Abstract
Strains of the plant-pathogenic bacterium Xanthomonas axonopodis pv. citri are differentiated into two groups with respect to aggressiveness (normal and weak) on Citrus grandis cultivars but not on other Citrus species such as Citrus sinensis. Random mutagenesis using the transposon Tn5 in X. axonopodis pv. citri strain KC21, which showed weak aggressiveness on a C. grandis cultivar, was used to isolate mutant KC21T46, which regained a normal level of aggressiveness on the cultivar. The gene inactivated by the transposon, hssB3.0, was shown to be responsible for the suppression of virulence on C. grandis. Sequence analysis revealed it to be a new member of the pthA homologs, which was almost identical in sequence to the other homologs except for the number of tandem repeats in the central region of the gene. hssB3.0 appears to be a chimera of other pthA homologs, pB3.1 and pB3.7, and could have been generated by recombination between these two genes. Importantly, in X. axonopodis pv. citri, hssB3.0 was found in all of the tested isolates belonging to the weakly aggressive group but not in the isolates of the normally aggressive group. Isolation of the virulence-deficient mutant KC21T14 from KC21, in which the pathogenicity gene pthA-KC21 was disrupted, showed that hssB3.0 induces a defense response on the host but partially interrupts canker development elicited by the pathogenicity gene in this bacterium.
Members of the avrBs3/pthA (avirulence and pathogenicity) gene family are widely distributed in phytopathogenic Xanthomonas species. Most members of the family have been isolated according to Flor's gene-for-gene hypothesis (6), in which a single dominant avirulence (avr) gene of the pathogen is recognized only in the cultivar that has a single cognate dominant resistance (R) gene. The plant R-gene-mediated recognition of an avr gene effector leads to the induction of plant defense reactions that usually include the hypersensitive response (HR), a rapid localized cell death associated with the arrest of pathogen ingress. Thus, avr genes restrict the pathogen's host range, an effect that is deleterious to the pathogen and is therefore unlikely to be their primary function (14). A few avrBs3/pthA family members have dramatic effects on the virulence of the Xanthomonas strains that harbor them. Among them, pthA of Xanthomonas axonopodis pv. citri (Hasse) (25) is essential for full virulence on the host plants, the Citrus species.
The avrBs3/pthA gene products share unique structural features: nearly identical repeats of 34 amino acids in their central portion, a leucine zipper, three nuclear localization signal sequences, and an acid transcriptional activation domain in the C terminus (13, 16, 33). The exact number and arrangement of their repeat units differ (6, 14) and contribute to function and specificity during the elicitation of resistance and virulence on the respective host species in the absence of resistance genes (9, 29).
These structural features suggest that AvrBs3/PthA proteins are trafficked via a type III secretion system into the plant cytoplasms, and some of these contain nuclear localization signals responsible for the translocation of these proteins into the nucleus, where they regulate the expression of genes required for genotype-specific HR or pathogenicity (13, 16, 23). For example, the transient expression of pthA in the leaves of Citrus species causes citrus canker symptoms including cell hypertrophy, division, and, finally, death (4). AvrBs3 from Xanthomonas campestris pv. vesicatoria activates the genes responsible for hypertrophy on susceptible solanaceaous plants (15).
Some xanthomonads contain multiple homologs of avrBs3/pthA family members. In X. campestris pv. malvacearum, it was shown that multiple avrBs3/pthA genes contribute additively to the water-soaking ability of the pathogen on cotton (32). All strains of Xanthomonas oryzae pv. oryzae harbor more than 12 avrBs3/pthA homologs, although only one or two homologs are major pathogenicity genes required for full virulence in rice. These genes include avrXa7, pthXo1, pthXo2, and pthXo3 (27). The presence of multiple homologs appears to facilitate the rapid generation of new pathogenicity genes by recombination in the event of host recognition or avoidance (27). Strains of X. axonopodis pv. citri also contain at least three avrBs3/pthA homologs (11). Among them, only apl1 (a pthA homolog) has the hallmark virulence function of canker formation, while the other homolog functions were negligible or not measurable (11).
Recently, two levels of aggressiveness have been found within X. axonopodis pv. citri with respect to pathogenesis towards pummelo cultivars of Citrus grandis Osbeck (19). The strains of the normally aggressive group increased to a greater number and caused larger lesions on cultivar leaves than those of the weakly aggressive group, although both groups elicited canker symptoms (19). This finding spurs interest in what determines such a host-specific interaction, since no race has been found in X. axonopodis pv. citri.
