencing project (http://genome.jgi-psf.org/draft_microbes/bur08/bur08.home.html).
Restriction endonuclease sites incorporated into primers are underlined.
Cloning of quorum-sensing genes from G4.
The cepR, bviI, and bviR genes and open reading frame 7350 were PCR amplified from G4 and cloned into the broad-host-range vector pUCP28T (51) as follows. The cepR clone pRM282 was constructed by amplifying a 1,547-bp fragment with the G4cepRKOF and G4cepRHind primers. The bviI clone pRM285 was constructed by amplifying a 997-bp fragment with the bviIKOF and bviIKOR primers. The bviR clone pRM284 was constructed by amplifying an 1,108-bp fragment with the bviRKOF and bviRKOR primers. The open reading frame 7350 clone pRM292 was constructed by amplifying a 1,426-bp fragment with the 7350F and 7350R primers. A 6.8-kb BamHI fragment from PC259 containing cepIR was shotgun cloned into pUCP26 (70). A positive clone was identified by colony hybridization (71) with a 627-bp cepI probe amplified using the cepIF and cepIR primers and designated pRM6.8.
Analysis of quorum-sensing genes in B. vietnamiensis strains.
The presence of the bviIR genes in seven B. vietnamiensis strains was determined by PCR with primers RTbviIF and RTbviIR for bviI and RTbviRF and RTbviRR for bviR. The bviIR loci from strains FC466, FC441, C2822, and G4 were cloned by PCR amplification of a 2.6-kb fragment from genomic DNA using the bviIKOR and bviRKOR primers. The nucleotide sequence for the G4 bviIR locus was confirmed with the incomplete B. vietnamiensis G4 sequencing project (http://www.jgi.doe.gov/), which contained only 1.2 kb of the G4 bviIR locus. Since the complete cepIG4 sequence was not available through the JGI sequencing project, the cepI gene was amplified on an 826-bp fragment with the G4cepIF and G4cepIR primers and sequenced. The presence of the open reading frame 7349 and 7350 LuxRI homologue genes in seven B. vietnamiensis strains was determined by PCR with primers IN7349F and IN7349R and IN7350F and IN7350R, respectively.
Construction of cepI and cepR mutants.
To construct a cepI::cat mutant of B. vietnamiensis G4, a 2.2-kb PstI fragment from pRM6.8 containing cepI was blunt ended and cloned into the EcoRV site of pCR2.1Topo (Invitrogen). A SmaI fragment from p35S-Cm (14) harboring a Cm resistance cassette was inserted into the MluI site of cepI, and the resulting fragment was subcloned into pEX18Tc (27) with BamHI and XbaI, resulting in pRM2X2-Cm, which was transferred into G4 by conjugation. Transconjugants were plated onto Pseudomonas isolation agar (Difco) plates with Cm to select for single-crossover events. Attempts to identify a cepI::cat mutant by screening Cm-resistant colonies for Tc sensitivity and loss of the plasmid were unsuccessful. Attempts were made to construct the cepI mutant with pEXCEPI (32) and other vectors with various amounts of flanking DNA and different resistance cassettes. None were successful; therefore, the G4 cepI mutant (G4cepI) is a merodiploid. The mutation was confirmed by PCR with the internal cepI primers UnicepIF and UnicepIR.
To construct a cepR::cat mutant of G4, a 2.4-kb fragment containing cepR was PCR amplified using the G4cepRKOF and G4cepRKOR primers and cloned into pEX18Tc (27), resulting in pRM2X1. The cepR open reading frame was disrupted at the PstI site by the Cm resistance cassette from p34S-Cm (14), resulting in pRM2X1-Cm. Attempts to construct a cepR::cat mutant were carried out as outlined above, again resulting in the merodiploid, G4cepR. The mutation was confirmed by PCR with the primers cepRF and cepRR. The cepR mutant of PC259 (PC259cepR) was constructed using the allelic-exchange vector pEXCEPR (33). The mutation was confirmed by PCR with the cepRF and cepRR primers and Southern hybridization using a 650-bp cepR probe PCR amplified with the same primers.
Construction of cepI and cepR insertion mutants of G4.
The cepI and cepR genes in B. vietnamiensis G4 were inactivated by insertion of pGSV3 (15). A 426-bp internal cepI fragment was PCR amplified using the UnicepIF and UnicepRR primers, and the resulting fragment was cloned into pCR2.1Topo. An EcoRI fragment containing the cepI fragment was cloned into pGSV3 (15) to construct pRMSVI. The cepR insertional activation construct pRMSVR was constructed in a similar manner following PCR amplification of a 647-bp cepR fragment with the cepRF and cepRR primers. The pRMSVR and pRMSVI constructs were mobilized into G4 by conjugation using pRK2013 (21). Transconjugants were plated onto Pseudomonas isolation agar (Difco) plates containing Gm. Insertions were confirmed by PCR with the G4cepIF and G4cepIR primers for the cepI mutant (G4cepIGSV) and with the G4cepRKOF and G4cepRKOR primers for the cepR mutant (G4cepRGSV).
Construction of luxCDABE transcriptional fusions.
Promoter regions were predicted in silico using SoftBerry BPROM. Promoter fragments were amplified by PCR and cloned upstream of the luxCDABE operon in the XhoI/BamHI promoter cloning site of pMS402 (18) as follows. The cepI-luxCDABE transcriptional fusion plasmid pRM463 was constructed by amplifying a 266-bp promoter fragment using the primers R2Cla3RE and ExcepI (33). The cepR-luxCDABE transcriptional fusion plasmid pRM462 was constructed by amplifying a 227-bp cepR promoter fragment with the cepRProF and cepRProR primers. The bviR-luxCDABE transcriptional fusion plasmid pRM464 was constructed by amplifying a 695-bp promoter fragment with the bviRUF and bviRUR primers. The bviI-luxCDABE transcriptional fusion plasmid pRM465 was constructed by amplifying a 983-bp fragment with the bviIproX and bviIproB primers. The bviI-luxCDABE transcriptional fusions pRM455, pRM475, pRM485, pRM495, and pRM415 were constructed using the respective genomic DNA (PC259, FC466, FC441, FC369T, and DBO1) as a template.
The cepI-luxCDABE transcriptional fusion plasmid pRM453 was constructed by amplifying a 769-bp promoter fragment from PC259 DNA with the INcepI and ExcepI primers (33) The cepR-luxCDABE transcriptional fusion plasmid pRM452 was constructed as previously described (39), using PC259 genomic DNA as the template.
