Abstract
The phosphopantetheinyl transferases (PPTs) are a superfamily of essential enzymes required for the synthesis of a wide range of compounds, including fatty acids, polyketides, and nonribosomal peptide metabolites. These enzymes activate carrier proteins in specific biosynthetic pathways by transfer of a phosphopantetheinyl moiety. The diverse PPT superfamily can be divided into two families based on specificity and conserved sequence motifs. The first family is typified by the Escherichia coli acyl carrier protein synthase (AcpS), which is involved in fatty acid synthesis. The prototype of the second family is the broad-substrate-range PPT Sfp, which is required for surfactin biosynthesis in Bacillus subtilis. Most cyanobacteria do not encode an AcpS-like PPT, and furthermore, some of their Sfp-like PPTs belong to a unique phylogenetic subgroup defined by the PPTs involved in heterocyst differentiation. Here, we describe the first functional characterization of a cyanobacterial PPT based on a structural analysis and subsequent functional analysis of the Nodularia spumigena NSOR10 PPT. Southern hybridizations suggested that this enzyme may be the only PPT encoded in the N. spumigena NSOR10 genome. Expression and enzyme characterization showed that this PPT was capable of modifying carrier proteins resulting from both heterocyst glycoplipid synthesis and nodularin toxin synthesis. Cyanobacteria are a unique and vast source of bioactive metabolites; therefore, an understanding of cyanobacterial PPTs is important in order to harness the biotechnological potential of cyanobacterial natural products.
Phosphopantetheinyl transferases (PPTs) are enzymes that are required for the activation of carrier proteins in the pathways for synthesis of fatty acids and a wide range of diverse metabolites, including nonribosomal peptides and polyketides (24). The PPT superfamily can be separated into two families of enzymes based on sequence and substrate specificity. The first family includes the acyl carrier protein (ACP) synthase (AcpS)-type PPTs (120 amino acids), which are involved in activating carrier proteins involved in primary metabolism, including carrier proteins involved in fatty acid synthesis (FAS). The majority of microorganisms harbor an AcpS-type PPT, which typically has a limited range of specificity for carrier proteins involved in secondary metabolism. The second group of PPTs is the Sfp-like family (230 amino acids), which exhibit similarity to the Bacillus subtilis PPT Sfp, which is responsible for the activation of carrier proteins in the biosynthetic pathway for surfactin (24, 34). Microorganisms which require activation of carrier proteins involved in secondary metabolism pathways, such as carrier proteins involved in nonribosomal peptide synthetase (NRPS) or polyketide synthase (PKS) pathways, require the activity of an Sfp-like PPT. Some members of the Sfp-like family, which can be further divided into the W/KEA and F/KES subfamilies (11), exhibit a wide range of activity with noncognate carrier proteins. This activity has been harnessed in diverse applications, including the Sfp labeling of carrier proteins (40). In organisms such as Pseudomonas aeruginosa PAO1 (13), Haemophilus influenzae, and Synechocystis sp. strain PCC 6803, an Sfp-like PPT acts in both primary metabolism and secondary metabolism due to the absence of an AcpS-type PPT in their genomes. Biosynthetic gene clusters often have a colocalized Sfp-like PPT, like that present in the gene cluster responsible for the synthesis of the telomerase inhibitor griseorhodin A in Streptomyces sp. strain JP95 (25). However, the majority of cyanobacterial biosynthetic clusters do not have a proximally associated PPT.
Cyanobacteria have been recognized as a useful and vast source of secondary metabolites with important pharmaceutical functions (3). These metabolites include the dolastatin family of antitumor compounds, several derivatives of which are currently in phase I and II trials (26, 27), and the cancer cell toxin curacin A, which is now in preclinical cancer trials (9). However, the production of cyanobacterial compounds in sustainable, easily manipulated heterologous hosts remains elusive. The requirement for PPTs to activate carrier proteins in biosynthetic pathways is critical for successful host selection.
Despite the plethora of screening approaches and promising natural product leads for cyanobacteria, few synthesis pathways have been characterized, and the majority of the pathways that have been characterized are involved in toxin production. These pathways include microcystin pathways in Microcystis aeruginosa PCC 7806 (41), Anabaena sp. strain 90 (38), and Planktothrix agardhii CYA 126 (10), a nostocyclopeptide pathway (20) identified in Nostoc sp. strain GSV 224, and a nostopeptolide pathway identified in Nostoc sp. strain ATCC 53789 (1).