In this study, we applied transposon mutagenesis to a normally aggressive strain (KC20) and a weakly aggressive strain (KC21) of X. axonopodis pv. citri in order to characterize the gene involved in the specific interaction between the bacterium and pummelo cultivars. This experiment identified a new member of the Xanthomonas avrBs3/pthA family from the weakly aggressive bacterial strain, which was responsible for host-specific suppression of virulence. Sequence analysis strongly suggested that the gene evolved through a recent recombination event among the pthA homologs of X. axonopodis pv. citri. In addition, we demonstrated that this gene induces a defense response on the host but partially interrupts canker development elicited by the pathogenicity gene in this bacterium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The strains of Escherichia coli and X. axonopodis pv. citri and the plasmids used in this study are listed in Table 1. E. coli DH5α, which was used as the cloning host throughout this study, was cultured in LB medium at 37°C. The strains of X. axonopodis pv. citri were cultured in YP medium (19) at 27°C. For solid medium, 1.5% agar was added to these media. Antibiotic selection was carried out using 50 μg/ml kanamycin, 30 μg/ml tetracycline, and 100 μg/ml ampicillin.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| X. axonopodis pv. citri | ||
| KC15 | Wild-type strain; normally aggressive to Otachibana | 19 |
| KC17 | Wild-type strain; normally aggressive to Otachibana | 19 |
| KC18 | Wild-type strain; normally aggressive to Otachibana | 19 |
| KC20 | Wild-type strain; normally aggressive to Otachibana | 19 |
| KC22 | Wild-type strain; normally aggressive to Otachibana | 19 |
| KC24 | Wild-type strain; normally aggressive to Otachibana | 19 |
| KC25 | Wild-type strain; normally aggressive to Otachibana | 19 |
| KC31 | Wild-type strain; normally aggressive to Otachibana | 19 |
| KC21 | Wild-type strain; less aggressive to Otachibana | 19 |
| KC30 | Wild-type strain; less aggressive to Otachibana | 19 |
| KC32 | Wild-type strain; less aggressive to Otachibana | 19 |
| KC33 | Wild-type strain; less aggressive to Otachibana | 19 |
| KC34 | Wild-type strain; less aggressive to Otachibana | 19 |
| KC35 | Wild-type strain; less aggressive to Otachibana | 19 |
| KC39 | Wild-type strain; less aggressive to Otachibana | 19 |
| KC40 | Wild-type strain; less aggressive to Otachibana | 19 |
| KC21T46 | Tn5 inserted mutant of KC21; normally aggressive to Otachibana | This study |
| KC21T14 | Tn5 inserted mutant of KC21; less virulence to citrus plants | This study |
| E. coli, DH5α | F−recA φ80dlacZΔM15 | TAKARA BIO Inc. |
| Plasmids | ||
| pSUP2021 | Suicide vector containing Tn5, 11.7 kb, ColE1 Ori; Apr Cmr Kmr | 20 |
| pLAFR3 | Broad-host-range cosmid cloning vector, 21.7 kb, RP4 Ori; Tcr | 21 |
| pUC18 | ColE1; lacZ Apr | TAKARA BIO Inc. |
| pUC19 | ColE1; lacZ Apr | TAKARA BIO Inc. |
| pNKBH1 | pBluescript clone of 13-kb DNA fragment containing apl1 from X. campestris pv. citri strain NA1 | 11 |
| pLpthAposi1 | pLAFR3 clone of 26.2-kb DNA fragment containing the 3.0-kb and 3.7-kb BamHI fragments hybridized with the 102-bp repeat unit of apl1 | This study |
| pLpB3.0 | Subclone of pLpthAposi1 containing the 3.0-kb BamHI fragments hybridized with the 102-bp repeat unit of apl1 | This study |
| pLpthAposi3 | pLAFR3 clone of 25.3-kb DNA fragment containing the 3.3-kb BamHI fragments hybridized with the 102-bp repeat unit of apl1 | This study |
| pLpthAposi5 | pLAFR3 clone of 22.1-kb DNA fragment containing the 3.1-kb BamHI fragments hybridized with the 102-bp repeat unit of apl1 | This study |
Recombinant DNA techniques.
All DNA manipulations, including isolation of total DNA, the alkaline method of plasmid purification, DNA purification by equilibrium centrifugation in a CsCl-ethidium bromide gradient, alkaline phosphatase treatment, ligation, gel electrophoresis, and size fractionation in a sucrose gradient were performed as described previously (1, 18).
Transformation into X. axonopodis pv. citri.
Transformation into X. axonopodis pv. citri was performed using electroporation as described previously (28). Briefly, 1 μg plasmid DNA was added to a 100-μl suspension of competent bacteria and subjected to electroporation in 2-mm gapped cuvettes at 12.5 kV/cm with a fixed capacitance of 25 μF and a resistance of 600 Ω. Electroporated cells were added to fresh YP liquid medium and incubated at 27°C for at least 1 h. Transformants were selected on YP agar medium containing appropriate antibiotics.
Transposon mutagenesis and pathogenicity screening.
Plasmid pSUP2021 (Table 1) containing Tn5 was introduced into strains KC20 (normally aggressive) and KC21 (weakly aggressive) of X. axonopodis pv. citri by electroporation as described above. Kanamycin-resistant mutants were picked from YP agar plates using sterile insect pins and screened for pathogenicity and aggressiveness by inoculating mature attached leaves of C. grandis Osbeck (Otachibana) with pin pricks. The inoculated plant was grown in a greenhouse under a temperature regimen of 28°C during the day and 25°C at night. Forty days after inoculation, the aggressiveness of each mutant was evaluated by measuring the diameters of the circular lesions, including the cork tissue and water-soaked margins.
Putative clones that altered the aggressiveness on Otachibana were further tested on the cultivar as well as on Citrus sinensis Osbeck (navel orange) as previously reported (19). Approximately 3 μl of bacterial suspension at a concentration of 1 × 108 cells per ml in 0.85% NaCl was placed onto attached mature leaves of both plants. The leaves were pricked through the bacterial suspension with an insect pin. The aggressiveness of each clone was evaluated as described above, 40 days after inoculation.
Construction of the DNA libraries.
DNA from strains KC21, KC21T14, and KC21T46 was extracted, partially digested with Sau3AI, and size fractionated using 10 to 45% sucrose density gradient centrifugation. DNA fragments between 20 and 30 kb were used to construct cosmid libraries in vector pLAFR3 as previously described (11). The recombinant linear fragments were packaged into phage heads with Gigapack III XL (Stratagene, La Jolla, CA) and used to infect E. coli DH5α cells according to the manufacturer's instructions. Individual clones were selected on LB agar containing tetracycline and stored in LB broth containing 25% glycerol at −80°C.
Colony hybridization and Southern blotting.
Colonies grown on LB agar were transferred to a Hybond N+ nylon membrane (GE Healthcare, Little Chalfont, United Kingdom) and lysed with denaturing solution (0.4 M NaOH, 1.5 M NaCl). The probes for both colony hybridization and Southern blot analysis were synthesized using plasmid pNKBH1 containing a 2.3-kb SphI fragment of apl1, which is almost identical to pthA (11), and a PCR DIG probe synthesis kit (Roche Diagnostics, Basel, Switzerland). Signal detection was performed using a DIG luminescent detection kit (Roche Diagnostics) according to the manufacturer's instructions.