Luminescence assays.
Overnight cultures were subcultured to an initial optical density at 600 nm (OD600) of 0.02 in 20 ml medium. At selected times, 100-μl aliquots were removed and the luminescence in counts per second and turbidity at an OD600 or OD620 were measured using a Wallac Victor2 multilabel counter (Perkin Elmer Life Sciences, Woodbridge, Ontario, Canada) or a MicroBeta TriLux microplate scintillation and luminescence counter (Perkin Elmer Life Sciences). The samples were read in black, clear-bottom, 96-well microtiter plates (Corning, Inc., Corning, NY). The level of promoter expression is reported as the ratio of luminescence to turbidity or relative luminescence. Luminescence assays for screening for gene expression in a 96-well plate format were performed as described above, with overnight cultures being subcultured (1/100) into 150 μl of medium.
RT-PCR.
Overnight cultures were subcultured (1/100) into 20 ml medium and grown for 24 h. Total RNA was isolated from approximately 1 × 109 cells with the RiboPure-Bacteria RNA isolation kit (Ambion, Austin, TX). RNA was treated with amplification-grade DNase I (Invitrogen) before use. Reverse transcription (RT)-PCR was performed using a Titan one-tube RT-PCR kit (Roche) according to the manufacturer's instructions. For each reaction, 50 ng of RNA was used. cDNA was synthesized by RT at 50°C for 40 min. Denaturation was performed for 2 min at 96°C, followed by 35 cycles of PCR as suggested by the manufacturer. A final elongation step at 68°C for 7 min was conducted.
The sigA gene encodes the principal sigma factor (8) and was used as a control gene. A homologue of B. cepacia sigA was identified in the B. vietnamiensis G4 sequencing project (http://www.jgi.doe.gov/. B. vietnamiensis G4 SigA is 89.93% identical to SigA of B. cepacia (accession no. AAD03549) (8).
Three primer sets were designed to internally amplify bviI, bviR, and sigA to yield 297-bp, 297-bp, and 347-bp products, respectively. The annealing temperatures and primers used are as follows: for bviI, 58°C with RTbviIF and RTbviIR; for bviR, 64°C with RTbviRF and RTbviRR; and for sigA, 62°C with RTsigAF and RTsigAR. To ensure that there was no DNA contamination in the RNA samples, PCR was performed on the RNA samples using the RTsigAF and RTsigAR primers with Platinum Taq polymerase (Invitrogen). RTbviRF and RTbviRR primers yielded an approximately 745-bp nonspecific contaminating band (data not shown). This band was sequenced, and a BLAST (3) search indicated that the fragment has homology to B. vietnamiensis LMG10929T 23S rRNA and is not related to bviR.
TLC-AHL bioassays.
AHLs were extracted from 40 ml of culture supernatants with equal volumes of acidified ethyl acetate as described elsewhere (33). Thin-layer chromatography and AHL extraction (TLC-AHL) bioassays were performed as described previously using Agrobacterium tumefaciens A136(pCF218)(pCF372) as a reporter strain (33). This reporter strain is able to identify AHLs with 3-oxo, 3-hydroxy, and 3-unsubstituted side chains ranging from 6 to 12 carbons in length (53). Synthetic HHL, OHL, and DHL (Sigma-Aldrich) were used as reference standards.
Phenotypic characterization.
Siderophore activity present in the culture supernatant fluid was measured by chrome azurol S assays (32, 52). Biofilm formation was determined by staining the cellular matter with crystal violet as described by Tomlin et al. (59), except that biofilms were formed on the polystyrene pegs of a 96-peg replica plate lid inserted in a 96-well plate containing the culture (Nalge Nunc International, Rochester, NY). Swarming motility was evaluated using semisolid agar (0.5%) motility assays as previously described (34). Virulence studies with the alfalfa infection model were performed as previously described (6, 7), with 40 sprouts per group and incubation of the sprouts at 30°C for 7 days. The API 20NE (bioMerieux, St. Laurent, Quebec, Canada) were used for testing basic biochemical attributes as per the instructions of the manufacturer. The ability to fix atmospheric nitrogen was tested by determining growth in the nitrogen-free medium BAz (20) after 48 h of growth at 30°C under anaerobic conditions. Antibiotic production was determined by disk diffusion as previously described (44) on lawns of Micrococcus luteus ATCC 9341.
Statistics.
Statistical analyses, including unpaired t tests and analyses of variance, were performed with INSTAT software (GraphPad Software, San Diego, CA). A P value of <0.05 was considered significant.
Nucleotide sequence accession numbers.
The G4, FC466, FC441, and C2822 bviIR sequences were deposited into the NCBI database with the respective accession nos. EF032807, EF032808, EF032809, and EF032810. In addition, the cepI gene sequence was deposited into the NCBI database with the accession no. EF212890.
RESULTS
Transcriptional analysis of bviIR and cepIR.
To determine whether BviR is involved in the regulation of the bviIR system, the levels of expression of bviI and bviR-luxCDABE transcriptional fusion constructs were compared between G4 and a G4 bviR mutant. The expression of bviI was reduced to almost background levels in the bviR mutant, indicating that BviR is required for the expression of bviI (Fig. 1a). The levels of expression of bviR were similar in G4 and the G4 bviR mutant, demonstrating that BviR is not involved in autoregulation (data not shown).
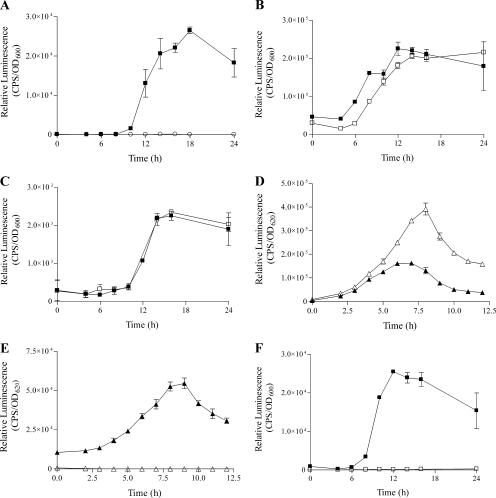
FIG. 1.
