Abstract
DNA microarrays revealed that expression of ycfR, which encodes a putative outer membrane protein, is significantly induced in Escherichia coli biofilms and is also induced by several stress conditions. We show that deletion of ycfR increased biofilm formation fivefold in the presence of glucose; the glucose effect was corroborated by showing binding of the cyclic AMP receptor protein to the ycfR promoter. It appears that YcfR is a multiple stress resistance protein, since deleting ycfR also rendered the cell more sensitive to acid, heat treatment, hydrogen peroxide, and cadmium. Increased biofilm formation through YcfR due to stress appears to be the result of decreasing indole synthesis, since a mutation in the tnaA gene encoding tryptophanase prevented enhanced biofilm formation upon stress and adding indole prevented enhanced biofilm formation upon stress. Deleting ycfR also affected outer membrane proteins and converted the cell from hydrophilic to hydrophobic, as well as increased cell aggregation fourfold. YcfR seems to be involved in the regulation of E. coli K-12 biofilm formation by decreasing cell aggregation and cell surface adhesion, by influencing the concentration of signal molecules, and by interfering with stress responses. Based on our findings, we propose that this locus be named bhsA, for influencing biofilm through hydrophobicity and stress response.
DNA microarrays show that hundreds of genes are differentially expressed in Escherichia coli biofilms (4, 60, 69) and that their expression is temporal (14). These genes are involved in many aspects of cellular physiology, from metabolism to signal transport. Many of these differentially expressed genes have unknown functions; hence, another round of DNA microarrays to compare differential gene expression in the biofilm for isogenic mutants relative to the wild-type strain has helped to determine some of the molecular roles of the uncharacterized proteins in biofilm formation. For example, differential gene expression was used to determine that MqsR regulates E. coli biofilm formation through the quorum-sensing signal autoinducer 2 (AI-2) (27a), that TqsA is involved in transporting AI-2 (29), and that BssR/BssS regulate biofilms by influencing the biofilm signals AI-2 and indole (15).
Indole is an interspecies extracellular biofilm signal (J. Lee, A. Jayaraman, and T. K. Wood, submitted for publication) that represses the biofilm formation of E. coli (15) through its interaction with SdiA (Lee et al., submitted). It is generated from tryptophanase (encoded by tnaA), which converts tryptophan to indole, ammonia, and pyruvate (33). tnaA is induced in the stationary phase (61) and by high pH (7) and has been shown to regulate gabT, astD, and tnaB (78). Transcription of tnaA is under the control of cyclic AMP (cAMP) and the cAMP receptor protein (CRP) (78); hence, catabolite repression is an important regulatory mechanism involved in indole synthesis and biofilm formation.
In addition to the AI-2 and indole signals, bacterial surface components, such as flagella, fimbriae, proteins, and surface hydrophobicity, also influence biofilm formation in E. coli. Flagellum-mediated motility is important for E. coli cell surface contact to initiate biofilm formation and bacterial spreading along surfaces (54). Type 1 fimbriae are required for E. coli abiotic surface attachment to initiate biofilm formation (54), and aggregative fimbriae (curli) are required for E. coli to form three-dimensional biofilms (40). The prominent surface protein antigen 43 (Ag43) of E. coli is a self-recognizing adhesin that promotes cell aggregation (41) and increases biofilm formation (11). Conjugative-plasmid-encoded fimbriae enhance biofilm formation (26) and mask the importance of flagella, type 1 fimbriae, Ag43, and curli (58). In addition, hydrophobic interactions between cells and the abiotic surface mediate bacterial attachment (17) and thereby may initiate biofilm development. Therefore, changes in cell surface components caused by mutation or environmental factors influence biofilm development.
YcfR belongs to the YhcN family, which contains nine paralogous low-molecular-weight proteins (YcfR, YahO, YbiJ, YbiM, YdgH, YhcN, YjfN, YjfO, and YjfY) with unknown functions in E. coli and other bacteria (66). Since most members share a common motif in their N termini and C termini and a predicted signal peptide, the YhcN family may have evolved from a common ancestor that is thought to have played roles in self-identification or colony organization by cell-cell contacts or intercellular signaling (66). Previously, we found that ycfR is induced 12-fold in E. coli biofilm cells compared to planktonic cells (60). YcfR (85 amino acids [aa]) is also involved in the general cellular stress response, since ycfR is induced in the presence of heavy metals [25-fold at 25 μM Cd(II)] (19), during drastic pH changes (2.6-fold for pH 8.7 to 5) (45), heat shock (12-fold after 7 min at 50°C) (63), sodium salicylate treatment (9-fold at 5 mM of NaSal) (53), and 1 mM hydrogen peroxide (26-fold) (83). This gene has also been linked to the global regulator CRP and cellular-catabolite repression (9); however, aside from its identification in these various microarray studies, its relationship to these phenotypes has not been investigated previously. Here, we sought to investigate the relationship between stress response and biofilm formation by comparing differential gene expression within a biofilm upon deleting ycfR.
MATERIALS AND METHODS
Bacterial strains, media, growth conditions, and growth rate assay.
The strains and plasmids used in the present study are listed in Table 1. Wild-type E. coli K-12 BW25113 and the isogenic mutants were obtained from the Genome Analysis Project in Japan for E. coli K-12 (46). Plasmid pCA24N ycfR, carrying ycfR under tight regulation via the lacIq repressor, was obtained from the Genomic Analysis Project in Japan (46). Expression of ycfR was induced by isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma, St. Louis, Mo.).
TABLE 1.
Strains and plasmids used
| Strains and plasmids | Genotype/relevant characteristicsa | Source |
|---|---|---|
| E. coli K-12 strains | ||
| BW25113 | lacIqrrnBT14ΔlacZWJ16hsdR514 ΔaraBADAH33ΔrhaBADLD78 | 12 |
| BW25113 ΔycfR | K-12 BW25113 ΔycfR Ω Kmr | 3 |
| BW25113 ΔmelR | K-12 BW25113 ΔmelR Ω Kmr | 3 |
| BW25113 ΔsoxS | K-12 BW25113 ΔsoxS Ω Kmr | 3 |
| BW25113 ΔtnaA | K-12 BW25113 ΔtnaA Ω Kmr | 3 |
| BW25113 ΔtrpE | K-12 BW25113 ΔtrpE Ω Kmr | 3 |
| BW25113 ΔhspQ | K-12 BW25113 ΔhspQ Ω Kmr | 3 |
| BW25113 ΔgatB | K-12 BW25113 ΔgatB Ω Kmr | 3 |
| BW25113 ΔcspB | K-12 BW25113 ΔcspB Ω Kmr | 3 |
| BW25113 ΔcspG | K-12 BW25113 ΔcspG Ω Kmr | 3 |
| BW25113 ΔompW | K-12 BW25113 ΔompW Ω Kmr | 3 |
| BW25113 ΔompX | K-12 BW25113 ΔompX Ω Kmr | 3 |
| Plasmid | ||
| pCA24N ycfR | CmrlacIq pCA24N PT5-lac::ycfR+ | 3 |
Kmr and Cmr are kanamycin and chloramphenicol resistance, respectively.
Luria-Bertani (LB) medium was used to preculture all of the E. coli cells (67). LB medium and LB medium supplemented with 0.2% (wt/vol) glucose (LB glu) were used for the crystal violet biofilm, aggregation, indole, and specific-growth-rate experiments. LB glu medium was also used for the glass wool biofilm DNA microarray and glucose consumption experiments. Kanamycin (50 μg/ml) was used for preculturing the isogenic knockouts. Chloramphenicol (30 μg/ml) was used for selecting plasmid pCA24N ycfR. Cells were precultured at 37°C with shaking (250 rpm) for the indole assay, growth rate assay, cell aggregation assay, cell surface hydrophobicity, glass wool biofilm DNA microarray, and glucose consumption experiments. The specific growth rates of the E. coli wild type and the ycfR mutant were determined by measuring the cell turbidity at 600 nm of two independent cultures of each strain as a function of time, using values of less than 0.7.
Crystal violet biofilm assay.
A static biofilm formation assay was performed in 96-well polystyrene plates as reported previously (54). Briefly, cells were inoculated with an initial turbidity at 600 nm of 0.05 at 37°C in LB or LB glu medium for 24 h without shaking, and then cell growth, biofilm at the liquid-plastic interface, and total biofilm were measured using crystal violet staining. Each data point was averaged from at least 12 replicate wells (6 wells from each of two independent cultures). For evaluating the effects of Cd(II), H2O2, and acid on biofilm formation, after inoculation, cells were incubated for 6 h at 37°C, and then either Cd(II) (25 μg/ml in the form of CdCl2), H2O2 (4 and 20 mM), HCl (10 mM), or Cd(II) (25 μg/ml) plus indole (500 μM, in order to study the effect of indole addition on stress-induced biofilm formation) was added, and the plates were incubated for another 18 h at 37°C. For evaluating the effect of low temperature, cells were incubated at 22°C for 24 h. Each data point was averaged from 12 replicate wells (6 wells from each of two independent cultures).