DNA sequencing.
A 3-kb BamHI fragment of hssB3.0 was isolated from pLpB3.0 (Table 1) and subcloned into pUC18 and pUC19. From these subclones, a deletion series of the 3-kb BamHI fragment was constructed using a Kilosequencing deletion kit (TAKARA BIO Inc., Otsu, Japan) and used to determine the primary structure of the DNA. Sequencing was performed with an ABI Prism 310 genetic analyzer (Applied Biosystems Japan, Tokyo, Japan) using a BigDye terminator cycle sequencing FS ready reaction kit (Applied Biosystems Japan) and the vector-based primers M13M4 (3′-GTTTTCCCAGTCACGAC-5′) and M13RV (3′-CAGGAAACAGCTATGAC-5′). A BigDye kit was used to sequence the regions of hssB3.0 outside the BamHI fragment from the DNA of pLpB3.0 and custom-synthesized oligonucleotide primers (Operon Biotechnologies Inc., Tokyo, Japan). The nucleotide sequences of other avrBs3/pthA family members from strain KC21 were determined in the same way.
Bacterial growth in planta.
To monitor bacterial growth in planta, the bacteria suspended in 0.85% NaCl were inoculated into mature attached leaves of Otachibana and navel oranges as described above. The inoculated plants were grown in a greenhouse under the temperature regimens described above. The number of bacterial populations present in the lesions at 0, 1, 2, 3, 4, 8, and 16 days after inoculation was determined by removing the lesions with a cork borer, macerating them in 0.85% NaCl, and plating the suspension onto semiselective XCSM medium (19).
Determination of phenylalanine ammonia-lyase (PAL) gene transcript levels in citrus leaves using real-time quantitative reverse transcription (RT)-PCR.
The inoculated leaves of Otachibana and navel orange were wiped with cotton soaked in a bacterial suspension of KC20, KC20/pLpB3.0, KC21, KC21T46, and KC21T46/pLpB3.0 at a concentration of 1 × 108 cells per ml. Distilled water was also inoculated into the leaves as a control. The inoculated plants were grown in a greenhouse under the temperature regimens described above.
Leaf disks including 78 lesions were removed with a cork borer 1, 4, 8, and 12 days after pin prick inoculation. Total RNA was then isolated with a FastRNA Pro Green kit (Qbiogene, Inc., Irvine, CA) according to the manufacturer's instructions. RNA samples were treated with DNase I (QIAGEN, Hilden, Germany) and purified using an RNeasy Plant Mini kit (QIAGEN), according to the manufacturer's instructions, in order to eliminate any traces of contaminating genomic DNA. DNase-treated RNA samples were mixed into the 10-μl reaction mixture supplied by TAKARA BIO Inc. (Otsu, Japan) containing 5 mM MgCl2, 1× RT buffer, 1 mM deoxynucleoside triphosphates, 10 U RNase inhibitor, 2.5 U AMV Reverse Transcriptase XL (TAKARA BIO Inc., Otsu, Japan), and 0.125 μM oligo(dT) adaptor primer. The reaction mixture was incubated at 51°C for 30 min, followed by heat inactivation at 99°C for 5 min. The resulting first-strand cDNA was diluted to a final volume of 50 μl, and SYBR green-labeled PCR fragments were amplified using PAL-F (5′-GCAGCCATTCCAAACAGG-3′) and PAL-R (5′-TCAAGTTGACTACACAACATGG-3′) primers designed from the transcribed region of the Citrus limon PAL gene (GenBank accession no. U43338) (17) using GENETYX (Tokyo, Japan) software.
Quantitative RT-PCR was performed using the ABI PRISM 7700 sequence detection system (Applied Biosystems, Darmstadt, Germany) with SYBR Premix EX Taq (TAKARA BIO Inc.), gene-specific primers at a final concentration of 0.2 μM each, and 1 μl cDNA as a template. PCR cycling started with the initial polymerase activation at 95°C for 10 s followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. Primer specificity and the formation of primer dimers were monitored by dissociation curve analysis and agarose gel electrophoresis on a 1.5% (wt/vol) gel. The expression levels of histone H4 genes were used as internal standards, and fragments were amplified using primers Histone H4-F (5′-AGGCAAGGGATTGGGAAAGG-3′) and Histone H4-R (5′-AGAGCGTAAACGACGTCCATC-3′) designed from the transcribed region of the Citrus jambhiri histone H4 gene (GenBank accession no. AB050889) (7). The comparative threshold method was used to calculate the relative mRNA level with respect to the corresponding transcript in uninfected leaves of the respective plant (equaling 1). All real-time quantitative RT-PCRs were performed in triplicate.
Nucleotide sequence accession numbers.
The nucleotide and amino acid sequence data for the hssB3.0, pthA-KC21, pB3.1, and pB3.7 regions can be found at the GenBank database under accession no. AB175482, AB206388, AB206387, and AB206389, respectively.
RESULTS
Mutant KC21T46 alters the level of aggression specifically on C. grandis.
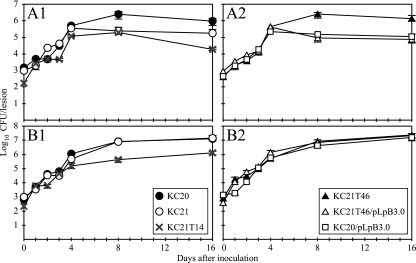
The single mutant X. axonopodis pv. citri KC21T46 was obtained from 760 kanamycin-resistant transformants derived from strain KC21, which had shown weak aggressiveness specifically on C. grandis. KC21T46 demonstrated higher aggressiveness on C. grandis (cv. Otachibana) than KC21. The KC21T46 mutant, as well as its parent strain, KC21, elicited canker symptoms on Otachibana within 4 days of inoculation. However, the lesions caused by KC21T46 on the cultivar were larger than those caused by KC21 and were the same as those caused by the normally aggressive strain KC20 40 days after inoculation (Table 2). The in planta growth of KC21T46 also resembled that of KC20 on Otachibana (Fig. 1A). By contrast, no isolate demonstrating suppression of aggressiveness in Otachibana was obtained out of 2,522 kanamycin-resistant transformants derived from strain KC20. The lesion expansion and in planta growth of KC21T46 and KC21 in C. sinensis (navel orange) did not differ throughout the experiments (Table 2 and Fig. 1B).