Bioinformatic analyses have revealed the absence of AcpS enzymes in almost all cyanobacterial genomes available for analysis (11). Therefore, it is likely that Sfp-like PPTs, which are typically responsible for the activation of secondary metabolite carrier proteins, are also involved in FAS. A recent phylogenetic analysis placed all cyanobacterial PPTs in the diverse W/KEA subfamily. In this subfamily a specific, heterocyst type of PPT is associated with synthesis of the heterocyst glycolipids (11). Heterocysts are specialized nitrogen fixation cells (29) with glycolipid membranes that are made up of complex lipid and carbohydrate residues. The glycolipid membrane prevents the entry of oxygen into the cell, which would otherwise inhibit the nitrogenase enzyme. Several heterocyst genetic loci have been identified, one of which is the hetMNI locus associated with heterocyst glycolipid synthesis (7). The hetMNI locus can be examined in the published genome of Nostoc punctiforme ATCC 29133 and the publicly available Nostoc sp. strain PCC 7120, Nodularia spumigena CCY 9414, and Anabaena variabilis ATCC 29413 genomes. A PPT (HetI) gene is in this locus, which also encodes an aryl carrier protein (ArCP) in an iterative polyketide synthase (HetM) (7) and an oxidoreductase (HetN) (6). N. spumigena NSOR10 is also able to form heterocysts; however the hetMNI locus in this species has not been characterized previously.
Algal blooms of N. spumigena occur mainly in the estuaries and coastal waters of southern Australia (18) but have also been reported in the Baltic Sea (38), New Zealand (8), and North America (28). Many blooms are highly toxic due to the nonribosomal production of the protein phosphatase-inhibiting hepatotoxin nodularin. The biosynthesis of nodularin has been characterized, and the gene cluster has been sequenced (30). The large hybrid NRPS/PKS biosynthetic gene cluster encodes four ACPs and five peptidyl carrier proteins (PCPs). The carrier proteins require a PPT for activity, and the gene encoding this PPT is not located in or adjacent to the biosynthetic cluster. A 220-bp fragment of a PPT gene from N. spumigena NSOR10 was recently identified by degenerate PCR-based screening (11). This PPT was designated PPTNs (N. spumigena phosphopantetheinyl transferase) in this study.
Due to the association of PPTNs with the heterocyst cluster, it was hypothesized that this PPT phosphopantetheinylates the HetM ArCP. Furthermore, due to the general absence of genes encoding multiple PPTs in cyanobacterial genomes, it was likely that this PPT also participates in the biosynthesis of the hepatotoxin nodularin. In this study, our aims were to express and characterize the cyanobacterial PPT PPTNs of N. spumigena NSOR10 and to determine this enzyme's potential role in both glycolipid synthesis and nodularin synthesis.
MATERIALS AND METHODS
Media and culturing.
N. spumigena NSOR10 was cultured at room temperature with a cycle consisting of 12 h of light and 12 h of darkness in ASM media supplemented with 1.5% NaCl (33). DNA was extracted as previously described (4).
DNA amplification, sequencing, and analysis.
PCR and sequencing reactions were carried out as previously described (11). Panhandle-based gene walking with adaptor-mediated and specific primers (Table 1) (30) was utilized to amplify the unknown genomic regions flanking the N. spumigena PPT fragment. The outputs from BLAST (Basic Local Alignment Search Tool), Pileup from GCG, and the multiple-sequence alignment tool in CLUSTAL X (12) were utilized for analysis and alignment of sequences. Automated sequencing was performed using the PRISM Big Dye cycle sequencing system and a model 373 sequencer (Applied Biosystems) at the Automated DNA Analysis Facility, University of New South Wales. Sequence analysis was performed using the Applied Biosystems Autoassembler software.
TABLE 1.