Transcriptional analysis of B. vietnamiensis quorum-sensing genes. All values are means ± standard deviations of the results of triplicate assays. ▪, G4; ○, G4 bviR mutant (RMT-14); □, G4cepR (PC259); ▴, PC259; ▵, PC259cepR. (A) Effect of BviR on bviI expression (bviI-luxCDABE, pRM465). (B) Effect of CepR on cepI expression (cepI-luxCDABE, pRM463). (C) Effect of CepR on cepR expression (cepR-luxCDABE, pRM462). (D) Effect of CepR on cepR expression (cepR-luxCDABE, pRM452). The expression of pRM452 was significantly greater in PC259cepR from 3 to 12 h (P < 0.05, t test). (E) Effect of CepR on cepI expression (cepI-luxCDABE, pRM453). (F) Effect of CepR on bviI expression (bviI-luxCDABE, pRM465). CPS, counts per second.
To determine whether CepR is involved in the regulation of cepIR in B. vietnamiensis, transcriptional analysis of cepI and cepR-luxCDABE fusions in G4 and a G4 cepR mutant was performed. Unexpectedly, the levels of expression of cepI and cepR were similar in both the parent and the cepR mutant (Fig. 1B and C), indicating that CepR does not regulate either cepI or cepR in B. vietnamiensis G4. To determine whether the lack of CepR autoregulation was common in B. vietnamiensis, similar comparative expression studies were performed with strain PC259. The expression of cepR was significantly greater in the cepR mutant than in the parent strain from hours 3 to 12 (P < 0.05, t test) (Fig. 1D), indicating that CepR is involved in negative autoregulation. The expression of cepI was reduced to almost background levels in the PC259 cepR mutant (Fig. 1E), indicating that CepR is required for the expression of cepI.
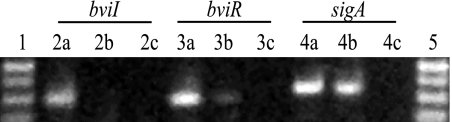
To determine whether there is a regulatory relationship between the cepIR and bviIR systems, comparative transcriptional analysis of cepI and cepR-luxCDABE transcriptional fusions in G4 and the G4 bviR mutant as well as analysis of the expression of bviI and bviR-luxCDABE in G4 and G4cepR were performed. The levels of expression of cepR and cepI were similar in G4 and the G4 bviR mutant (data not shown), indicating that BviR is not involved in the regulation of the cepIR system. The levels of expression of bviR were also similar between the parent and the cepR mutant (data not shown); however, the expression of bviI in the G4 cepR mutant was reduced to almost background levels (Fig. 1F), indicating that CepR positively regulates bviI in G4. To determine whether CepR positively regulates bviI in PC259, RT-PCR was performed to amplify bviI and bviR from PC259 and the PC259 cepR mutant total RNA (Fig. 2). RT-PCR was employed since bviI expression was not detected in PC259 using the bviI- luxCDABE fusion plasmid pRM455 (Table 3). There was a smaller amount of the bviR product in the PC259 cepR mutant than in PC259 (Fig. 2, compare lanes 3a and 3b) and no detectable bviI product in the PC259 cepR mutant (Fig. 2, lane 2b), indicating that CepR positively regulates the bviIR system in PC259. The sigA gene was used as a control, and there was no observable difference in expression between PC259 and the PC259 cepR mutant (Fig. 2, compare lanes 4a and 4b).
FIG. 2.
Effect of CepR on bviIR expression in PC259 by RT-PCR. Products were electrophoresed on a 1.0% gel. Lane 1, ladder; lanes 2a to 2c, product amplified by RTbviIF and RTbviIR; lanes 3a to 3c, product amplified by RTbviRF and RTbviRR; lanes 4a to 4c, product amplified by RTsigAF and RTsigAR; lane 5, ladder. Lanes a, PC259 RNA; lanes b, PC259cepR RNA; lanes c, no-template control.
TABLE 3.
Summary of AHL production, bviIR gene presence, and bviIR expression in clinical and environmental B. vietnamiensis strainsi
| Strain | Source of isolate | HHL productiona | OHL productiona | DHL productiona | Presence of bviIR genesb | Expression of bviI | Expression of bviRg | Presence of 7349 and 7350h |
|---|---|---|---|---|---|---|---|---|
| PC259 | Clinical | + | + | + | + | +c/−d,e | + | − |
| FC466 | Clinical | (+) | (+) | − | + | −c,d,e | (+) | + |
| FC441 | Clinical | + | + | − | + | −c,d,e | (+) | + |
| C2822 | Clinical | + | + | − | + | −c,e | (+) | − |
| FC369T | Environmental | + | + | + | + | +c,d,f | ND | + |
| DBO1 | Environmental | + | + | + | + | +d,f | ND | + |
| G4 | Environmental | + | + | + | + | +c,d,f | + | + |
Production of AHLs was determined by AHL-TLC bioassays using A. tumefaciens A136(pCF218)(pCF372) as the reporter strain.
The presence of the bviI and bviR genes was determined by PCR.
The expression of bviI was assessed by RT-PCR.
The expression of bviI was analyzed by transcriptional analysis of bviI-luxCDABE fusion constructs (pRM455, pRM465, pRM475, pRM485, pRM495, pRM415) in their respective host backgrounds.
The expression of bviIG4 was analyzed by transcriptional analysis of the bviIG4-luxCDABE fusion construct (pRM465).
The expression of bviIPC259 was analyzed by transcriptional analysis of the bviIPC259-luxCDABE fusion construct (pRM465).
The expression of bviR was assessed by RT-PCR with primers internal to bviR.
The presence of the 7349 and 7350 genes was determined by PCR.
+, positive; (+), weakly positive; −, negative; ND, not determined.
Phenotypic characterization of B. vietnamiensis quorum-sensing mutants.