TABLE 2.
Lesion expansion for wild-type strains, a Tn5 mutant, and the mutant complemented by cloned wild-type DNA on pLpB3.0 of X. axonopodis pv. citri on navel orange and Otachibanaa
| Strain | Mean diam (mm) ± SE of lesions on:
|
|
|---|---|---|
| Otachibana | Navel orange | |
| KC20 | 2.44 ± 0.28a | 2.38 ± 0.17a |
| KC20/pLpB3.0 | 1.62 ± 0.23b | 2.35 ± 0.26a |
| KC20/pLAFR3 | 2.33 ± 0.24a | 2.37 ± 0.06a |
| KC21 | 1.62 ± 0.29b | 2.40 ± 0.10a |
| KC21T46 | 2.57 ± 0.41a | 2.28 ± 0.17a |
| KC21T46/pLpB3.0 | 1.81 ± 0.38b | 2.37 ± 0.09a |
| KC21T46/pLAFR3 | 2.65 ± 0.23a | 2.35 ± 0.11a |
Each value represents the mean diameters (mm) and standard errors of 16 lesions from each strain 40 days after inoculation by pricking the attached leaves of each species. Data followed by unlike letters differ significantly at an α of 0.01 according to the Tukey-Kramer honestly significant difference test.
FIG. 1.
Time course of bacterial growth in the leaves of the citrus plants (A) Otachibana and (B) navel orange. Strains KC20, KC21, and KC21T14 (shown in panel 1 in A and B) and strain KC21T46 and transformants KC21T46/pLpB3.0 and KC20/pLpB3.0 (shown in panel 2 in A and B) were used to inoculate citrus plants by pricking. Leaves were sampled 16 days after inoculation. Data represent means from three repetitions, and vertical bars represent standard errors.
Hybridization of EcoRI-digested DNA from KC21T46 with the Tn5 probe revealed a single insertion site (data not shown). A clone containing the Tn5 inserted region was isolated from the genomic library of KC21T46, which was constructed using vector pLAFR3, and was sequenced from the end of Tn5. Approximately 500 bp of KC21T46 sequence at the 5′ end of the transposon demonstrated high homology with the 102-bp repeat unit of pthA. As at least seven regions homologous to pthA, apl1, apl2, apl3, pthA1, pthA2, pthA3, and pthA4, have been found in X. axonopodis pv. citri (3, 11), KC21T46 and KC21 DNA was subjected to Southern blot analysis to identify the region of the transposon insertion.
Southern blot analysis of KC21 showed that four BamHI fragments (3 kb, 3.1 kb, 3.3 kb, and 3.7 kb) hybridized with the probe corresponding to a 2.3-kb SphI fragment of apl1, which indicates that the strain harbors at least four pthA homologs (Fig. 2A). All of the BamHI fragments were detected from not only the total DNA but also the plasmid DNA of the strain, which indicates that they are located on the latter (Fig. 2A and B). The 3-kb BamHI fragment was absent in KC21T46 DNA; however, 4.9-kb and 3.9-kb fragments containing the Tn5 fragment were detected (Fig. 2A and C). The 3-kb fragment was absent in DNA from normally aggressive strains (KC15, KC17, KC18, KC20, KC22, KC24, KC25, and KC31) but was detected in DNA prepared from all of the weakly aggressive strains tested (KC21, KC30, KC32, KC33, KC34, KC35, KC39, and KC40) (Fig. 2A). This suggests that the 3-kb fragment is involved in the suppression of virulence in C. grandis.
FIG. 2.
(A) Southern blotting analysis of plasmid and total DNA from various strains of X. axonopodis pv. citri. The blot was probed with a digoxigenin-labeled, 2.3-kb internal SphI fragment of apl1 (pthA homolog). Lanes 1 and 2, intact and BamHI-digested plasmid DNA from strain KC21; lanes 3 to 22, BamHI-digested total DNA from various strains; lanes 3 and 4, KC21T46 and its transformant with pLpB3.0; lanes 5 and 6, KC21T14 and its transformant with pLpthAposi3; lanes 7 to 14, less aggressive strains KC21, KC30, KC32, KC33, KC34, KC35, KC39, and KC40; lanes 15 to 22, normally aggressive strains KC15, KC17, KC18, KC20, KC22, KC24, KC25, and KC31. (B) Agarose gel electrophoresis of plasmid and total DNA from various strains of X. axonopodis pv. citri shown in A. (C) Schematic representation of Tn5 insertion into hssB3.0 and pthA-KC21 from mutants KC21T46 and KC21T14. Black boxes represent the leucine zipper-like region (LZ), nuclear location signals (NLSs), and an acidic transcriptional activation domain (AD). Restriction enzyme cleavage sites are shown with a B for BamHI and an S for SphI.
Mutant KC21T14 demonstrates reduced nonspecific virulence on Citrus species.