Indole, aggregation, and cell surface hydrophobicity assays.
Extracellular and intracellular indole concentrations of the E. coli wild type and the ycfR mutant cultured in LB and LB glu media at 37°C with shaking (250 rpm) were measured spectrophotometrically, as described previously (15). The extracellular indole concentration of each stationary-phase planktonic culture was measured at 7 h, 15 h, and 24 h. Intracellular indole concentrations in biofilm cells were measured at 7 h and 15 h. Each experiment was performed twice with two independent cultures for each strain.
Cell aggregation was measured as described previously (27). Briefly, each bacterial culture was incubated for 20 h at 37°C with shaking (250 rpm) to stationary phase and then washed and diluted in 3 ml LB or LB glu medium (turbidity at 600 nm, 2.5) in 14-ml sterile tubes; after the tubes were incubated quiescently at 37°C for 15 h, the absorbance 5 mm beneath the surface was used to gauge aggregation. To detect coaggregation between the wild-type strain and the ycfR mutant, 1.5 ml of each of the diluted cultures were mixed together in one tube, followed by brief vortexing. Each experiment was performed twice with two independent cultures for each strain.
The cell surface hydrophobicity was measured as published previously by extracting the stationary-phase cells with organics (65), except that a mixture of linear hexane isomers (H302-4; Fisher Scientific Co., Pittsburgh, PA) was used to generate the hydrophobic fractions. The mixtures were vortexed thoroughly for 1 min. After standing for 15 min at room temperature for phase separation, the aqueous phase was removed and was measured to determine the cell density in the phase. Each experiment was performed twice with two independent cultures for each strain.
Glucose and curli assay.
Glucose concentrations in LB glu planktonic-cell cultures were analyzed enzymatically with a glucose assay kit (GAHK-20; Sigma). Each culture was sampled after 3 h, 7 h, and 15 h of incubation at 37°C with shaking (250 rpm), and two independent experiments were performed (the 7-h and 15-h samples were from the stationary phase). LB agar medium containing 20 μg/ml Congo red (Sigma), 10 μg/ml Coomassie brilliant blue (Sigma), and 15 g/liter agar was used as described previously (59) to visualize E. coli curli expression by inspecting the red color intensity, which is proportional to the curli concentration after 16 h of incubation at both 37°C and 30°C.
Acid resistance assay.
The acid resistance assay was adapted (44). Overnight cultures grown for 18 h at 37°C in LB medium were regrown either to mid-log phase in LB (turbidity at 600 nm, 1) or for 24 h at 37°C in LB glu medium (to validate our microarray acid resistance gene induction data), and then the cultures were diluted 40-fold in phosphate-buffered saline (pH 7.2) or 37°C LB medium (pH 2.5). E. coli in LB medium (pH 2.5) was incubated for 1 h at 37°C without shaking. The percentage of cells surviving the acid treatment was calculated as the number of CFU/ml remaining after acid treatment divided by the initial CFU/ml at time zero. At least two independent experiments were conducted.
Hydrogen peroxide resistance assay.
Overnight cultures grown for 18 h at 37°C in LB medium were regrown to mid-log phase in LB medium (turbidity at 600 nm, 1), and 1 ml of each culture was incubated with H2O2 at a final concentration of 20 mM at 37°C for 15 min without shaking. The percentage of cells surviving the H2O2 treatment was calculated as the number of CFU/ml remaining after H2O2 treatment divided by the initial CFU/ml at time zero. At least two independent experiments were conducted.
Cadmium resistance assay.
Overnight cultures grown for 18 h at 37°C in LB medium were regrown to mid-log phase in LB medium (turbidity at 600 nm, 1), and 1 ml of each culture was incubated with a final concentration of 200 μg/ml CdCl2 at 37°C for 20 min without shaking. The percentage of cells surviving the Cd(II) treatment was calculated as the number of CFU/ml remaining after Cd(II) treatment divided by the initial CFU/ml at time zero. At least two independent experiments were conducted.
Heat shock resistance assay.
Overnight cultures grown for 18 h at 37°C in LB medium were regrown to mid-log phase in LB medium (turbidity at 600 nm, 1); 1 ml of each culture was transferred to a water bath and incubated at 65°C for 20 min without shaking. The percentage of cells surviving the heat treatment was calculated as the number of CFU/ml remaining after the heat treatment divided by the initial CFU/ml at time zero.
Biofilm total RNA isolation for DNA microarrays.
Wild-type and ycfR mutant strains were precultured overnight in LB medium and LB medium with kanamycin (50 μg/ml), respectively. From each of these cultures, 2.5 ml was used to inoculate 250 ml of fresh LB glu medium with 10 g of submerged glass wool (Corning Glass Works, Corning, NY) for forming biofilm. After incubation for 15 h at 37°C with shaking (250 rpm), the glass wool was carefully and quickly removed and rinsed with 100 ml of sterile 0.85% NaCl solution at 0°C. Biofilm cells were removed by sonicating the glass wool in 200 ml of sterile 0.85% NaCl solution at 0°C, and then the total RNA was isolated as described previously (60).
DNA microarrays.
The E. coli Genechip antisense genome array (part no. 900381; Affymetrix, Santa Clara, CA) was used to analyze the complete E. coli transcriptome as described previously (27a). Based on the manufacturer's guidelines, each array contains probes for more than 4,200 open reading frames. Each open reading frame is covered by 15 probe pairs consisting of a perfect-match and a mismatch pair. The expression of each gene is evaluated by comparing the intensity of the perfect-match probe and the mismatch probe in each of the 15 probe pairs, leading to reliable gene expression profiles. The DNA microarray procedures are described in the Gene Expression Technical Manual (Affymetrix). Individual strain reports for both the wild-type strain and the ycfR mutant cDNA samples, as well as comparison reports of the ycfR mutant to the wild type, were obtained by using the GeneChip operating software (Affymetrix). The data quality was assessed based on the manufacturer's guidelines (GeneChip Expression Analysis: Data Analysis Fundamentals; Affymetrix) and also based on the expected signals of the E. coli K-12 BW25113 and the ycfR mutant genotypes (e.g., both the wild type and the ycfR mutant had a low signal for deleted genes araA and rhaA, while the ycfR mutant had no signal for ycfR). The total signal intensity was scaled to an average value of 500. Genes were identified as differentially expressed if both the P value and the corrected P value based on the Benjamini and Hochberg false-discovery rate method (5) were less than 0.05 (the corrected P value was adapted in this analysis to reduce incorrectly identified differentially expressed genes) and if the expression ratio was greater than threefold, since the standard deviation for the expression ratio for all of genes in the data was 2.3. The gene functions were obtained from the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/; 18) and the database of SRI International, the Institute for Genomic Research, the University of California at San Diego, and UNAM (http://ecocyc.org/; 39).
EMSA.
CRP protein was synthesized using the EasyXpress Linear Template Kit Plus (QIAGEN, Valencia, CA). The ycfR promoter region (262 bp, consisting of 259 bp upstream and 3 bp 5′ of ycfR) was amplified by PCR from genomic DNA of the wild-type strain BW25113 with the primers 5′-GTG TTG AGT CAG TTG CCA-3′ and 5′-CAT AAT AGT GGC CTT ATG-3′; the PCR product was gel purified with a QIAquick Gel Extraction Kit (QIAGEN) and then labeled with biotin using the Biotin 3′ End DNA Labeling Kit (Pierce Biotechnology, Rockford, IL). After the binding reaction, samples were loaded on a 6% DNA retardation gel (Invitrogen, Carlsbad, CA), and electrophoresis was carried out at 100 V for 2.5 h at 4°C. The samples were transferred to a nylon membrane (Roche Diagnostics GmbH, Mannheim, Germany) using a Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad Laboratories, Hercules, CA). 3′-Biotin-labeled DNA was detected with the LightShift Chemiluminescent EMSA kit (Pierce). In vitro-synthesized CRP protein (1 μl) was incubated with biotin-labeled ycfR promoter (8 ng) and nonspecific competitor DNA [poly(dI-dC), 1 μg] in a 20-μl binding reaction system supplied in the electrophoretic mobility shift assay (EMSA) kit. A final concentration of 1 mM cAMP (Sigma) was applied to each reaction for CRP-DNA probe binding. For the competition assay, unlabeled ycfR promoter from 40 ng to 1,200 ng was used to confirm the specificity of protein-DNA binding. As an additional negative control, biotin-labeled gadA promoter (8 ng, 294 bp, consisting of 285 bp upstream and 9 bp 5′ of gadA) lacking a CRP binding site was also amplified by PCR (primers 5′-GAT GTG GAT GAT ATC GTA-3′ and 5′-CTG GTC CAT TTC GAA CTC-3′) and incubated with in vitro-synthesized CRP (1 μl) in the presence of 1 mM cAMP.
Microarray data accession number.
The expression data have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE5904 (18).
RESULTS
Deletion of ycfR increases biofilm formation in LB glu medium.