Primers used to amplify the unknown genomic region surrounding the PPTNs gene
| Primer | Sequence (5′-3′) |
|---|---|
| Panhandle Roko1 | GTGGTAAACCAGTATTAGCGG |
| Panhandle Roko2 | GTCTAGTCCACAAGGCTTTAAG |
| Panhandle Roko3 | CCATTGTTATTCCTGGTC |
| Panhandle Roko4 | CATTAAGGCCGCATAACCGAT |
| Panhandle Roko5 | GCATCGTGAATCACAGTGCC |
| Panhandle Roko6 | CAATAGTAGTGGCGTGGCAA |
| Panhandle Roko7 | CTAAATGGCCAGCATAAGCA |
| Panhandle Roko8 | GCTTATGCTGGCCATTTAG |
| Panhandle Roko9 | GATGAATAGCCGGGTCAAG |
| Panhandle Roko10 | GCGTGATGCTGTAAATGC |
| Panhandle Roko11 | CTTGACCCGGCTATTCATC |
| Panhandle Thorne1 | GTGATAATTCCCGGCACACAGAGC |
| Panhandle Thorne 2 | CCGTTATTGCACTTGTAAAGAGG |
| T7P Adapter | CCCCTATCCACCCTTACACCTATC |
| T7Pr2 Adapter | CCTTACACCTATCCCTCCGTAATAC |
Southern hybridization.
Pure genomic DNA samples (∼10 μg) were digested overnight with XbaI or XmnI used according to the manufacturer's recommendations (Promega, Australia). Digests and positive controls (0.5 to 1.0 ng of linearized plasmid) were separated by electrophoresis on 0.8% agarose gels at 60 mV for approximately 2.5 h and vacuum blotted onto a nylon membrane (Amersham). A DIG-High Prime DNA labeling kit (Roche, Australia) was utilized for Southern hybridization. Specific PPT probes were created by amplification with specific primers. Primers slrup (5′-TGTTTAAACTCACCTGTG-3′) and slrdn (5′-CCCAAGGTAACGAAACGA-3′) were used to amplify slr0495 from Synechocystis sp. strain PCC 6803. Primers npunf (5′-GGATCCGCGATCGCCAGTCTGAGTTC-3′) and npunr (5′-GAGCTCTTTGTGTAGTAGCGAATTATC-3′) were utilized to amplify a PPT from N. punctiforme ATCC 29133. Primers npptf (5′-CATGAAAGATATCACGGCGCTT-3′) and npptr (5′-GAAGATAACAAGCTTGTATTGCC-3′) were used to amplify the full-length PPTNs open reading frame. Probes were labeled by PCR with digoxigenin, and their efficiency was tested by using concentrations ranging from 100 fg to 10 ng. Hybridization was performed overnight at 40°C, stringency washes were performed with 0.5% SSC-0.1% sodium dodecyl sulfate at 65°C (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and signals were analyzed by chemiluminescent detection with disodium 3-(4-methoxy-spiro{1,2-dioxetane-3,2′-(5′-chloro) tricyclo[3.3.1.13,7]decan}-4-yl) phenyl phosphate (CPSD) substrate by using an LAS-3000 (FUJIFILM).
Creation of expression plasmids.
The N. spumigena NSOR10 PPTNs gene (accession no. AY836561) was amplified with primers npptf and npptr. The 720-bp PCR-amplified product was cloned into pET30 (Novagen) to obtain the expression plasmid pPPTNs. The N. punctiforme ATCC 29133 hetM gene was amplified from the heterocyst hetMNI locus encoding the ArCP/ketoreductase HetM (accession no. ZP_00107100). Primers hetmf (5′-GCCATGGCTATAAAACAGTCTTTC-3′) and hetmr (5′-GGGATCCGAGATTCAAGAAACC-3′) were used to amplify a 1.7-kb fragment, which was cloned into pET30 to create pHetM. The N. punctiforme ATCC 29133 pArCP vector for expression of N. punctiforme ArCP (ArCPNp) was constructed in a similar manner utilizing primers hetmf and arcpr (5′-TAGCTCGAGAACCATCTTGCAC-3′) to amplify and clone the 260-bp ArCP domain of HetM and create pArCP.
The N. spumigena NSOR10 PCP and ACP (designated PCPNs and ACPNs) sequences were amplified from the hybrid NRPS/PKS ndaC gene (accession no. AAO64404) of the ndaS gene cluster responsible for production of the hepatotoxin nodularin (30). Primers nspcpf (5′-CTCGAGCAGCCTCTACAACTGCA-3′) and nspcpr (5′-GGATCCGCCAGGAGAACGGCGG-3′) and primers nsacpf (5′-GGAGCTCTTTTCCAAACATTCT-3′) and nsacpr (5′-GGGATCCTCTAAGCATTCCATCAGTC-3′) were utilized. The resulting fragments were manipulated as described above to obtain pNsACP and pNsPCP, respectively.