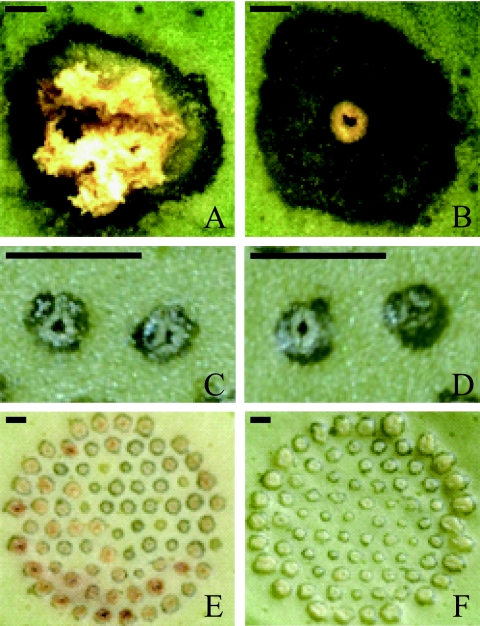
Among the kanamycin-resistant mutants derived from KC21, KC21T14 demonstrated a loss of the ability to elicit hyperplastic canker symptoms nonspecifically on citrus plants (Fig. 3A and B). This mutant retained the ability to cause water-soaked lesions and was able to increase the number of lesions on the leaves of citrus plants. However, its in planta growth was reduced by approximately 5 and 18 times in Otachibana and navel orange, respectively, compared with its parent strain, KC21 (Fig. 1A, panel 1, and B, panel 1).
FIG. 3.
Canker symptoms on abaxial leaf surfaces of navel oranges (A and B) and Otachibana (C to F). (A and B) Symptoms developed 40 days after inoculation of strain KC21 (A) and its derivative pthA-KC21-deficient mutant strain KC21T14 (B). (C to F) Comparisons of symptoms developed after inoculation of strain KC21 (C and E) and its derivative hssB3.0-deficient mutant strain KC21T46 (D and F) after 4 (C and D) and 12 (E and F) days. The attached leaves were pricked with single pins (A and B) or 78 fasciculate insect pins (C to F). The bar represents 0.5 mm.
Southern blot analysis of KC21T14 revealed that the 3.3-kb BamHI fragment hybridized by the 2.3-kb SphI fragment probe was absent and that a 5.8-kb fragment was detected instead (Fig. 2A). Approximately 400 bp of KC21T14 sequence at the 5′ end of the transposon demonstrated high homology with the N-terminal coding region of pthA (Fig. 2C). The 3.3-kb BamHI fragment was detected in all other strains investigated, which suggests that it might correspond to the region of pthA required for full virulence of the bacterium (Fig. 2A).
Complementation of KC21T46 and KC21T14.
The cosmid clone pLpthAposi1 containing the 3-kb and 3.7-kb BamHI fragments hybridized with the 2.3-kb SphI fragment probe was selected from the genomic library of KC21. Subclone pLpB3.0, which contained only the 3-kb BamHI fragment, was obtained from pLpthAposi1 and was used to complement isolate KC21T46 in the inoculation of Otachibana. The lesions produced by KC21T46/pLpB3.0 after 40 days of inoculation were similar to those caused by the weakly aggressive strain KC21 (Table 2). Moreover, in planta populations of KC21 and KC21T46/pLpB3.0 increased at a similar rate for 16 days after inoculation (Fig. 1A). No significant difference was observed in lesion expansion and in planta growth on navel oranges among the strains that were tested (Table 2 and Fig. 1B). These results suggest that the gene contained the 3-kb BamHI fragment that is homologous to pthA (called hssB3.0 hereafter) confers a host-specific suppression of virulence.
The function of hssB3.0 in the suppression of virulence was also confirmed using a transformant derived from the normally aggressive strain KC20 with plasmid pLpB3.0. Inoculation of KC20/pLpB3.0 onto Otachibana resulted in a level of lesion development similar to that of KC21 (Table 2). In addition, in planta populations of KC20/pLpB3.0 and KC21T46/pLpB3.0 increased at similar rates throughout the study (Fig. 1A2), which suggests that hssB3.0 confers lower aggressiveness on the inherently normally aggressive strains of X. axonopodis pv. citri.
Complementation of isolate KC21T14 with cosmid clone pLpthAposi3 restored the ability of the isolate to elicit hyperplastic canker symptoms on citrus plants (data not shown). This result indicates that the gene containing this 3.3-kb BamHI fragment (called pthA-KC21 hereafter) is required for full virulence of the bacterium.
Enhancement of PAL gene transcript accumulation by the strain harboring hssB3.0 of X. axonopodis pv. citri.
PAL is a key enzyme in the phenylpropanoid pathway, which leads to the production of phytoalexins or reactive compounds during plant defense responses to incompatible pathogens (8). To determine whether hssB3.0 affects the defense response in Otachibana, the level of accumulation of the PAL gene transcript in the plant after the inoculation of X. axonopodis pv. citri was examined using real-time quantitative RT-PCR.
Four days after inoculation of all Xanthomonas strains, PAL gene transcription had not increased (Fig. 4A), although pustules consistent with canker symptoms had already developed (Fig. 3C and D). At 8 and 12 days after inoculation, the level of transcript accumulation was significantly different between the strains (Fig. 4A). PAL gene transcript levels were 2.5 to 4.6 times higher 12 days after inoculation of Xanthomonas strains carrying hssB3.0 (KC21, KC20/pLpB3.0, and KC21T46/pLpB3.0) than after inoculation of strains lacking the gene (KC20 and KC21T46) (Fig. 4A). Most of the lesions caused by the strains with hssB3.0 turned brown during the 12 days after inoculation, which might indicate necrosis (Fig. 3E and F show symptoms induced by KC21 and KC21T46, respectively). Control inoculation of distilled water did not increase levels of PAL gene transcripts and did not lead to the development of visible symptoms. These results suggest that hssB3.0 increases the accumulation of PAL gene transcripts in Otachibana, although this enhancement begins after the development of primary canker symptoms. In navel oranges, no significant difference was observed in the level of PAL gene transcripts following the inoculation of all Xanthomonas strains or distilled water (Fig. 4B).
FIG. 4.