To investigate how YcfR controls biofilm formation, biofilm formation by the ycfR deletion mutant was measured using the 96-well crystal violet assay at 37°C. Upon deleting ycfR, total biofilm (at both the air-liquid and liquid-solid interfaces) in LB glu medium after 24 h was (5 ± 1)-fold greater than that of the wild-type strain. A consistent three- to fourfold-larger cell pellet was observed for the ycfR mutant after 15 h of incubation on glass wool during the LB glu microarray assay. In addition, the biofilm at the liquid-solid interface in LB glu medium after 24 h was (3.0 ± 0.6)-fold greater than that of the wild-type strain (data not shown); hence, the ycfR mutation elicited significant biofilm at the liquid-plastic interface. The specific growth rates of the ycfR mutant in LB and LB glu media were not significantly different from those of the wild-type strain (1.42 ± 0.05 versus 1.4 ± 0.1/h in LB medium and 1.3 ± 0.2 versus 1.44 ± 0.03/h in LB glu medium). These results indicate that the presence of YcfR leads to a decrease in biofilm formation for E. coli K-12 in LB glu medium after 24 h at 37°C (especially at the liquid-solid interface) and that the effect is not due to a difference in growth. The biofilm formation of the ycfR mutant could be complemented (diminished) to that of the wild-type strain by expressing YcfR from pCA24N ycfR under the induction of 4 mM IPTG; hence, YcfR reduces E. coli K-12 biofilm formation in LB glu medium.
Unlike in LB glu medium, total biofilm formation of the ycfR mutant in LB medium was only 25 to 50% greater than that of the wild-type strain. This suggests that YcfR leads to a decrease in biofilm formation by affecting glucose uptake and metabolism. Measurement of the glucose consumption of the ycfR mutant and the wild type in LB glu medium indicated that deleting ycfR significantly reduced glucose consumption; after incubation for 3 h, the wild-type strain consumed 55% ± 5% glucose in the medium, while the ycfR mutant consumed only 31% ± 7% glucose (cf. 100% ± 0% glucose consumption of the wild type versus 88% ± 2% after 7 h of incubation). The glucose consumption of the ycfR mutant could be increased to that of the wild-type strain by expressing YcfR from pCA24N ycfR (data not shown).
Time, temperature, and medium dependence of YcfR-mediated biofilm formation.
Beloin et al. (4) reported that deleting ycfR in E. coli K-12 TG1 (which carries a conjugation plasmid) resulted in reduced biofilm formation at 30°C in flow cells with minimal glucose medium, whereas we found that deleting ycfR in E. coli BW25113 at 37°C in LB glu medium resulted in large increases in biofilm formation. An obvious difference is the absence of the conjugation plasmid in our study (conjugation plasmids dramatically affect biofilm formation [26]), but we investigated this discrepancy further by examining the effects of time, temperature, and medium on biofilm formation with the ycfR mutation. In the absence of glucose at lower temperatures (30°C in LB medium), deleting ycfR results in a (16 ± 4)-fold decrease in biofilm at 15 h, a (4 ± 2)-fold decrease at 24 h, and a (1.4 ± 0.2)-fold increase at 48 h; hence, the time of biofilm quantification is important with this mutant. Furthermore, in LB glu medium after 24 h, there was (2.7 ± 0.1)-fold-greater biofilm at 30°C but there was (1.7 ± 0.2)-fold less biofilm at 22°C; hence, the temperature at which the biofilm is measured is also important for this mutant.
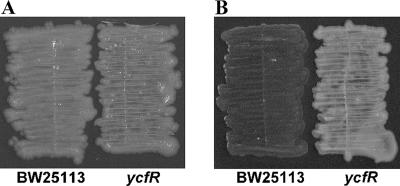
It appears that the effect of temperature on curli production is part of the reason for these effects, since deleting ycfR did not affect curli production by BW25113 at 37°C in LB medium (both the ycfR mutant and the wild-type BW25113 produce curli at low levels, as shown in Fig. 1A), but at 30°C, deleting ycfR dramatically decreased curli production compared to that of the wild-type strain (Fig. 1B). Thus, the 16-fold-diminished biofilm production of the ycfR mutant at 30°C after 15 h may be linked with the dramatic decrease in curli formation at this temperature. Clearly, the effect of ycfR deletion on biofilm formation is complex and depends on temperature, time, and medium composition.
FIG. 1.
Curli production by wild-type E. coli BW25113 and the ycfR mutant after 16 h in LB medium at 37°C (A) and at 30°C (B) as indicated by Congo red staining.
Deletion of ycfR induces acid response genes in biofilms.
To investigate the mechanism of biofilm increase caused by the ycfR deletion in glucose-containing (LB glu) medium, DNA microarrays were performed to explore differential gene expression in a biofilm as a result of deletion of ycfR. At 15 h, 1.8% of the E. coli genes were differentially expressed in the glass wool biofilm using a 3-fold cutoff based on the 2.3-fold standard deviation, which includes 48 induced genes (Table 2) and 28 repressed genes (Table 3).
TABLE 2.
E. coli genes induced more than threefold (P < 0.05) in an LB glu biofilm after 15 h at 37°C upon deleting ycfR
| Group and gene | b no.a | Description | Expression ratio |
|---|---|---|---|
| tRNA | |||
| metW | b2815 | tRNA | 3.2 |
| leuX | b4270 | tRNA | 3.0 |
| metY | b3171 | tRNA | 3.0 |
| metZ | b2814 | tRNA | 3.0 |
| RNA related | |||
| rnpB | b3123 | RnpB RNA, catalytic subunit of RNase P | 3.7 |
| rrlC | b3758 | 23S rRNA | 3.5 |
| rrlD | b3275 | 23S rRNA | 3.5 |
| rrfH | b0205 | 5S rRNA | 3.0 |
| Regulator | |||
| ybgS | b0753 | Putative regulator, not classified, putative homeobox protein | 4.9 |
| yiaG | b3555 | Putative transcriptional regulator | 3.2 |
| Stress related | |||
| gadA | b3517 | Glutamate decarboxylase A subunit, acid resistance protein | 6.5 |
| gadB | b1493 | Glutamate decarboxylase B subunit, acid resistance protein | 4.9 |
| gadC | b1492 | Putative transporter, acid resistance protein | 6.5 |
| gadE | b3512 | GadE transcriptional activator, acid resistance protein | 3.0 |
| slp | b3506 | Outer membrane constituents, starvation lipoprotein | 4.3 |
| nhaA | b0019 | Na+/H antiporter, pH dependent | 5.7 |
| hdeB | b3509 | Acid resistance protein | 4.0 |
| hdeA | b3510 | Acid resistance protein, possible chaperone, subunit of HdeA dimer | 3.5 |
| hdeD | b3511 | Protein involved in acid resistance | 5.3 |
| osmY | b4376 | Hyperosmotically inducible periplasmic protein | 4.3 |
| osmB | b1283 | Osmotically inducible lipoprotein, adaptation to osmotic pressure | 3.5 |
| bssS (yceP) | b1060 | Regulator of biofilm through signal secretion | 3.5 |
| uspB | b3494 | Ethanol tolerance protein | 3.2 |
| dnaK | b0014 | Chaperone Hsp70, autoregulated heat shock proteins | 3.0 |
| sodC | b1646 | Superoxide dismutase precursor (Cu-Zn), detoxification | 3.0 |
| ompX | b0814 | Outer membrane protein X, adhesion | 3.7 |
| hspQ | b0966 | Hemimethylated DNA-binding protein | 3.5 |
| Metabolism | |||
| yohC | b2135 | Predicted GTP-binding transport protein, essential for E. coli growth | 5.3 |
| prpB | b0331 | Putative carboxyphosphonoenolpyruvate mutase | 4.6 |
| prpD | b0334 | 2-Methyl citrate dehydratase | 3.0 |
| ybaY | b0453 | Glycoprotein/polysaccharide metabolism, predicted outer membrane lipoprotein | 3.7 |
| yfeP | b2392 | High-affinity manganese transporter | 3.5 |
| ynhG | b1678 | Putative ATP synthase subunit | 4.0 |
| pykF | b1676 | Pyruvate kinase I monomer, subunit of pyruvate kinase I | 3.5 |
| yjgA | b4234 | Putative ABC superfamily transport protein | 3.5 |
| deoA | b4382 | Thymidine phosphorylase | 3.2 |
| deoC | b4381 | Deoxyribose-phosphate aldolase | 3.2 |
| deoD | b4384 | Purine-nucleoside phosphorylase | 3.0 |
| pyrG | b2780 | Subunit of CTP synthetase | 3.2 |
| yfhN | b2529 | Scaffold protein involved in iron-sulfur cluster assembly | 3.0 |
| yfhO | b2530 | Cysteine desulfurase | 3.0 |
| Unknown function | |||
| ybiM | b0806 | Hypothetical protein | 5.7 |
| ybaA | b0456 | Hypothetical protein | 4.6 |
| yceK | b1050 | Hypothetical protein | 4.6 |
| yjbJ | b4045 | Highly abundant nonessential protein | 4.3 |
| yjdN | b4107 | Hypothetical protein | 3.7 |
| ygaM | b2672 | Conserved hypothetical protein | 3.2 |
| ymgE | b1195 | Predicted inner membrane protein | 3.0 |
b no., a unique numeric identifier assigned to each E. coli gene.