The M. aeruginosa PCP (MPCP) sequence was amplified from the hybrid NRPS/PKS mcyG gene (accession no. AAX73195) of the mcyS gene cluster responsible for production of the hepatotoxin microcystin (41). PCP primers mpcpf (5′-GGATCCTGAACAGGGA-3′) and mpcpr (5′-CTCGAGATGGCGACGGCTCC-3′) were used to construct the expression vector pMPCP, as described above.
The gene encoding the PKS N. punctiforme ACP (designated ACPNp) was amplified from the putative nostopeptolide NosB gene cluster of N. punctiforme ATCC 29133. This species has been shown to produce nostopeptolide (21), and the gene cluster is similar to the characterized nostopeptolide gene cluster in Nostoc sp. strain GSV224 (20). NosB (accession no. ZP_00110898) is a modular type 1 PKS encoded by a gene in this cluster. The gene encoding C-terminal NosB ACP was amplified utilizing primers npacpbf (5′-GGATCCTAAAATCTAGGCTAG-3′) npacpsr (5′-GAGCTCAAATTGTTATTTCTT-3′) and was cloned as described above to obtain pNpACP.
Protein expression and purification and enzyme activity analysis.
Plasmids pNsPPT, pArCP, and pMPCP were utilized for expression by incubating them at 30°C for 4 h with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for induction. ACPNp was expressed at 22 to 24°C with 0.1 mM IPTG for 6 h. HetM was expressed at 18°C overnight with 0.1 mM IPTG. ArCPNp was expressed at 37°C for 2 h with 1 mM IPTG. After expression, cells were pelleted by centrifugation at 4,000 × g and frozen at −80°C overnight. Pellets were thawed on ice, resuspended in 5 ml of 50 mM HEPES (Sigma)-150 mM NaCl (pH 7.4), and either subjected to three passages through a cooled French pressure cell at 1,000 lb/in2 or sonicated at 4°C and 30% amplitude for 25 s with 0.5-s pulses. The soluble fraction was collected after centrifugation at 20,000 × g for 30 min at 4°C. A HiTrap chelating column (Amersham) was utilized for purification of the recombinant proteins. Fractions containing the desired protein, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, were pooled, desalted, and snap frozen in 50 mM HEPES-150 mM NaCl-8% glycerol (pH 7.4) for storage at −80°C. Protein concentrations were determined using the calculated extinction coefficient of each protein, as follows; PPTNs, 37,650 cm−1 M−1; HetM, 67,430 cm−1 M−1; ArCPNp, 8,250 cm−1 M−1; MPCP, 6,970 cm−1 M−1; and ACPNp, 13,940 cm−1 M−1.
PPT assays were carried out as previously described (13). In brief, 100- to 400-μl reaction mixtures containing 5 mM Tris-HCl (pH 7.4), 12.5 mM MgCl2, 0.5 mM coenzyme (CoA), 2 μM dithiothreitol, carrier proteins at concentrations of 10 to 100 μM, and 300 nM (final concentration) PPT were incubated at 37°C for 30 min. The reactions were terminated by addition of 1 ml of 10% trichloroacetic acid. Assay mixtures were precipitated overnight at −20°C before centrifugation at 4°C for 15 min at 16,000 × g. Protein pellets were analyzed by electrospray ionization mass spectrometry. Spectra were acquired using an API QStar Pulsar i hybrid tandem mass spectrometer (Applied Biosystems). Samples (∼200 to 400 fmol) were dissolved in water-acetonitrile-formic acid (50:49:1) and loaded (1 μl) into nanospray needles (Proxeon), and the tip was positioned ∼10 mm from the orifice. Nitrogen was used as the curtain gas, and a 900-V potential was applied to the needle. A time of flight mass spectrometry scan was acquired (m/z 550 to 2000, 1 s) and accumulated for ∼1 min in a single file. Spectra were deconvoluted using the Bayesian reconstruction method contained in the Analyst QS software.
RESULTS
Sequencing and analysis of the hetMNI locus from N. spumigena NSOR10.