Accumulation of PAL gene transcript in the leaves of Otachibana (A) and navel orange (B) after inoculation with X. axonopodis pv. citri strain KC20 and its transformant with pLpB3.0, KC21T46 and its transformant with pLpB3.0, and KC21. Distilled water (DW) was used as the control inoculum. Total RNA was isolated from the lesions 1, 4, 8, and 12 days after inoculation and used for real-time quantitative RT-PCR. The relative mRNA level was calculated with respect to the level of the corresponding transcript in uninfected leaves of the respective plant (equaling 1). Bars represent standard errors (n = 3).
The fragment amplified with the primer set PAL-F and PAL-R from mRNA of both Otachibana and navel oranges was identical to that from the transcribed region of the C. limon PAL gene (17).
Sequence analysis of avrBs3/pthA family members from strain KC21.
All four distinct avrBs3/pthA family members, hssB3.0, pthA-KC21, pB3.1, and pB3.7, were isolated and characterized from strain KC21 of X. axonopodis pv. citri. pB3.1 and pB3.7 were found to contain the 3.1-kb and 3.7-kb BamHI fragments, respectively (Fig. 2A). The nucleotide sequences of all genes were determined bidirectionally using the deletion series approach.
The open reading frames were determined to be 3.2 kb, 3.3 kb, 3.5 kb, and 3.9 kb for hssB3.0, pB3.1, pthA-KC21, and pB3.7, respectively. The 5′ and 3′ ends of the open reading frame loci were defined by 62-bp long terminal repeats (LTRs), although the 3′ region of the hssB3.0 locus was interrupted with the Tn5045 resolvase gene, tnpR, that carries the insertion sequence ISXc5 (22) 27 bp away from the 3′ end of the LTR, as seen in pthA2 (3). The nucleotide sequences from the 5′ LTR to the start codons, the N-terminal coding regions, and the C-terminal coding regions were almost identical between all genes. The C-terminal coding regions contained the same leucine zipper-like motif, three putative nuclear localization signals, and an acidic transcriptional activation domain (Fig. 2C).
The central region of all genes was characterized by tandem, 102-bp direct repeats that differed in number according to the gene. Most avrBs3/pthA family members characterized have half a repeat at the 3′ end of the tandem repeats, after which the sequence breaks off. Thus, hssB3.0 contained 14.5 repeats, pB3.1 contained 15.5, pthA-KC21 contained 17.5, and pB3.7 contained 21.5 (Fig. 2C and 5). Little variation was seen between the deduced amino acid sequences of each repeat, except at the 4th, 12th, and 13th amino acid residues within each repeat (Fig. 5B), as previously observed by Yang and Gabriel (30). In repeat units G and N, the 24th and 11th residues, respectively, differed from those in the other units (Fig. 5B).
FIG. 5.
Comparison of central repeat units within the deduced amino acid sequences of avrBs3/pthA family members from strain KC21 of X. axonopodis pv. citri (A). Each central repeat unit is displayed as a distinctive capital letter as seen in B. The 1st to the 6th repeats of hssB3.0 were identical to the 1st to the 6th repeats of pB3.1, while the 7th to the 12th repeats were identical to the 2nd to the 7th repeats of pB3.7. The 13th to the last repeat was also identical to the 14th to the last repeat of pB3.1. The central region of pthA-KC21 was similar in arrangement to that of pthA, although displacements were observed at the 6th, 13th, and 15th repeats. GenBank accession numbers are as follows: pthA, U28802; hssB3.0, AB175482; pB3.1, AB206387; pthA-KC21, AB206388; pB3.7, AB206389.
Remarkably, the regions of hssB3.0 from the 1st to the 6th and from the 7th to the 12th central repeats were identical to the 1st to the 6th repeats of pB3.1 and from the 2nd to the 7th repeats of pB3.7, respectively (Fig. 5). The region of hssB3.0 from the 13th to the last repeat was also identical to that from the 14th to the last repeat of pB3.1 (Fig. 5). hssB3.0 therefore appears to be a chimera consisting of pB3.1 and pB3.7 (Fig. 5A). The central region of pthA-KC21 was similar to that of pthA in the repeat unit arrangement, although displacements at the 6th, 13th, and 15th repeats were observed (Fig. 5A).
DISCUSSION
We report here on the isolation and characterization of a new member of the Xanthomonas avrBs3/pthA gene family, hssB3.0, from X. axonopodis pv. citri that confers host-specific suppression of virulence and might control the bacterial elicitation of resistance in C. grandis. hssB3.0 reduces the ability of X. axonopodis pv. citri to multiply in the Citrus species, enhances the accumulation of PAL gene transcripts, and, consequently, reduces lesion expansion. The 3.0-kb BamHI fragment of hssB3.0 has not been found in normally aggressive Xanthomonas strains but has been found in all less aggressive strains investigated thus far. Our findings also indicate that pthA-KC21 functions as a pathogenicity gene. hssB3.0 induces a defense response on the host; however, this gene partially interrupts canker symptom development elicited by pthA-KC21. Usually, avirulence genes are associated with the onset of the HR by the host plant, and thus, the infection is stopped, and there is not a further development of symptoms. In this regard, hssB3.0 is hardly considered to be an avirulence gene.
The structure of hssB3.0 is typical of the Xanthomonas avrBs3/pthA family members. It has a central domain containing a series of 102-bp direct repeats (6, 14), three nuclear localization signals (31), and a eukaryotic transcriptional activation domain (33) and is flanked by terminal inverted repeats (11). The N-terminal coding region of hssB3.0 is also identical to that of pB3.1. The number of direct repeats in the HssB3.0 protein differs from those of other AvrBs3/PthA family members, although every repeat unit is represented in all members.
Interestingly, the central repeat region of hssB3.0 appears to be a chimera of pB3.1 and pB3.7. This finding suggests that recombination and transposition among these genes have created a new functional gene, as previously demonstrated by Yang and Gabriel (30). The chimerical structure of hssB3.0 also provides evidence that multiple homologs of avrBs3/pthA family members in individual Xanthomonas strains can serve as a reservoir for potential recombinatorial alleles (26, 27).