TABLE 3.
E. coli genes repressed more than threefold (P < 0.05) in an LB glu biofilm after 15 h at 37°C upon deleting ycfR
| Group and gene | b no.a | Description | Expression ratio |
|---|---|---|---|
| tRNA | |||
| thrT | b3979 | tRNA | −3.0 |
| valX | b2402 | tRNA | −3.2 |
| valZ | b0746 | tRNA | −3.0 |
| Regulator | |||
| cspB | b1557 | CspA family of cold shock protein | −5.7 |
| cspG | b0990 | Homolog of Salmonella cold shock protein | −3.0 |
| lrhA | b2289 | NADH dehydrogenase transcriptional regulator, LysR family | −3.2 |
| marA | b1531 | Regulator, drug/analog sensitivity | −3.0 |
| putA | b1014 | Bifunctional enzyme, as well as a transcriptional repressor of the put (proline utilization) regulon | −4.0 |
| Transport and metabolism | |||
| atpB | b3738 | Membrane-bound ATP synthase, F0 sector, subunit A | −3.5 |
| atpE | b3737 | Membrane-bound ATP synthase, F0 sector, subunit C | −3.0 |
| atpF | b3736 | ATP synthase, F0 complex, subunit B | −3.0 |
| atpH | b3735 | Membrane-bound ATP synthase, F1 sector, delta subunit | −3.2 |
| feoA | b3408 | Ferrous iron transport protein A | −3.2 |
| gatA | b2094 | GatA, subunit of EIIGat, galactitol PTS permease | −3.0 |
| gatB | b2093 | Transport, transport of small molecules: carbohydrates, organic acids, alcohols | −4.9 |
| gatC | b2092 | Transport, transport of small molecules: carbohydrates, organic acids, alcohols | −3.7 |
| gatD | b2091 | Enzyme, degradation of small molecules: carbon compounds | −4.0 |
| nmpC | b0553 | Outer membrane porin | −6.5 |
| putP | b1015 | Sodium/proline symporter responsible for the uptake of proline | −4.3 |
| rbsD | b3748 | d-Ribose high-affinity transport system; membrane-associated protein | −3.5 |
| rbsA | b3749 | ATP-binding component of d-ribose high-affinity transport system | −3.5 |
| rbsC | b3750 | d-Ribose high-affinity transport system | −3.5 |
| sdaC | b2796 | Probable serine transporter | −4.0 |
| ompW | b1256 | Outer membrane protein W; colicin S4 receptor; putative transport protein | −6.0 |
| Metabolism | |||
| tnaA | b3708 | Tryptophanase | −4.6 |
| tnaL (tnaC) | b3707 | Tryptophanase leader peptide | −4.9 |
| yfiD | b2579 | Putative formate acetyltransferase | −4.3 |
| Unknown function | |||
| yeeI | b1976 | Conserved hypothetical protein | −3.2 |
b no., a unique numeric identifier assigned to each E. coli gene.
The microarray analysis indicated that deleting ycfR induced a group of acid resistance genes in the biofilm; for example, gadABC, which encode two glutamate decarboxylase polypeptides (GadA and GadB) (73) and a putative gamma-aminobutyrate antiporter (GadC) (13), were induced five- to sixfold. gadE, which encodes a transcriptional activator of GadABC (31), was also induced three-fold. hdeABD, which encode two periplasmic acid resistance proteins (HdeA and HdeB) (74) and an acid resistance-related protein (HdeD) (44), were induced four- to fivefold. slp, which encodes an outer membrane starvation-inducible lipoprotein (1), was induced fourfold. Slp, HdeABD, GadE, and GadABC are involved in the E. coli YdeO-induced acid resistance regulatory network (44).
To investigate why the ycfR mutation induced acid resistance genes in the LB glu biofilm, we measured the culture pH. After 7 h, the pH of the cultures of both the ycfR mutant and the wild type dropped to 5. After 15 h and 24 h, the pH of the culture of the ycfR mutant was significantly lower than that of the wild-type strain (4.9 versus 7.6 and 5.0 versus 8.6, respectively); hence, the acid resistance genes were identified in the microarrays as induced by deleting ycfR, since the ycfR cells experienced a much lower pH. To further verify our microarray results, the survival of 24-h LB glu-grown planktonic ycfR cells after 1 h of incubation at pH 2.5 was investigated; ycfR cells were (6 ± 2)-fold more resistant to the acid incubation than the wild-type strain (due to previous induction of the acid resistance genes), which validates our microarray results.
Deletion of ycfR induces stress response genes in biofilms and increases sensitivity to stress.
Along with the acid resistance genes, other stress response genes were induced three- to fourfold in the biofilm by the ycfR deletion, including three osmotic-stress-inducible genes, osmY, which encodes a hyperosmotically inducible periplasmic protein (82); osmB, which encodes an osmotically inducible outer membrane lipoprotein (36); and ompX, which encodes an outer membrane protein regulated by osmolarity and pressure (47). Other stress genes that were induced include sodC, which encodes a periplasmic superoxide dismutase and protects bacteria from oxidation (28); uspB, which encodes an inner membrane ethanol tolerance protein (22); bssS, which encodes a global regulator involved in stress response, as well as regulation of biofilm formation (15); and dnaK, which encodes a heat shock protein maintaining DNA structure against thermal stress (50). Therefore, acid, osmotic, oxidative, and heat stress response genes were induced upon deletion of ycfR in the biofilm, and taking this together with the fact that the transcription of ycfR in suspension cells is induced under various stress conditions [e.g., H2O2 treatment (83), Cd(II) (19), pH (45), heat shock (63), and sodium salicylate treatment (53)], it is likely that YcfR may be involved in a global stress resistance response and that the ycfR mutant biofilm contends with higher stress levels than the wild-type biofilm. Therefore, we tested the sensitivity of the ycfR mutant to the following stresses in LB medium: low pH, heat, H2O2, and high Cd(II) concentration. For all the tested stress conditions [1 h at pH 2.5, 20 min at 65°C, 15 min with 20 mM H2O2, and 20 min with 200 μg/ml Cd(II)], the ycfR mutant had diminished survival compared to the wild-type strain; deleting ycfR caused (10 ± 2)-fold more sensitivity to acidic pH, (14 ± 3)-fold more sensitivity to heat, (66 ± 7)-fold more sensitivity to H2O2, and (16 ± 4)-fold more sensitivity to Cd(II) than for the wild-type strain (Fig. 2). Therefore, YcfR is a multiple stress resistance protein.
FIG. 2.
Survival percentages of the wild-type strain and the ycfR mutant in LB medium after addition of hydrogen peroxide (A), heat (B), acid (C), and cadmium (D). The experiments were repeated at least two times (one representative data set shown), and 1 standard deviation is shown.
Deletion of ycfR decreases indole synthesis.
In E. coli, indole is synthesized from tryptophan by tryptophanase (encoded by tnaA) and is exported by multidrug exporters, such as ArcEF (38). Our microarray analysis indicated that deleting ycfR repressed tnaA 5-fold at 15 h in LB glu biofilm; this was validated by the (2.3 ± 0.2)-fold reduction in the extracellular indole concentration for the stationary-phase planktonic ycfR culture at 15 h and (5 ± 1)-fold reduction at 24 h in LB glu medium. The extracellular indole concentrations were corroborated by examining the intracellular indole concentrations of LB glu biofilm cells (on glass wool); as expected, the intracellular indole concentration for the ycfR biofilm cells was (4.3 ± 1)-fold lower at 7 h and (4.8 ± 0.6)-fold lower at 15 h. Since indole acts as an extracellular signal that represses biofilm formation in E. coli K-12 (15), our finding that deleting ycfR decreased indole is consistent with the observation that the ycfR mutation increased biofilm formation in LB glu medium. In contrast, in LB medium, the extracellular indole concentrations of the ycfR mutant after 15 h and 24 h of incubation were 75% ± 32% and 85% ± 32% of that of the wild-type strain, respectively, which is consistent with the observation that deletion of ycfR did not significantly affect biofilm formation in LB medium.
Stress increases biofilm formation by decreasing indole.
Since we found that YcfR protects suspension cells from different kinds of stresses [low pH, heat, H2O2, and Cd(II)] and that the ycfR deletion induced stress genes (the acid resistance genes gadABC, gadE, and hdeABD; the DNA-binding heat shock gene hspQ; the starvation lipoprotein gene slp; the osmotic-stress-induced genes osmBY; the ethanol-resistance protein uspB; the periplasmic superoxide dismutase precursor gene sodC; and the chaperone protein gene dnaK involved in the protection of cells against heat shock and oxidative stress), we hypothesized that the biofilm formed by the ycfR mutant would be defective in coping with stress and that this elevated stress level might stimulate biofilm formation in LB glu medium. This hypothesis is corroborated by the fact that the deletion of genes that are involved in stress tolerance (e.g., the stress regulator oxyR in E. coli [11], ropA in Streptococcus mutans [80], spx in Staphylococcus aureus [52], and soxS in E. coli in this work) increases biofilm 0.3- to 5-fold. Further evidence of this relationship is the fact that stress-associated genes are induced in biofilms (4, 14, 60, 69) and that biofilm has been shown to increase in response to stress conditions (e.g., sublethal concentrations of aminoglycoside antibiotics in Pseudomonas aeruginosa and E. coli [30], osmotic stress in S. aureus [56], and high metal concentration, extreme pH and temperature, and the addition of xenobiotics, antibiotics, and oxygen in the archaebacterium Archaeoglobus fulgidus [42]).