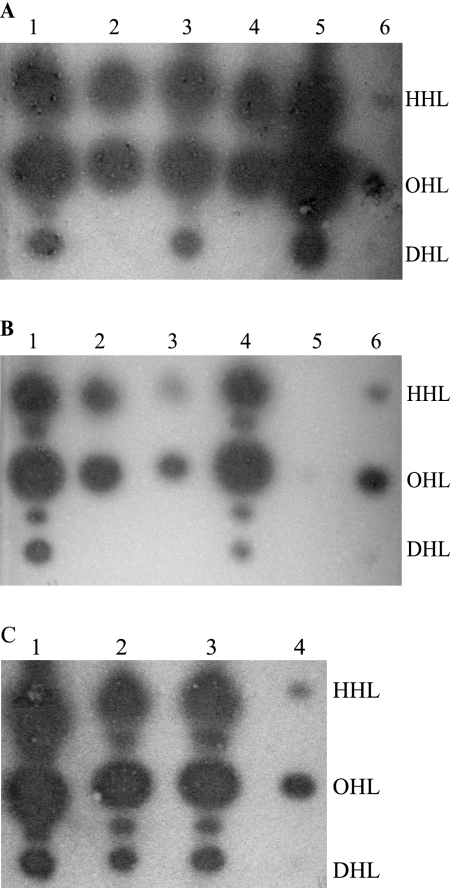
TLC-AHL bioassays of the G4 bviI and bviR mutants were performed (Fig. 3A). Ethyl acetate extracts of spent culture supernatants were chromatographed and AHLs were visualized using an A. tumefaciens A136(pCF218)(pCF372) AHL reporter strain agar overlay. G4 produced detectable amounts of HHL, OHL, and DHL (Fig. 3A, lane 1). DHL production was not detected for the G4 bviI or bviR mutant (Fig. 3A, lanes 2 and 4). The production of DHL was restored when bviI and bviR were present in trans (Fig. 3A, lanes 3 and 5), thus corroborating the role of BviR in the positive regulation of bviI. Considerable amounts of HHL and OHL remained in the AHL production profiles of the bviIR mutants (Fig. 3A, lanes 2 and 4), thereby confirming that the bviIR system is not required for the expression of cepIR.
FIG. 3.
AHL production profiles of the quorum-sensing mutants using TLC-AHL bioassays with the A. tumefaciens(pCF218)(pCF372) reporter strain. (A) AHL production profile of G4 bviI and bviR mutants. Lane 1, G4(pUCP28T); lane 2, G4 bviR(pUCP28T); lane 3, G4 bviR(pRM612) containing the bviR gene; lane 4, G4 bviI(pUCP28T); lane 5, G4 bviI(pRM611) containing the bviI gene; lane 6, synthetic standards. (B) AHL production profiles of cepIR mutants. Lane 1, G4; lane 2, G4cepR; lane 3, G4cepI; lane 4, PC259; lane 5, PC259cepR; lane 6, synthetic standards. (C) AHL production profiles of the G4 cepI and G4 cepR insertion mutants. Lane 1, G4; lane 2, G4cepIGSV; lane 3, G4cepRGSV; lane 4, synthetic standards.
The G4 cepR mutant produces marginally less HHL and OHL than the parent but no detectable amounts of DHL (Fig. 3B, compare lanes 1 and 2), confirming the role of CepR in the positive regulation of bviI and in the negative regulation of cepI. The AHL production profile of the G4 cepI mutant has considerably less HHL and OHL than the parent (Fig. 3B, compare lanes 1 and 3) and no detectable DHL, indicating that cepI is expressed and encodes a functional AHL synthase in G4. The AHLs that remain in the G4 cepI mutant supernatant are presumed to be due to the functional BviI synthase. PC259 produces detectable amounts of HHL, OHL, and DHL (Fig. 3B, lane 4). The AHL production profile for the PC259 cepR mutant was devoid of detectable AHL (Fig. 3B, lane 5), confirming that CepR is involved in the positive regulation of the AHL synthase genes cepI and bviI in this strain.
The discrepancies between the G4 cepR merodiploid and the PC259 cepR haploid mutant, with regard to the regulation of cepIR and AHL production, prompted a second mutant construction strategy for G4. Mutant construction has proven successful in Burkholderia mallei and Burkholderia thailandensis by insertion of pGSV3 (15, 42). G4 cepI and cepR mutants were constructed with this approach and were designated G4cepIGSV and G4cepRGSV, respectively. PCR analysis indicated that the mutant cultures had a mixture of wild-type and mutated cepI or cepR genes (data not shown), indicating that the mutation was not stable. The presence of bacteria with wild-type genes decreased with increasing Gm concentrations (data not shown), although there were always revertants in the population. The AHL production profiles of G4cepIGSV and G4cepRGSV were unaffected (Fig. 3C, compare lanes 2 and 3 with lane 1); therefore, these mutants were not studied further.
Assays, including siderophore biosynthesis, biofilm formation, antibiotic production, nitrogen fixation, and swarming motility assays (12, 29, 32-34, 44), were performed to determine whether the bviIR quorum-sensing system is involved in phenotypes known to be quorum-sensing regulated. Extracellular protease production and expression of the AHL-dependent gene aidA were not investigated, since B. vietnamiensis does not produce proteases and G4 does not contain aidA (25, 26, 72). G4 did not exhibit swarming motility, was not virulent in the alfalfa model of infection, and was unable to grow on nitrogen-free media. Previously, G4 AHL production was correlated with antibacterial activity that inhibited the growth of M. luteus (44); however, we found that M. luteus was not sensitive to G4-secreted products. The levels of siderophore biosynthesis in the G4 bviI and bviR mutants were indistinguishable from that in wild-type G4 (data not shown). The bviI and bviR mutants exhibited marginally less biofilm formation; however, this decrease was not restored when bviI and bviR were added in trans (data not shown). The bviI and bviR mutants did not exhibit a phenotype distinguishable from that of the wild type in any of the API 20 NE biochemical tests (data not shown).
Analysis of differences in AHL production in clinical and environmental strains of B. vietnamiensis.
AHL production in the BCC is strain dependent with respect to quantity and type of AHL produced (11, 26, 72). Since the regulation of the quorum-sensing systems appeared to be strain dependent, further investigation into the differences in AHL production by B. vietnamiensis strains was pursued. The AHL production profiles of seven B. vietnamiensis strains were determined by TLC-AHL bioassays (Table 3). Only one of four clinical strains but all three environmental strains produced DHL, suggesting that the bviIR genes may be less expressed in clinical strains than in environmental strains, since OHL and HHL could be produced by CepI in these strains.
The presence of the bviI and bviR genes in the seven B. vietnamiensis strains was confirmed by PCR analysis with primers that amplified DNA fragments internal to bviI and bviR (Table 3). To determine whether bviI and bviR are expressed in the clinical strains that do not produce DHL, RT-PCR was performed (Table 3). There was a decrease in amplification of the bviR RT-PCR products and no detectable bviI RT-PCR products from the non-DHL-producing strains, indicating that the bviI and bviR genes are poorly expressed in these three strains. The bviI RT-PCR data were confirmed for the majority of strains by transcriptional analysis of bviI-luxCDABE transcriptional fusions (Table 3). The only discrepancy was that the expression of bviI in PC259 was almost at background levels, whereas a bviI RT-PCR product was obviously amplified.