The C terminus of HssB3.0 differs with respect to PthA-KC21 at two residues (data not shown). However, the HincII-SphI regions of the C termini are identical, which is considered to be essential for their virulence activity on citrus (10). These findings suggest that the central domain of hssB3.0 directly contributes to host specificity, which is consistent with previous observations of avrBs3/pthA family members (6).
Citrus canker disease is highly dependent on the presence of pthA (2), while four naturally occurring avrBs3/pthA family members, avrXa7, pthXo1, pthXo2, and pthXo3, which were identified in X. oryzae pv. oryzae strains, act as major virulence effectors in rice (27). Similar features were found among the central repeat regions of these genes, which differed in terms of number and arrangement. In contrast, the genes identified as being essential for full virulence on citrus plants, pthA, apl1, and pthA-KC21, have 17.5 uniform direct repeats within their central regions, and the arrangements of the repeat units are very similar. This highly conserved central region indicates that full virulence on citrus plants requires an appropriate protein structure encoded by avrBs3/pthA genes.
Prior to the enhancement of PAL gene transcript accumulation in lesions elicited by KC21 inoculation, canker symptoms such as hypertrophy had already developed on the Otachibana leaf. This correlates with the findings of a previous study in which HR-induced necrosis was not observed following the infiltration of a Xanthomonas strain into a citrus leaf at high cell densities despite the onset of canker symptoms (19). The avrBs3 gene of X. campestris pv. vesicatoria, which induces HR in pepper plants carrying the resistance gene Bs3, elicited AvrBs3-dependent hypertrophy of the mesophyll tissue on susceptible plants. Genes upregulated in the host by AvrBs3, which plays a putative role in cell enlargement, were activated even in resistant Bs3-positive plants before the onset of HR (15). This could explain how canker symptoms developed in the present study after the inoculation of KC21 onto citrus leaves, although the contribution of hssB3.0 to virulence has not been completely elucidated.
All avrBs3/pthA genes identified in strain KC21 in the present study exist in plasmid DNA of this strain. Partial sequencing of cosmid clone pLpthAposi1 suggests that the inserted DNA in this clone could correspond almost exactly to part of plasmid pXAC33 (3), which was characterized in X. axonopodis pv. citri A strain “306” from Brazil (data not shown). Intriguingly, hssB3.0 and pB3.7 from strain KC21 appear to be located at the same positions as pthA2 and pthA1, respectively, on plasmid pXAC33. In addition, the flanking 5′ and 3′ LTRs of pB3.1 and pthA-KC21 loci were identical in sequence to those of pthA3 and pthA4, respectively, on plasmid pXAC64 from strain “306” (3). The relative sizes and endonuclease cleavage sites of these two plasmids from the Brazilian strain “306” correspond almost perfectly to two plasmids previously characterized and reported from another Japanese strain, X. axonopodis pv. citri XAS4501 (24). These results suggest not only that the avrBs3/pthA family members are plasmid borne but also that the plasmids might be conserved within these distantly isolated bacterial strains.
PAL gene transcript accumulation was observed in navel oranges 4 days after inoculation, but this did not suppress bacterial growth in planta or limit canker symptom development (Fig. 4B). The temporary increase in PAL gene transcripts could be associated with a response to the wound caused by pin prick inoculation, as observed in a previous study (12). Recently, it was revealed that bacterial effectors encoded by genes of the avrBs3/pthA family might suppress the plant defense response (5). The decrease in PAL gene transcripts from 4 days after inoculation of navel orange in the present study might depend on pthA-KC21 activity in suppressing the host defense response. Inoculation of Otachibana, however, elicited a moderate increase in PAL gene transcript levels from 8 days after inoculation, even with strains KC20 and KC21T46, which lack hssB3.0. The transient expression of pthA in Citrus spp. induced raised cankers followed by cell death (4). In Otachibana, it is possible that programmed cell death elicited by pthA-KC21 occurs earlier than in navel orange. It is also possible that another factor from X. axonopodis pv. citri is involved in the elicitation of moderate PAL gene transcript increases. This might be responsible for the differences in bacterial growth and durability in planta between Otachibana and navel oranges (Fig. 1 and 4).
According to avirulence principles, the gene product of hssB3.0 might be recognized by a cognate resistance gene in C. grandis. However, the analysis of citrus lines remains to be completed, and further studies will need to be carried out to elucidate the C. grandis resistance gene.