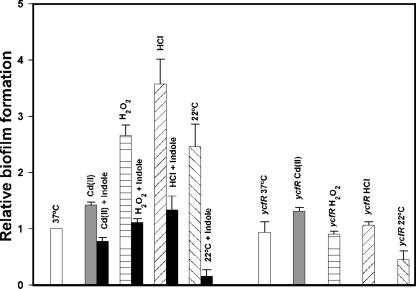
In order to test this hypothesis, we performed a series of crystal violet biofilm assays under stress conditions, initiated after 6 h of normal growth at 37°C in LB medium, by adding 25 μg/ml Cd(II), 20 mM H2O2, or 10 mM HCl; also, low-temperature stress was evaluated by incubating cells continuously at 22°C (Fig. 3). Except for 22°C, these conditions only slightly affected the total growth of both the wild-type strain and the ycfR mutant; therefore, the results were not due to differences in growth. These stresses stimulated biofilm formation by the wild-type strain by (0.50 ± 0.02)-fold for Cd(II) addition, (3.0 ± 0.2)-fold for the H2O2 treatment, (3.0 ± 0.3)-fold for the HCl treatment, and (2.0 ± 0.8)-fold for low-temperature incubation. Under the same conditions, these stresses did not stimulate biofilm formation by the ycfR mutant, except for the addition of Cd(II), which increased its biofilm slightly (0.3-fold) (Fig. 3). Hence, biofilm formation in E. coli is induced by oxidative, acid, low-temperature, and heavy metal stresses, and YcfR is required for these responses.
FIG. 3.
Relative biofilm formation of the wild-type strain and the ycfR mutant after addition of 25 μg/ml of cadmium, 20 mM hydrogen peroxide, and 10 mM hydrochloric acid to LB medium at 37°C and after incubation at 22°C. The effect of the addition of 500 μM indole on wild-type biofilm formation under the same conditions is also shown. All the biofilm formation values were normalized to that of the wild type at 37°C. The experiments were repeated at least two times (one representative data set shown), and 1 standard deviation is shown.
Based in the observation that the induction of sigma E factor (rpoE), which controls the expression of an important stress resistance regulon, drastically represses the transcription of the tnaA gene (7-fold in stationary-phase cells and 33-fold in exponential-phase cells) (37), we hypothesized that the induction of rpoE by the accumulation of unfolded proteins produced by stress (37) probably reduces the indole production via tnaA repression and that the decreased indole concentration enhances biofilm formation. To further explore this proposed mechanism, we tested the Cd(II) biofilm induction phenotype of the tnaA and trpE deletion mutants (each has 10-fold less indole than the wild-type strain [Lee et al., submitted]) and found that both mutants repressed their biofilm formation upon Cd(II) addition (tnaA, [0.8 ± 0.07]-fold, and trpE, [0.5 ± 0.14]-fold) rather than increasing it like the wild-type strain and other non-biofilm-related deletion mutants (e.g., melR). Additionally, deleting tnaA prevented an increase in biofilm formation upon H2O2 and HCl addition but, surprisingly, not upon low temperature (22°C) (data not shown). These results indicate that decreasing indole could also be a mechanism that enhances biofilm formation in response to Cd(II), H2O2, and acidification. Furthermore, adding 500 μM indole to the wild-type strain simultaneously with Cd(II), H2O2, or HCl or during the 22°C incubation rendered the cells incapable of increasing biofilm formation (Fig. 3); hence, indole concentrations are important for controlling biofilm formation upon stress in E. coli.
Deletion of ycfR increases aggregation and cell surface hydrophobicity.
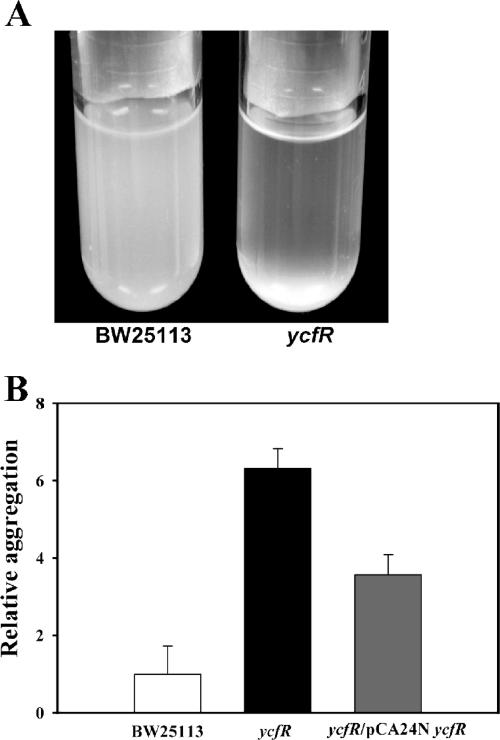
Deleting ycfR caused (4 ± 1)-fold-greater aggregation than that of the wild-type strain in LB medium (Fig. 4A and B) and (5.1 ± 0.1)-fold greater aggregation in LB glu medium (data not shown). Furthermore, this phenotype could be partially complemented by expressing ycfR from a multicopy plasmid (Fig. 4B). These results indicate that YcfR impedes cell aggregation. To further examine the mechanism by which YcfR influences cell aggregation, a coaggregation test was performed by mixing the wild-type strain with the ycfR mutant; no apparent coaggregation was observed (data not shown). Resuspending wild-type bacterial cells in the supernatant of the ycfR mutant LB or LB glu culture did not increase aggregation of the wild-type cells either (data not shown).
FIG. 4.
(A) Cell aggregation in LB medium at 37°C after 15 h upon deleting ycfR. (B) Relative aggregation of the wild-type, ycfR, and ycfR/pCA24N ycfR cultures in LB medium (1 mM IPTG added to ycfR/pCA24N ycfR to induce expression of YcfR). Two to 10 replicates were used, and 1 standard deviation is shown.
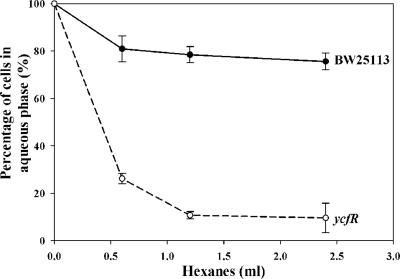
Deleting ycfR also caused the cells to become dramatically more hydrophobic (Fig. 5), which explains the increased cell aggregation. The increase in hydrophobicity also explains our observation that deleting ycfR significantly increased biofilm formation at the liquid-solid interface in 96-well plates, since hydrophobic interactions between the hydrophobic bacterial cell surface and the hydrophobic surface of the plastic well mediates attachment to plastic (17), which appears to enhance biofilm formation. It was also noticed that although deleting ycfR increased cell aggregation in both LB and LB glu media in similar ways, the increase of biofilm by deleting ycfR in LB glu medium is much greater than that in LB medium. This suggests that an increase in cell surface hydrophobicity is only one of several effects on biofilm formation caused by this mutation.
FIG. 5.
Hydrophobicity of the wild-type strain and the ycfR mutant after growth in LB medium at 37°C. The experiments were repeated twice, and 1 standard deviation is shown.
The ycfR biofilm microarray data also indicated that deleting ycfR differentially induced a large group of genes encoding cell surface proteins (33% of the induced genes), including outer membrane proteins (e.g., OmpX, OsmB, Slp, and YbaY), periplasmic proteins (e.g., OsmY, SodC, HdeA, HdeB, and YbiM), and some inner membrane-associated proteins (e.g., GadC, HdeD, NhaA, YfeP, YgaM, YhiO, and YohC). Additionally, 60% of the repressed genes encoded proteins on the cell surface, including the outer membrane proteins OmpW and NmpC, and membrane-associated transporters, such as AtpBEF and RrbsDAC. Hence, deletion of ycfR critically affects bacterial cell surface properties, including cell surface hydrophobicity, aggregation, and, ultimately, biofilm formation.
Differentially expressed genes due to ycfR deletion are biofilm related.
In order to further explore the importance of ycfR for biofilm formation, we assayed biofilm formation for six isogenic mutants based on their differential gene expression (hspQ, gatB, cspBG, and ompWX) in both LB and LB glu media after 24 h at 37°C. Deletion of all the genes but hspQ and ompW significantly altered biofilm formation in LB (cspBG and ompX reduced biofilm formation by two- to threefold, whereas gatB increased biofilm twofold) and in LB glu (cspBG, gatB, and ompX increased biofilm formation twofold) media. These results are consistent with our conclusion that ycfR plays an important role in E. coli biofilm formation and also show that the previously identified biofilm genes cspBG (14) control biofilm formation.