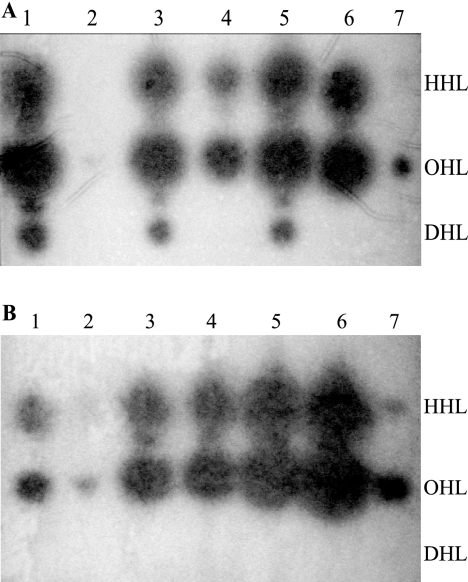
Given that BviR is implicated in the positive regulation of bviI and that the expression of bviR was lower in the three non-DHL-producing clinical strains, heterologous expression of bviRG4 was performed to determine whether the absence of bviI expression in these strains was due to a lack of induction by BviR (Fig. 4A). The presence of bviRG4 in trans restored DHL production in each of the strains (Fig. 4A, lanes 3, 5, and 7), suggesting that the absence of bviI expression in the non-DHL-producing strains is due to a hindered induction of bviI by BviR.
FIG. 4.
Heterologous expression of bviRG4 and cepRG4 in non-DHL-producing clinical strains using TLC-AHL bioassays with the A. tumefaciens(pCF218)(pCF372) reporter strain. (A) AHL production profile of non-DHL-producing strains complemented with bviRG4 in trans. Lane 1, FC466(pRM284); lane 2, FC466(pUCP28T); lane 3, FC411(pRM284); lane 4, FC441(pUCP28T); lane 5, C2822(pRM284); lane 6, C2822(pUCP28T); lane 7, synthetic standards. (B) AHL production profile of non-DHL-producing strains with cepRG4 in trans. Lane 1, FC466(pRM282); lane 2, FC466(pUCP28T); lane 3, FC411(pRM282); lane 4, FC441(pUCP28T); lane 5, C2822(pRM282); lane 6, C2822(pUCP28T); lane 7, synthetic standards.
The bviIR loci of the three non-DHL-producing strains and G4 were sequenced to determine whether there were any sequence differences in the non-DHL-producing strains that could lead to a lack of DHL synthesis. The sequence of bviIR from DBO1 (accession no. AF296284) (36) was included in the comparison. The BviI sequences were identical in all six strains (data not shown), demonstrating that each strain encodes a functional AHL synthase. The BviR sequences were identical at all but residue 106 (data not shown). This difference was not related to the ability to produce DHL, since C2822 and G4 contained a glycine and FC466 and FC411 contained aspartate at this position. Therefore, bviR encodes a functional transcriptional regulator in all strains. There are nucleotide differences in the intergenic region proximal to the bviI start codon. The −10 and −35 promoter elements did not contain sequence differences that correlate with bviI expression levels. A lux box-type consensus sequence, the same in all five strains, was predicted to be centered 56 bp upstream of the bviI start codon. A B. cenocepacia cep box sequence (69) was predicted to be centered 337 bp upstream of the bviI start codon. The predicted cep boxes were identical in all strains except for G4.
To determine whether the absence of bviI expression in the non-DHL-producing strains is due to sequence differences upstream of bviI, expression of bviIG4-luxCDABE in PC259, FC466, FC441, and C2822 and expression of bviIPC259-luxCDABE in FC369T, DBO1, and G4 were examined (Table 3). There was expression of the bviIPC259-luxCDABE fusion in FC369T, DBO1, and G4, but there was no detectable bviIG4-luxCDABE fusion expression in FC466, FC441, and C2822, indicating that the lack of bviI expression in these three clinical strains is not due to mutations in the bviI promoter region.
It is possible that the lack of DHL production in these three clinical strains may be due to CepR not inducing bviI expression, since CepR positively regulates bviI in both G4 and PC259. Heterologous expression of cepRG4 in FC466, FC441, and C2822 was performed to determine whether CepR is able to induce bviI (Fig. 4B). The presence of cepRG4 increased the production of HHL and OHL in FC466 (Fig. 4B, compare lanes 1 and 2), but it did not restore DHL production in FC466, FC441, or C2822 (Fig. 4B, lanes 1, 3, and 5), indicating that the lack of DHL is not due to a lack of CepR functioning as a positive regulator of bviI in these strains.
Additional LuxI and LuxR homologues were identified in silico by a BLAST search (3) of the incomplete draft G4 genome sequence (http://www.jgi.doe.gov/) with Vibrio fischeri LuxI (accession no. 1403259B) (17), LuxR of V. fischeri (accession no. 1403259A) (17), B. vietnamiensis PC259 CepI (accession no. AAK70355) (36), and B. vietnamiensis PC259 CepR (accession no. AAK70352) (36). There are at least three LuxI homologues and five LuxR homologues present in the G4 genome draft sequence. There are three sets of AHL synthase/transcriptional regulator pairs, including cepIR, bviIR, and an additional system designated open reading frames 7349 and 7350 (http://www.jgi.doe.gov/). There are two LuxR homologues that do not have a proximal AHL synthase and are designated open reading frames 6095 and 3039 (http://genome.ornl.gov/microbial/bcep_1808/).
To determine whether open reading frame 7350 encodes a functional AHL synthase, the 7350 open reading frame was cloned and expressed in E. coli. There were no AHLs detected in the culture supernatant of E. coli(pRM292), containing 7350, when the A. tumefaciens TLC-AHL bioassay was performed under the conditions used to visualize AHLs produced by both cepI and bviI (data not shown). The presence of the 7349/7350 luxIR homologues in other B. vietnamiensis strains was determined by PCR with primers that amplified DNA fragments internal to 7349 and 7350. All of the strains except for PC259 and C2822 contained these genes (Table 3).