Acknowledgments
This work was supported by Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 2.Brunings, A. M., and D. W. Gabriel. 2003. Xanthomonas citri: breaking the surface. Mol. Plant Pathol. 4:141-157. [DOI] [PubMed] [Google Scholar]
- 3.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with different host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 4.Duan, Y. P., A. L. Castaneda, G. Zhao, G. Erdos, and D. W. Gabriel. 1999. Expression of a single, host-specific, bacterial pathogenicity gene in plant cells elicits division, enlargement and cell death. Mol. Plant-Microbe Interact. 12:556-560. [Google Scholar]
- 5.Fujikawa, T., H. Ishihara, J. E. Leach, and S. Tsuyumu. 2006. Suppression of defense response in plants by the avrBs3/pthA gene family of Xanthomonas spp. Mol. Plant-Microbe Interact. 19:342-349. [DOI] [PubMed] [Google Scholar]
- 6.Gabriel, D. W. 1999. The Xanthomonas avr/pth gene family, p. 39-55. In G. Stacey and N. T. Keen (ed.), Plant-microbe interactions, vol. 4. APS Press, St. Paul, MN. [Google Scholar]
- 7.Gomi, K., H. Yamamoto, and K. Akimitsu. 2002. Characterization of a lipoxygenase gene in rough lemon induced by Alternaria alternata. J. Gen. Plant Pathol. 68:21-30. [Google Scholar]
- 8.Hammond-Kosack, K. E., and J. D. G. Jones. 1996. Resistance gene-dependent plant defense responses. Plant Cell 8:1773-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbers, K., J. Conrads-Strauch, and U. Bonas. 1992. Race-specificity of plant resistance to bacterial spot disease determined by repetitive motifs in a bacterial avirulence protein. Nature 356:172-174. [Google Scholar]
- 10.Ishihara, H., G. Ponciano, J. E. Leach, and S. Tsuyumu. 2003. Functional analysis of the 3′ end of avrBs3/pthA genes from two Xanthomonas species. Physiol. Mol. Plant Pathol. 63:329-338. [Google Scholar]
- 11.Kanamori, H., and S. Tsuyumu. 1998. Comparison of nucleotide sequences of canker-forming and non-canker-forming pthA homologues in Xanthomonas campestris pv. citri. Ann. Phytopath. Soc. Jpn. 64:462-470. [Google Scholar]
- 12.Koizumi, M. 1983. Relationship between wound-healing process of citrus leaf tissues and successful infection through wounds by Xanthomonas campestris pv. citri (Hasse) Dye. Ann. Phytopath. Soc. Jpn. 49:352-360. [Google Scholar]
- 13.Lahaye, T., and U. Bonas. 2001. Molecular secrets of bacterial type III effector proteins. Trends Plant Sci. 6:479-485. [DOI] [PubMed] [Google Scholar]
- 14.Leach, J. E., and F. F. White. 1996. Bacterial avirulence genes. Annu. Rev. Phytopathol. 34:153-179. [DOI] [PubMed] [Google Scholar]
- 15.Marois, E., G. Van den Ackerveken, and U. Bonas. 2002. The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol. Plant-Microbe Interact. 15:637-646. [DOI] [PubMed] [Google Scholar]
- 16.Ponciano, G., H. Ishihara, S. Tsuyumu, and J. E. Leach. 2003. Bacterial effectors in plant disease and defense: keys to durable resistance? Plant Dis. 87:1272-1282. [DOI] [PubMed] [Google Scholar]
- 17.Prieto, H., M. Chiong, D. Seelenfreund, J. Garrido, A. Quaas, and L. M. Perez. 1997. Citrus limon seedlings without functional chloroplasts are unable to induce phenylalanine ammonia-lyase in response to inoculation with Alternaria alternata. J. Plant Physiol. 150:645-651. [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Shiotani, H., S. Tsuyumu, and K. Ozaki. 2000. Pathogenic interactions between Xanthomonas axonopodis pv. citri and cultivars of pummelo (Citrus grandis). Phytopathology 90:1383-1389. [DOI] [PubMed] [Google Scholar]
- 20.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 21.Stascawicz, B., D. Daulbeck, N. T. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swarup, S., Y. Yang, M. T. Kingsley, and D. W. Gabriel. 1992. A Xanthomonas citri pathogenicity gene, pthA, pleiotropically encodes gratuitous avirulence on nonhosts. Mol. Plant-Microbe Interact. 5:204-213. [DOI] [PubMed] [Google Scholar]
- 23.Szurek, B., O. Rossier, G. Hause, and U. Bonas. 2002. Type III-dependent translocation of the Xanthomonas AvrBs3 protein into the plant cell. Mol. Microbiol. 46:13-23. [DOI] [PubMed] [Google Scholar]
- 24.Tu, J., H. R. Wang, S. F. Chang, Y. C. Charng, R. Lurz, B. Dobrinksi, and W. C. Wu. 1989. Transposable elements of Xanthomonas campestris pv. citri originating from indigenous plasmids. Mol. Gen. Genet. 217:505-510. [Google Scholar]
- 25.Vauterin, L., B. Hoste, K. Kersters, and J. Swings. 1995. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 45:472-489. [Google Scholar]
- 26.Yang, B., A. Sugio, and F. F. White. 2005. Avoidance of host recognition by alterations in the repetitive and C-terminal regions of AvrXa7, a type III effector of Xanthomonas oryzae pv. oryzae. Mol. Plant-Microbe Interact. 18:142-149. [DOI] [PubMed] [Google Scholar]
- 27.Yang, B., and F. F. White. 2004. Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol. Plant-Microbe Interact. 17:1192-1200. [DOI] [PubMed] [Google Scholar]
- 28.Yang, M., W. Su, and T. Kuo. 1991. Highly efficient transfection of Xanthomonas campestris by electroporation. Bot. Bull. Acad. Sinca. 32:197-203. [Google Scholar]
- 29.Yang, Y., R. De Feyter, and D. W. Gabriel. 1994. Host-specific symptoms and increased release of Xanthomonas citri and X. campestris pv. malvacearum from leaves are determined by the 102-bp tandem repeats of pthA and avrb6, respectively. Mol. Plant-Microbe Interact. 7:345-355. [Google Scholar]
- 30.Yang, Y., and D. W. Gabriel. 1995. Intergenic recombination of a single plant pathogen gene provides a mechanism for the evolution of new host specificities. J. Bacteriol. 177:4963-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, Y., and D. W. Gabriel. 1995. Xanthomonas avirulence/pathogenicity gene family encodes functional plant nuclear targeting signals. Mol. Plant-Microbe Interact. 8:627-631. [DOI] [PubMed] [Google Scholar]
- 32.Yang, Y., Q. Yuan, and D. W. Gabriel. 1996. Watersoaking function(s) of XcmH1005 are redundantly encoded by members of the Xanthomonas avr/pth gene family. Mol. Plant-Microbe Interact. 9:105-113. [Google Scholar]
- 33.Zhu, W., B. Yang, J. M. Chittoor, L. B. Johnson, and F. F. White. 1998. AvrXa10 contains an acidic transcriptional activation domain in the functionally conserved C terminus. Mol. Plant-Microbe Interact. 11:824-832. [DOI] [PubMed] [Google Scholar]