YcfR is a putative membrane protein, and the ycfR promoter has CRP and SoxS binding sites.
We investigated the possibility that YcfR is a membrane protein by using multiple-protein analysis programs, such as the ExPASy server (24), SignalP (49), and PSORTb v.2.0 (62), all of which predicted that YcfR (85 aa) contains an N-terminal signal peptide with cleavage between Ala22 and Ala23; therefore, YcfR may have multiple localization sites in the membrane, in the periplasmic space, or in the extracellular space. Consistently, expression of YcfR protein from pCA24N ycfR and consequent analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis indicated that the YcfR protein was from the insoluble membrane fraction, not from the soluble cytosolic fraction (data not shown).
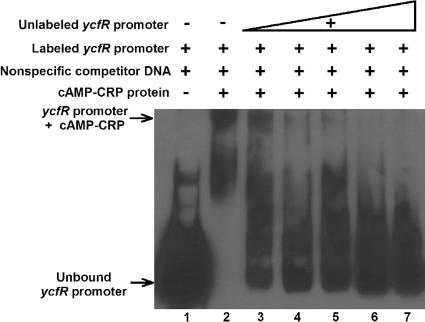
We hypothesized that the remarkably enhanced biofilm formation of the ycfR mutant when cultured in LB glu medium, but not in LB medium, may be due to regulation by CRP, and a putative CRP binding site (9) was identified between positions −70 and −83 in the ycfR promoter. Hence, we tested CRP binding to the ycfR promoter using an EMSA. We found that CRP binds the upstream region of the ycfR gene (Fig. 6). This binding is specific, since applying excess nonspecific DNA [poly(dI-dC)] does not affect CRP-ycfR promoter region binding while increasing unlabeled ycfR promoter DNA reverses the binding. An additional negative control, CRP and the gadA promoter (which lacks a CRP binding site), did not show CRP binding under the same conditions (data not shown).
FIG. 6.
EMSA to test the binding of cAMP-CRP to the ycfR promoter (ycfRp). Lane 1, labeled ycfRp; lane 2, cAMP-CRP protein and labeled ycfRp; lanes 3 to 7: cAMP-CRP protein, labeled ycfRp, and nonspecific competitor unlabeled ycfRp.
Further analysis of the ycfR promoter with BPROM, a bacterial promoter prediction program (SoftBerry, Mount Kisco, NY), showed the presence of a putative SoxS binding site between positions −59 and −66. Hence, ycfR may also be part of the stress response soxRS regulon, which protects the cell against superoxide (2) and H2O2 (70) stresses. Moreover, soxS is one of the most induced genes in E. coli K-12 biofilms (50-fold) (60), so it may be responsible for the concomitant ycfR induction. However, EMSA to confirm the SoxS protein binding ycfR was unsuccessful (data not shown), which was probably caused by the extremely short half-life of the SoxS protein (∼2 min) (71).
DISCUSSION
In this study, by investigating the profound changes induced by deletion of ycfR, we demonstrated that YcfR inhibits E. coli K-12 biofilm formation by repressing cell aggregation, by increasing the biofilm signal indole, by decreasing hydrophobicity, and by interfering with the acid stress response. We also showed that YcfR increases viability under stress conditions and that stress in general increases biofilm formation.
The development of biofilm is a complex and dynamic process that couples a cascade of responses to a variety of environmental signals in bacterial cells (14, 55). Cell surface adhesion and cell aggregation initiate bacterial biofilm formation (68). Our results demonstrate that deleting ycfR induces significant cell aggregation and also increases liquid/solid biofilm formation. This suggests that YcfR decreases biofilm formation by repressing cell-cell interaction and cell surface interaction. It is intriguing that it is the absence of this small membrane protein that induces rather than represses aggregation and shows that YcfR itself is not the cell surface protein that is directly involved in cell-cell adhesion. Further evidence of this is the fact that there is no coaggregation between wild-type cells and the ycfR mutant, and since coaggregation is mediated by specific interactions between cell surface lectin-like adhesin proteins and receptors on the surfaces of other cells (64), the aggregation caused by deleting ycfR should not be a consequence of changes in cell surface adhesins or receptors. Furthermore, resuspending wild-type cells in the supernatants of the ycfR mutant LB or LB glu cultures did not increase aggregation of the wild-type cells, which suggests that the aggregation of the ycfR mutant is not caused by extracellular signals secreted by the mutant. In E. coli, the adhesion protein Ag43, encoded by the flu gene, is a self-recognizing surface adhesin that confers cell aggregation (41) and is subject to phase variation (11). The fact that flu is not induced in the ycfR mutant in the LB glu biofilm implies that the aggregation is not mediated by overexpression of Ag43. Our hydrocarbon extraction experiment (Fig. 5) clearly demonstrated that the dramatic change of the E. coli K-12 cell surface from hydrophilic to hydrophobic is caused by the ycfR deletion. This is consistent with the observation that the ycfR mutant has increased bacterial aggregation and increased liquid-solid interface biofilm formation. However, this hydrophobicity change cannot be simply explained by loss of the YcfR protein, since the predicted isoelectric point of YcfR is 9, yet when the culture pH was 9, the wild-type strain did not show increased aggregation (data not shown). Therefore, most likely YcfR modulates bacterial cell surface hydrophobic properties by affecting the expression of other surface proteins.
Our differential gene expression analysis demonstrated that a previously identified aggregation-related gene, osmB, was significantly induced by deletion of ycfR. osmB encodes an osmotically inducible outer membrane lipoprotein (35). Interestingly, E. coli cells aggregate upon elevated osmolarity in an OsmB-dependent manner (35). Moreover, we demonstrated that deleting ycfR induces and represses about 30 outer membrane-associated, periplasmic, or inner membrane-associated protein-encoding genes. The differential expression of these genes may critically change bacterial cell surface properties. Taken together, these results suggest that YcfR affects bacterial cell surface structures and properties by affecting cell surface protein gene expression, which further affects cell aggregation and consequent biofilm formation. Further research on these differentially expressed genes may reveal the molecular mechanism for the change of the cell surface hydrophobicity and their roles in biofilm development. Our finding of the ycfR mutation blocking curli generation at low temperature is also interesting. Since curli expression is dependent on environmental cues, such as a low growth temperature (8), whether YcfR plays a role as a temperature sensor involved in E. coli curli generation deserves further investigation.
Another part of the mechanism by which YcfR decreases biofilm formation is through its induction of indole. Indole is an extracellular signal (Lee et al., submitted) that represses biofilm formation (15). Our analysis clearly demonstrates that deleting ycfR significantly reduces both extracellular and intracellular indole concentrations, and this result is consistent with our DNA microarray analysis that showed tnaA is repressed by fivefold in the ycfR mutant. The expression of tnaA is under cAMP and CRP regulation (78) and is also inducible by high pH (75). Indole production in the wild-type strain increased 10-fold from 7 h to 15 h after glucose in the medium was depleted and increased further when the pH turned alkaline. However, since the culture of the ycfR mutant remained acidic even after the glucose was consumed, the expression of the tnaA gene was repressed. Therefore, deletion of ycfR decreases indole generation by reducing glucose uptake and metabolism and by maintaining an acidic pH, which together lead to greater biofilm formation.
That the YcfR protein may be directly involved in the cellular transport of metabolites is suggested by its periplasmic or outer membrane nature; by the fact that glucose remains in the culture medium of the ycfR mutant (which suggests a lower glucose transport rate than the wild type); by the switch from acid to alkaline pH in the wild-type strain when grown in LB glu medium (the transmembrane H+ gradient and metabolite transport are interdependent [48]); by the fact that transport of several amino acids, like phenylalanine, tyrosine, and tryptophan, depends on the H+ gradient (10); and by the fact that acidification of the external medium of the ycfR mutant may be both cause and consequence of different electrochemical potentials between the cell interior and the external medium. The induction and repression of many transporter-encoding genes in our microarray and the capacity of the YcfR protein to provide the cells with multiple stress resistances also suggest that YcfR is involved in the transport of metabolites, since some stress resistance proteins function as efflux systems and pumps, like the MtsABC transporter in Streptococcus pyogenes (34) and the heavy metal transport ATPases of several species of bacteria (72). Moreover, a protein BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/) of YcfR showed that this protein has high identity with predicted cation transport ATPases (important in stress response) of Shigella boydii and several E. coli strains; however, the small size of YcfR (85 aa) and the apparent absence of transmembrane segments suggest that YcfR is part of a multimeric complex.