DISCUSSION
AHL-mediated quorum-sensing systems have complex regulatory organizations (68). The quorum-sensing systems of the BCC species are no exception, since the CepIR system is required for cciIR expression in B. cenocepacia (39) and we have now demonstrated that CepR is required for the expression of bviIR in B. vietnamiensis. The initiation of cepI expression at least 6 h before bviI expression is consistent with the idea that the CepIR system is required for the expression of bviI. Phylogenetic analysis established that the cepIR system is the ancestral quorum-sensing system for the BCC and that the bviIR system is distinct (39); therefore, the bviIR system is presumed to have been acquired independently and subsequently incorporated into the CepIR regulatory network. A principal role for the cepIR system in B. vietnamiensis quorum sensing is contrary to the previous hypotheses that indicated a role for bviIR as the principal system based on the AHL production profiles of the G4 bviI and bviR mutants (10). The differences in results and interpretation may be due to the difference in AHL reporters used to characterize the AHL production profile of the G4 bviI and bviR mutants in the two studies. Conway and Greenberg (10) used an AHL radiotracer assay and an E. coli-based AHL bioassay to examine the AHL production profiles of the bviI and bviR mutants. The radiotracer assay quantifies the amount of radiolabeled methionine incorporated into AHLs during synthesis and distinguishes the AHLs present by high-performance liquid chromatography (54). The E. coli biosensor (pHV200I−) is based on the V. fischeri luxIR system and is most sensitive to 3-oxo-HHL (46). The A. tumefaciens reporter used in the current study is most sensitive to 3-oxoacyl-homoserine lactones with side chains of 10 or 12 carbons and is able to detect unsubstituted homoserine lactones with side chains ranging from 6 to 12 carbons in length (53). If the assay employed by Conway and Greenberg (10) were as sensitive to HHL and OHL, it is likely that they would have detected greater AHL production by the bviIR mutants and a greater role for CepI in AHL production. The Conway and Greenberg study (10) and the current study are in agreement regarding the role of bviR in the positive regulation of bviI. Autoregulation is common in AHL-mediated quorum-sensing systems to achieve exponential activation of the system (68). Feedback also occurs for the cepIR system in PC259, since CepR positively regulates cepI and negatively regulates itself. The G4 cepIR system, however, did not exhibit autoregulation. There was a marked difference in the AHL production profiles for the cepR mutants in PC259 and G4 that is likely due to the differences in the roles of CepR in autoregulation. The difference in the AHL production profiles of the cepR mutants could also be due to the 7349/7350 system, which is present in G4 but not present in PC259. Although the 7350 AHL synthase was not shown to produce AHLs under the set of conditions tested, this system could be involved in the quorum-sensing regulatory hierarchy in G4. Another possibility is that G4cepR is a merodiploid and therefore could still produce functional copies of CepR that would obscure the effect of a cepR mutation.
The AHL production profiles of the quorum-sensing mutants confirmed the transcriptional-analysis experiments. The G4 cepI mutant AHL production profile exhibits less HHL and OHL than the wild type, presumably due to the lack of synthesis of these AHLs by CepI; however, the G4 cepI mutant did not produce detectable levels of DHL. With less AHL present in the environment of the cepI mutant, there would be less of the active AHL-bound form of CepR, leading to a decrease in bviI expression and the synthesis of DHL.
It is curious that all of the environmental isolates tested produced DHL but that all of the clinical isolates examined either did not produce DHL or had less bviI expression than G4. The ability of BCC strains to produce AHLs has been extensively surveyed, and a clear correlation between the quantity or the type of AHL produced and the origin of the strain has not been demonstrated (11, 26, 72). These three studies employed different methodologies but agree with the current study of the AHLs produced by all strains except FC441, for which one study reported DHL production (72). All 3 environmental isolates collectively assayed in four independent studies produced DHL, whereas 4 out of the 10 clinical isolates either did not produce DHL or did not produce DHL as their predominant AHL (11, 26, 72), suggesting less DHL production in B. vietnamiensis clinical strains.
It has been suggested that differences between environmental strains and those which cause infections may occur at the level of the regulation of genes rather than in their presence or absence (45). Heterologous expression of bviR from G4 in the clinical non-DHL-producing strains restored DHL production, implicating inefficient induction of bviI by BviR as the reason for the absence of DHL production in these three strains. Transcriptional analysis of the G4 bviI promoter in the non-DHL-producing strains and of the PC259 bviI promoter in the DHL-producing strains determined that the lack of bviI expression is not due to mutations in the bviI promoter region, suggesting that an unknown upstream regulatory element influences the expression of bviI. Heterologous expression of cepR from G4 in the clinical non-DHL-producing strains did not restore DHL production; therefore, CepR is not the affected upstream regulatory element. The distribution of the luxR homologue 7349 is inconsistent with that of the non-DHL-producing strains, and 7349 is not likely the involved upstream regulator. Like B. vietnamiensis G4, B. pseudomallei and B. thailandensis possess three complete AHL-mediated quorum-sensing systems and two additional luxR homologues (61, 63). B. mallei possesses two luxI and four luxR homologues (62). The regulatory networks of these systems have yet to be characterized, but each gene may play a role in the coordinate expression of quorum-controlled genes.
Environmental conditions influence the expression of quorum-sensing networks (50) and could be a factor in the expression of bviI and the production of DHL in the clinical non-DHL-producing strains. Expression of bviI and DHL production were greater when cultures were grown at 30°C than at 37°C (data not shown). Efforts were made to identify culture conditions for bviI expression in the non-DHL-producing strains, yet none were found. However, the possibility that bviI can be expressed in these strains under specific environmental conditions should be acknowledged.
This study has resulted in further characterization of the B. vietnamiensis quorum-sensing regulatory network, yet little is known about what genes are regulated by these systems. Since the G4 bviIR mutants retain the ability to produce HHL and OHL, it is believed that the cepIR system and potentially the 7349/7350 system are still functional in these mutants and compensate for mutations in bviIR, making it difficult to detect phenotypic differences in the mutants. It might be possible to detect quorum-sensing-regulated phenotypes by constructing double or triple mutants; however, the construction of such mutants of B. vietnamiensis proves difficult, as this species is less amenable to genetic manipulation than other BCC species. There is a possibility of analyzing the effect of the absence of AHLs in B. vietnamiensis strains by employing a quorum-quenching approach, as suggested by Wopperer et al. (72). With this approach, Wopperer et al. (72) determined that aidA, a cepIR-regulated gene of unknown function that is involved in nematode virulence (28), is also regulated by quorum sensing in B. vietnamiensis strains. The quorum-quenching approach, however, does not make it possible to distinguish which quorum-sensing system is involved in the regulation of aidA in B. vietnamiensis.
Quorum-sensing systems in B. cepacia and B. cenocepacia regulate many physiological processes (19, 65). Proteomic (48) and transcriptional (58) analyses of quorum-sensing mutants of B. cenocepacia have been successful in identifying numerous quorum-sensing-regulated genes. With further development of molecular tools for B. vietnamiensis, including a complete genome sequence, the role of quorum sensing in regulating environmentally beneficial phenotypes as well as pathogenic traits may be elucidated.
Acknowledgments
This study was supported by a grant from the Canadian Cystic Fibrosis Foundation to P. A. Sokol. R. J. Malott is the recipient of a studentship from the Alberta Heritage Foundation for Medical Research.
We thank T. Louie for providing M. luteus.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Aguilar, C., I. Bertani, and V. Venturi. 2003. Quorum-sensing system and stationary-phase sigma factor (rpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 69:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, C., A. Friscina, G. Devescovi, M. Kojic, and V. Venturi. 2003. Identification of quorum-sensing-regulated genes of Burkholderia cepacia. J. Bacteriol. 185:6456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1989. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, NY.
- 5.Baldwin, A., P. A. Sokol, J. Parkhill, and E. Mahenthiralingam. 2004. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernier, S. P., L. Silo-Suh, D. E. Woods, D. E. Ohman, and P. A. Sokol. 2003. Comparative analysis of plant and animal models for characterization of Burkholderia cepacia virulence. Infect. Immun. 71:5306-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernier, S. P., and P. A. Sokol. 2005. Use of suppression-subtractive hybridization to identify genes in the Burkholderia cepacia complex that are unique to Burkholderia cenocepacia. J. Bacteriol. 187:5278-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, H.-K., and G. J. Zylstra. 1998. Novel organization of the genes for phthalate degradation from Burkholderia cepacia DBO1. J. Bacteriol. 180:6529-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiarini, L., A. Bevivino, C. Dalmastri, S. Tabacchioni, and P. Visca. 2006. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol. 14:277-286. [DOI] [PubMed] [Google Scholar]
- 10.Conway, B.-A., and E. P. Greenberg. 2002. Quorum-sensing signals and quorum-sensing genes in Burkholderia vietnamiensis. J. Bacteriol. 184:1187-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conway, B.-A. D., V. Venu, and D. P. Speert. 2002. Biofilm formation and acyl homoserine lactone production in the Burkholderia cepacia complex. J. Bacteriol. 184:5678-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniels, R., D. E. De Vos, J. Desair, G. Raedschelders, E. Luyten, V. Rosemeyer, C. Verreth, E. Schoeters, J. Vanderleyden, and J. Michiels. 2002. The cin quorum sensing locus of Rhizobium etli CNPAF512 affects growth and symbiotic nitrogen fixation. J. Biol. Chem. 277:462-468. [DOI] [PubMed] [Google Scholar]
- 13.Dennis, J. J., and P. A. Sokol. 1995. Electrotransformation of Pseudomonas. Methods Mol. Biol. 47:125-133. [DOI] [PubMed] [Google Scholar]
- 14.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeShazer, D., D. M. Waag, D. L. Fritz, and D. E. Woods. 2001. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb. Pathog. 30:253-269. [DOI] [PubMed] [Google Scholar]
- 16.Detsika, M. G., J. E. Corkill, M. Magalhães, K. J. Glendinning, C. A. Hart, and C. Winstanley. 2003. Molecular typing of, and distribution of genetic markers among, Burkholderia cepacia complex isolates from Brazil. J. Clin. Microbiol. 41:4148-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devine, J. H., C. Countryman, and T. Baldwin. 1988. Nucleotide sequence of the luxI and luxR genes and structure of the primary regulatory region of the lux regulon of Vibrio fischeri ATCC 7744. Biochemistry 27:837-842. [Google Scholar]
- 18.Duan, K., C. Dammel, J. Stein, H. Rabin, and M. G. Surette. 2003. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50:1477-1491. [DOI] [PubMed] [Google Scholar]
- 19.Eberl, L. 2006. Quorum sensing in the genus Burkholderia. Int. J. Med. Microbiol. 296:103-110. [DOI] [PubMed] [Google Scholar]
- 20.Estrada-De Los Santos, P., R. Bustillos-Cristales, and J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:2790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 23.Fuqua, C., and S. C. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillis, M., T. Van Van, R. Bardin, M. Goor, P. Hebbar, A. Willems, P. Segers, K. Kersters, T. Heulin, and M. P. Fernandez. 1995. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int. J. Syst. Bacteriol. 45:274-289. [Google Scholar]
- 25.Gingues, S., C. Kooi, M. B. Visser, B. Subsin, and P. A. Sokol. 2005. Distribution and expression of the ZmpA metalloprotease in the Burkholderia cepacia complex. J. Bacteriol. 187:8247-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gotschlich, A., B. Huber, O. Geisenberger, A. Togl, A. Steidle, K. Riedel, P. Hill, B. Tummler, P. Vandamme, B. Middleton, M. Camara, P. Williams, A. Hardman, and L. Eberl. 2001. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 24:1-14. [DOI] [PubMed] [Google Scholar]
- 27.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 28.Huber, B., F. Feldmann, M. Köthe, P. Vandamme, J. Wopperer, K. Riedel, and L. Eberl. 2004. Identification of a novel virulence factor in Burkholderia cenocepacia H111 required for efficient slow killing of Caenorhabditis elegans. Infect. Immun. 72:7220-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 30.Kothe, M., M. Antl, B. Huber, K. Stoecker, D. Ebrecht, I. Steinmetz, and L. Eberl. 2003. Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell. Microbiol. 5:343-351. [DOI] [PubMed] [Google Scholar]
- 31.Larsen, G. Y., T. L. Stull, and J. L. Burns. 1993. Marked phenotypic variability in Pseudomonas cepacia isolated from a patient with cystic fibrosis. J. Clin. Microbiol. 31:788-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewenza, S., and P. A. Sokol. 2001. Regulation of ornibactin biosynthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J. Bacteriol. 183:2212-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewenza, S., M. B. Visser, and P. A. Sokol. 2002. Interspecies communication between Burkholderia cepacia and Pseudomonas aeruginosa. Can. J. Microbiol. 48:707-716. [DOI] [PubMed] [Google Scholar]