Besides the acid response genes, our differential gene expression analysis demonstrated that other stress-related genes, such as ompXY, omsB, sodC, yhiO, bssS, and dnaK, were induced by deletion of ycfR. The expression of ompX is induced by both acidic and basic conditions (75), and deletion of ompX increases cell surface adhesion of fimbriated strains of E. coli and decreases cell surface adhesion of nonfimbriated strains (51). Our observation that deletion of ycfR significantly increases biofilm at the liquid-solid interface in the crystal violet biofilm assay may be related to a YcfR-OmpX adhesion feedback. Both the lipoprotein OmsB (36) and the predicted periplasmic OsmY (82) are thought to function in osmotic-stress response, since their expression was induced by osmotic stress. Interestingly, the expression of dnaK, omsBY, and ompX responds to a global signal, acetyl phosphate, which functions during biofilm development (81). Whether YcfR influences biofilm development through acetyl phosphate requires further research. Recently, our laboratory identified a negative regulator of E. coli K-12 biofilm formation, bssS, which influences cell signaling (15). Our finding of induction of bssS in the ycfR mutant biofilm might indicate that the bacterial cells try to repress the abnormal increase of biofilm caused by deletion of ycfR.
Our study demonstrates that the ycfR gene, which is strongly induced under stress conditions, confers resistance to several kinds of stress, including low pH, heat shock, H2O2, and high Cd(II) concentration. Hence, ycfR induction may be related to a general effect of all the stresses or to the common mechanisms that the cells use to protect themselves against stress, as seen in plants (23), Saccharomyces cerevisiae (76), and bacteria (32); for example, it has been shown that both heavy metal stress (23) and heat shock stress (76) converge in the generation of oxygen-reactive species.
ycfR is one of the most induced genes in E. coli biofilms, since it is induced 12-fold at 7 h compared to planktonic cells (60) and 6.4-fold in mature (8-day) biofilm (4). These data suggest that ycfR helps cells to contend with the stress generated in the biofilm, and we have shown that YcfR is required for survival under some stress conditions (Fig. 2). The fact that stress genes are induced in biofilms is established; for example, (i) ycfR, the transcriptional regulator soxS, and the small heat shock proteins genes ibpAB are among the most induced genes in biofilms (60); (ii) genes involved in cellular processes, such as envelope stress responses, like pspABCDE, cpxP, spy, rpoE, and rseA, and other stress-associated genes, like recA and dinl, are induced in mature biofilms (4); and (iii) the cold shock protein genes cspABFGI are induced in young (4- and 7-h) E. coli K-12 biofilms (14). Moreover, it has been proposed that the Cpx regulon is a strategic signaling pathway for coping with adverse conditions necessary for biofilm communities (16). Therefore, a strong cellular response against stress is developed in biofilms. In fact, high stress levels are normal in microbial biofilms, since there is a large proportion of cells with injured membranes, as in Streptococcus oral biofilm (25), and there are reports of elevated cell death in the biofilm of P. aeruginosa (79) and Pseudomonas tunicate (43).
The relationship between stress and bacterial biofilm formation has been studied to some extent in eubacteria, particularly in S. aureus (56) and S. mutans (80), where it is established that osmotic and acid/oxidative stress, respectively, are related to biofilm induction. In E. coli, biofilm induction in response to subinhibitory concentrations of aminoglycoside antibiotics has been shown (30), but there is only indirect evidence about the general relationship between stress and biofilm formation in this organism (6). In this work, we showed that E. coli K-12 produces more biofilm as a defensive response against several stresses, including acidic pH, oxidative stress (H2O2), heavy metals [Cd(II)], and cold shock (22°C). In addition, we showed that this stress induction in the biofilm is related to a diminution of indole concentrations. In archaea, it has been demonstrated that the hyperthermophile A. fulgidus can produce enhanced biofilm in response to nonphysiological extremes (e.g., pH, temperature, metals, antibiotics, xenobiotics, and oxygen), and these biofilm cells show increased tolerance of adverse environmental conditions (42). Several other works have shown that biofilm cells resist stresses, like H2O2 (20), heavy metals (77), and antibiotics (30), better than planktonic cells. Our data also suggest that the E. coli biofilm tolerates H2O2 stress better than suspension cells, and the enhanced biofilm tolerance of stress is not surprising, since biofilm cells form a barrier that protects inner cells from the hazardous environment via a polysaccharide matrix capable of sequestering metals and dangerous compounds (42). The gradient in concentrations found in biofilms (57) is also expected to protect inner biofilm cells, and only the most metabolically active cells are more susceptible to antimicrobial agents (21); therefore, internal biofilm cells are less exposed to toxins. Taking into account all of the above, it is reasonable to surmise that biofilm induction in response to stress is a general strategy that prokaryotes (eubacteria and archaeobacteria) evolved in order to protect themselves against unfavorable environmental conditions. Although we are just beginning to explore the mechanism by which stress induces biofilm formation, at least for E. coli, our findings suggest that reducing the concentration of the biofilm inhibitor indole (15) is linked with increases in biofilm formation as a result of stress.
Acknowledgments
This research was supported by the NIH (EB003872-01A1) and the ARO (W911NF-06-1-0408).
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Alexander, D. M., and A. C. St John. 1994. Characterization of the carbon starvation-inducible and stationary phase-inducible gene slp encoding an outer membrane lipoprotein in Escherichia coli. Mol. Microbiol. 11:1059-1071. [DOI] [PubMed] [Google Scholar]
- 2.Amabile-Cuevas, C. F., and B. Demple. 1991. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 19:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. doi: 10.1038/msb4100060. [DOI] [PMC free article] [PubMed]
- 4.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. [DOI] [PubMed] [Google Scholar]
- 5.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289-300. [Google Scholar]
- 6.Bianco, C., E. Imperlini, R. Calogero, B. Senatore, A. Amoresano, A. Carpentieri, P. Pucci, and R. Defez. 2006. Indole-3-acetic acid improves Escherichia coli's defences to stress. Arch. Microbiol. 185:373-382. [DOI] [PubMed] [Google Scholar]
- 7.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brombacher, E., A. Baratto, C. Dorel, and P. Landini. 2006. Gene expression regulation by the Curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 188:2027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, C. T., and C. G. Callan, Jr. 2004. Evolutionary comparisons suggest many novel cAMP response protein binding sites in Escherichia coli. Proc. Natl. Acad. Sci. USA 101:2404-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosgriff, A. J., G. Brasier, J. Pi, C. Dogovski, J. P. Sarsero, and A. J. Pittard. 2000. A study of AroP-PheP chimeric proteins and identification of a residue involved in tryptophan transport. J. Bacteriol. 182:2207-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 14.Domka, J., J. Lee, T. Bansal, and T. K. Wood. 2007. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9:332-346. [DOI] [PubMed] [Google Scholar]
- 15.Domka, J., J. Lee, and T. K. Wood. 2006. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ Microbiol. 72:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorel, C., P. Lejeune, and A. Rodrigue. 2006. The Cpx system of Escherichia coli, a strategic signaling pathway for confronting adverse conditions and for settling biofilm communities? Res. Microbiol. 157:306-314. [DOI] [PubMed] [Google Scholar]
- 17.Dunne, W. M., Jr. 2002. Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 15:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egler, M., C. Grosse, G. Grass, and D. H. Nies. 2005. Role of the extracytoplasmic function protein family sigma factor RpoE in metal resistance of Escherichia coli. J. Bacteriol. 187:2297-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkins, J. G., D. J. Hassett, P. S. Stewart, H. P. Schweizer, and T. R. McDermott. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ Microbiol. 65:4594-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eng, R. H., F. T. Padberg, S. M. Smith, E. N. Tan, and C. E. Cherubin. 1991. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 35:1824-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farewell, A., K. Kvint, and T. Nystrom. 1998. uspB, a new σS-regulated gene in Escherichia coli which is required for stationary-phase resistance to ethanol. J. Bacteriol. 180:6140-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garnier, L., F. Simon-Plas, P. Thuleau, J. P. Agnel, J. P. Blein, R. Ranjeva, and J. L. Montillet. 2006. Cadmium affects tobacco cells by a series of three waves of reactive oxygen species that contribute to cytotoxicity. Plant Cell Environ. 29:1956-1969. [DOI] [PubMed] [Google Scholar]
- 24.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gelle, M. P., L. F. Jacquelin, and C. Choisy. 2003. Compared viability of planktonic bacteria and bacteria in biofilms by flow cytometry. Ann. Pharm. Fr. 61:243-252. [PubMed] [Google Scholar]