- 35.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell III, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 36.Lutter, E., S. Lewenza, J. J. Dennis, M. B. Visser, and P. A. Sokol. 2001. Distribution of quorum-sensing genes in the Burkholderia cepacia complex. Infect. Immun. 69:4661-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. W. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 39.Malott, R. J., A. Baldwin, E. Mahenthiralingam, and P. A. Sokol. 2005. Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia. Infect. Immun. 73:4982-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer, J. M., and M. A. Abdallah. 1978. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physico-chemical properties. J. Gen. Microbiol. 107:319-328. [Google Scholar]
- 41.Mohr, C. D., M. Tomich, and C. A. Herfst. 2001. Cellular aspects of Burkholderia cepacia infection. Microbes Infect. 3:425-435. [DOI] [PubMed] [Google Scholar]
- 42.Moore, R. A., S. Reckseidler-Zenteno, H. Kim, W. Nierman, Y. Yu, A. Tuanyok, J. Warawa, D. DeShazer, and D. E. Woods. 2004. Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect. Immun. 72:4172-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson, M. J. K., S. O. Montgomery, W. R. Mahaffey, and P. H. Pritchard. 1987. Biodegradation of trichloroethylene and involvement of an aromatic biodegradative pathway. Appl. Environ. Microbiol. 53:949-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park, J., I. Hwang, J. Kim, S. Lee, B. Conway, E. P. Greenberg, and K. Lee. 2001. Characterization of quorum-sensing signaling molecules produced by Burkholderia cepacia G4. J. Microbiol. Biotechnol. 11:804-811. [Google Scholar]
- 45.Parke, J. L., and D. Gurian-Sherman. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39:225-258. [DOI] [PubMed] [Google Scholar]
- 46.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reik, R., T. Spilker, and J. J. LiPuma. 2005. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J. Clin. Microbiol. 43:2926-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riedel, K., C. Arevalo-Ferro, G. Reil, A. Gorg, F. Lottspeich, and L. Eberl. 2003. Analysis of the quorum-sensing regulon of the opportunistic pathogen Burkholderia cepacia H111 by proteomics. Electrophoresis 24:740-750. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 50.Schuster, M., and E. P. Greenberg. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:73-81. [DOI] [PubMed] [Google Scholar]
- 51.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 52.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 53.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 55.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149:3649-3658. [DOI] [PubMed] [Google Scholar]
- 56.Speert, D. P., M. E. Campbell, D. A. Henry, R. Milner, F. Taha, A. Gravelle, A. G. Davidson, L. T. Wong, and E. Mahenthiralingam. 2002. Epidemiology of Pseudomonas aeruginosa in cystic fibrosis in British Columbia, Canada. Am. J. Respir. Crit. Care Med. 166:988-993. [DOI] [PubMed] [Google Scholar]
- 57.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subsin, B., C. E. Chambers, M. B. Visser, and P. A. Sokol. 22 November 2006. Identification of genes regulated by the cepIR quorum-sensing system in Burkholderia cenocepacia by high-throughput screening of a random promoter library. J. Bacteriol. doi: 10.1128/JB.01201-06. (Subsequently published, J. Bacteriol. 189:968-979, 2007.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomlin, K. L., R. J. Malott, G. Ramage, D. G. Storey, P. A. Sokol, and H. Ceri. 2005. Quorum-sensing mutations affect attachment and stability of Burkholderia cenocepacia biofilms. Appl. Environ. Microbiol. 71:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tran Van, V., O. Berge, S. Ngo, J. Balandreau, and T. Heulin. 2000. Repeated beneficial effects of rice inoculation with a strain of Burkholderia vietnamiensis on early and late yield components in low fertility sulphate acid soils of Vietnam. Plant Soil 218:273-284. [Google Scholar]
- 61.Ulrich, R. L., D. Deshazer, E. E. Brueggemann, H. B. Hines, P. C. Oyston, and J. A. Jeddeloh. 2004. Role of quorum sensing in the pathogenicity of Burkholderia pseudomallei. J. Med. Microbiol. 53:1053-1064. [DOI] [PubMed] [Google Scholar]
- 62.Ulrich, R. L., D. DeShazer, H. B. Hines, and J. A. Jeddeloh. 2004. Quorum sensing: a transcriptional regulatory system involved in the pathogenicity of Burkholderia mallei. Infect. Immun. 72:6589-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ulrich, R. L., H. B. Hines, N. Parthasarathy, and J. A. Jeddeloh. 2004. Mutational analysis and biochemical characterization of the Burkholderia thailandensis DW503 quorum-sensing network. J. Bacteriol. 186:4350-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 65.Venturi, V., A. Friscina, I. Bertani, G. Devescovi, and C. Aguilar. 2004. Quorum sensing in the Burkholderia cepacia complex. Res. Microbiol. 155:238-244. [DOI] [PubMed] [Google Scholar]
- 66.Walsh, P. S., D. A. Metzger, and R. Higuchi. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 10:506-513. [PubMed] [Google Scholar]
- 67.Walsh, T. A., and D. P. Ballou. 1983. Halogenated protocatechuates as substrates for protocatechuate dioxygenase from Pseudomonas cepacia. J. Biol. Chem. 258:14413-14421. [PubMed] [Google Scholar]
- 68.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 69.Weingart, C. L., C. E. White, S. Liu, Y. Chai, H. Cho, C. S. Tsai, Y. Wei, N. R. Delay, M. R. Gronquist, A. Eberhard, and S. C. Winans. 2005. Direct binding of the quorum sensing regulator CepR of Burkholderia cenocepacia to two target promoters in vitro. Mol. Microbiol. 57:452-467. [DOI] [PubMed] [Google Scholar]
- 70.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 71.Woods, D. 1984. Oligonucleotide screening of cDNA libraries. Focus 6:1-2. [Google Scholar]
- 72.Wopperer, J., S. T. Cardona, B. Huber, C. A. Jacobi, M. A. Valvano, and L. Eberl. 2006. A quorum-quenching approach to investigate the conservation of quorum-sensing-regulated functions within the Burkholderia cepacia complex. Appl. Environ. Microbiol. 72:1579-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu, J., J. W. Beaber, M. I. Moré, C. Fuqua, A. Eberhard, and S. C. Winans. 1998. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 180:5398-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]