- 26.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 27.González Barrios, A. F., R. Zuo, D. Ren, and T. K. Wood. 2006. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93:188-200. [DOI] [PubMed] [Google Scholar]
- 27a.González Barrios, A. F., R. Zuo, Y. Hashimoto, L. Yang, W. E. Bentley, and T. K. Wood. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol 188:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gort, A. S., D. M. Ferber, and J. A. Imlay. 1999. The regulation and role of the periplasmic copper, zinc superoxide dismutase of Escherichia coli. Mol. Microbiol. 32:179-191. [DOI] [PubMed] [Google Scholar]
- 29.Herzberg, M., I. K. Kaye, W. Peti, and T. K. Wood. 2006. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman, L. R., D. A. D'Argenio, M. J. MacCoss, Z. Zhang, R. A. Jones, and S. I. Miller. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171-1175. [DOI] [PubMed] [Google Scholar]
- 31.Hommais, F., E. Krin, J. Y. Coppee, C. Lacroix, E. Yeramian, A. Danchin, and P. Bertin. 2004. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology 150:61-72. [DOI] [PubMed] [Google Scholar]
- 32.Hrimpeng, K., B. Prapagdee, P. Banjerdkij, P. Vattanaviboon, J. M. Dubbs, and S. Mongkolsuk. 2006. Challenging Xanthomonas campestris with low levels of arsenic mediates cross-protection against oxidant killing. FEMS Microbiol. Lett. 262:121-127. [DOI] [PubMed] [Google Scholar]
- 33.Isaacs, H., Jr., D. Chao, C. Yanofsky, and M. H. Saier, Jr. 1994. Mechanism of catabolite repression of tryptophanase synthesis in Escherichia coli. Microbiology 140:2125-2134. [DOI] [PubMed] [Google Scholar]
- 34.Janulczyk, R., S. Ricci, and L. Bjorck. 2003. MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect. Immun. 71:2656-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung, J. U., C. Gutierrez, F. Martin, M. Ardourel, and M. Villarejo. 1990. Transcription of osmB, a gene encoding an Escherichia coli lipoprotein, is regulated by dual signals. Osmotic stress and stationary phase. J. Biol. Chem. 265:10574-10581. [PubMed] [Google Scholar]
- 36.Jung, J. U., C. Gutierrez, and M. R. Villarejo. 1989. Sequence of an osmotically inducible lipoprotein gene. J. Bacteriol. 171:511-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabir, M. S., D. Yamashita, S. Koyama, T. Oshima, K. Kurokawa, M. Maeda, R. Tsunedomi, M. Murata, C. Wada, H. Mori, and M. Yamada. 2005. Cell lysis directed by σE in early stationary phase and effect of induction of the rpoE gene on global gene expression in Escherichia coli. Microbiology 151:2721-2735. [DOI] [PubMed] [Google Scholar]
- 38.Kawamura-Sato, K., K. Shibayama, T. Horii, Y. Iimuma, Y. Arakawa, and M. Ohta. 1999. Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol. Lett. 179:345-352. [DOI] [PubMed] [Google Scholar]
- 39.Keseler, I. M., J. Collado-Vides, S. Gama-Castro, J. Ingraham, S. Paley, I. T. Paulsen, M. Peralta-Gil, and P. D. Karp. 2005. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33:D334-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kikuchi, T., Y. Mizunoe, A. Takade, S. Naito, and S. Yoshida. 2005. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 49:875-884. [DOI] [PubMed] [Google Scholar]
- 41.Klemm, P., L. Hjerrild, M. Gjermansen, and M. A. Schembri. 2004. Structure-function analysis of the self-recognizing Antigen 43 autotransporter protein from Escherichia coli. Mol. Microbiol. 51:283-296. [DOI] [PubMed] [Google Scholar]
- 42.Lapaglia, C., and P. L. Hartzell. 1997. Stress-induced production of biofilm in the hyperthermophile Archaeoglobus fulgidus. Appl. Environ Microbiol. 63:3158-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mai-Prochnow, A., F. Evans, D. Dalisay-Saludes, S. Stelzer, S. Egan, S. James, J. S. Webb, and S. Kjelleberg. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ Microbiol. 70:3232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 45.Maurer, L. M., E. Yohannes, S. S. Bondurant, M. Radmacher, and J. L. Slonczewski. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187:304-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mori, H., K. Isono, T. Horiuchi, and T. Miki. 2000. Functional genomics of Escherichia coli in Japan. Res. Microbiol. 151:121-128. [DOI] [PubMed] [Google Scholar]
- 47.Nakashima, K., K. Horikoshi, and T. Mizuno. 1995. Effect of hydrostatic pressure on the synthesis of outer membrane proteins in Escherichia coli. Biosci. Biotechnol. Biochem. 59:130-132. [DOI] [PubMed] [Google Scholar]
- 48.Newman, M. J., and T. H. Wilson. 1980. Solubilization and reconstitution of the lactose transport system from Escherichia coli. J. Biol. Chem. 255:10583-10586. [PubMed] [Google Scholar]
- 49.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 50.Ogata, Y., T. Mizushima, K. Kataoka, K. Kita, T. Miki, and K. Sekimizu. 1996. DnaK heat shock protein of Escherichia coli maintains the negative supercoiling of DNA against thermal stress. J. Biol. Chem. 271:29407-29414. [DOI] [PubMed] [Google Scholar]
- 51.Otto, K., and M. Hermansson. 2004. Inactivation of ompX causes increased interactions of type 1 fimbriated Escherichia coli with abiotic surfaces. J. Bacteriol. 186:226-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pamp, S. J., D. Frees, S. Engelmann, M. Hecker, and H. Ingmer. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188:4861-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pomposiello, P. J., M. H. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 55.Prüß, B. M., C. Besemann, A. Denton, and A. J. Wolfe. 2006. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J. Bacteriol. 188:3731-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramsing, N. B., M. Kuhl, and B. B. Jorgensen. 1993. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl. Environ Microbiol. 59:3840-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 59.Reisner, A., K. A. Krogfelt, B. M. Klein, E. L. Zechner, and S. Molin. 2006. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J. Bacteriol. 188:3572-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren, D., L. A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515-524. [DOI] [PubMed] [Google Scholar]
- 61.Ren, D., L. A. Bedzyk, R. W. Ye, S. M. Thomas, and T. K. Wood. 2004. Stationary-phase quorum-sensing signals affect autoinducer-2 and gene expression in Escherichia coli. Appl. Environ Microbiol. 70:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rey, S., M. Acab, J. L. Gardy, M. R. Laird, K. deFays, C. Lambert, and F. S. Brinkman. 2005. PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 33:D164-D168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rickard, A. H., S. A. Leach, C. M. Buswell, N. J. High, and P. S. Handley. 2000. Coaggregation between aquatic bacteria is mediated by specific-growth-phase-dependent lectin-saccharide interactions. Appl. Environ Microbiol. 66:431-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosenberg, M., D. Gutnick, and E. Rosenberg. 1980. Adherence of bacteria to hydrocarbons: a simple method for measuring cell surface hydrophobicity. FEMS Microbiol. Lett. 9:29-33. [Google Scholar]
- 66.Rudd, K. E., I. Humphery-Smith, V. C. Wasinger, and A. Bairoch. 1998. Low molecular weight proteins: a challenge for post-genomic research. Electrophoresis 19:536-544. [DOI] [PubMed] [Google Scholar]
- 67.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 68.Schembri, M. A., M. Givskov, and P. Klemm. 2002. An attractive surface: gram-negative bacterial biofilms. Sci. STKE 2002:RE6. [DOI] [PubMed] [Google Scholar]
- 69.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 70.Semchyshyn, H., T. Bagnyukova, and V. Lushchak. 2005. Involvement of soxRS regulon in response of Escherichia coli to oxidative stress induced by hydrogen peroxide. Biochemistry 70:1238-1244. [DOI] [PubMed] [Google Scholar]
- 71.Shah, I. M., and R. E. Wolf, Jr. 2006. Inhibition of Lon-dependent degradation of the Escherichia coli transcription activator SoxS by interaction with ‘soxbox’ DNA or RNA polymerase. Mol. Microbiol. 60:199-208. [DOI] [PubMed] [Google Scholar]
- 72.Silver, S. 1996. Bacterial resistances to toxic metal ions—a review. Gene 179:9-19. [DOI] [PubMed] [Google Scholar]
- 73.Smith, D. K., T. Kassam, B. Singh, and J. F. Elliott. 1992. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J. Bacteriol. 174:5820-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spory, A., A. Bosserhoff, C. von Rhein, W. Goebel, and A. Ludwig. 2002. Differential regulation of multiple proteins of Escherichia coli and Salmonella enterica serovar Typhimurium by the transcriptional regulator SlyA. J. Bacteriol. 184:3549-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stancik, L. M., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sugiyama, K., S. Izawa, and Y. Inoue. 2000. The Yap1p-dependent induction of glutathione synthesis in heat shock response of Saccharomyces cerevisiae. J. Biol. Chem. 275:15535-15540. [DOI] [PubMed] [Google Scholar]
- 77.Teitzel, G. M., and M. R. Parsek. 2003. Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ Microbiol. 69:2313-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, D., X. Ding, and P. N. Rather. 2001. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183:4210-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wen, Z. T., P. Suntharaligham, D. G. Cvitkovitch, and R. A. Burne. 2005. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect. Immun. 73:219-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolfe, A. J., D. E. Chang, J. D. Walker, J. E. Seitz-Partridge, M. D. Vidaurri, C. F. Lange, B. M. Pruss, M. C. Henk, J. C. Larkin, and T. Conway. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol. Microbiol. 48:977-988. [DOI] [PubMed] [Google Scholar]
- 82.Yim, H. H., and M. Villarejo. 1992. osmY, a new hyperosmotically inducible gene, encodes a periplasmic protein in Escherichia coli. J. Bacteriol. 174:3637-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]