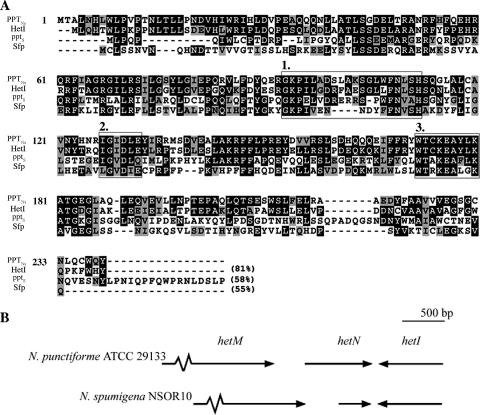
Flanking regions of the partial PPT gene fragment were amplified to allow sequencing of 3,450 bp of the N. spumigena NSOR10 hetMNI locus (accession no. AY836561). The sequence analysis revealed the 240-residue PPT PPTNs, a 27,555-Da protein with an isoelectric point of 6.1. PPTNs exhibited similarity to HetI from Nostoc sp. strain PCC 7120 (81%) and to Sfp from B. subtilis (55%). PPTNs is encoded in reverse orientation compared to the hetM and hetN genes, as observed in the hetMNI locus of the heterocyst-forming cyanobacteria N. punctiforme ATCC 29133 (Fig. 1), Nostoc sp. strain PCC 7120, and A. variabilis ATCC 29413.
FIG. 1.
(A) Alignment of PPT representatives. PPTNs from N. spumigena NSOR10 was aligned with HetI from Nostoc sp. strain PCC 7120 (accession no. P37695), pptS from Synechocystis sp. strain PCC 6803 (accession no. BAA10326), and Sfp from B. subtilis (accession no. P39135). PPTNs numbering is shown, and the levels of similarity to PPTNs are indicated in parentheses. Black shading indicates amino acid identity, and gray shading shows amino acid similarity. PPT motifs 1, 2, and 3 are enclosed in boxes and numbered. (B) Comparison of the hetMNI loci of N. punctiforme ATCC 29133 and N. spumigena NSOR10. A partial segment of the 1,520-bp hetM sequence is indicated by a broken arrow. The arrows indicate the direction of gene transcription.
The predicted protein products of the genes downstream of the PPTNs gene in N. spumigena NSOR10 were analyzed and compared to homologous proteins. The protein product of hetN is 126 amino acids long and exhibits 83% similarity to the C-terminal half of the corresponding protein in A. variabilis ATCC 29413. The partial hetM gene, encoding an iterative PKS in N. spumigena, exhibited 90% similarity to the gene encoding a homologous protein in N. punctiforme ATCC 29133, hetM (also designated hglB). No open reading frames were detected in the sequenced region extending 900 bp upstream from the PPTNs gene.
Southern hybridization.
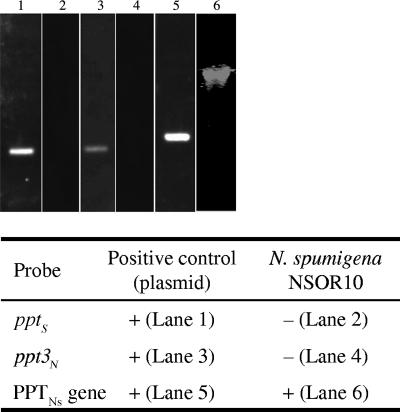
Southern analysis was utilized to ascertain the number of PPTs encoded in the N. spumigena NS0R10 genome. Probes were constructed from the genes encoding a diverse range of cyanobacterial PPTs, including the PPTNs gene, pptS in Synechocystis sp. strain PCC 6803 (accession no. BAA10326), and the gene encoding a PPT in N. punctiforme ATCC 29133 (accession no. ZP_00110892) designated ppt3N. These PPTs are produced by organisms belonging to distinct cyanobacterial phylogenetic clades (11). Hybridizations performed with the PPTNs gene probe revealed a single band when N. spumigena NSOR10 was used (Fig. 2), and no hybridization was detected with the pptS- or ppt3N-specific probes. Taken together, these results suggest that PPTNs may be the only genomically encoded PPT in N. spumigena NSOR10. However, the single band detected by Southern analysis did not exclude the slight possibility that there were two PPTs encoded by the single digested genomic fragment. The possibility that there was a more divergent PPT that was not detected by the specific Southern hybridization probes also could not be excluded.
FIG. 2.
Southern hybridization of PPT gene probes to digested N. spumigena NSOR10 genomic DNA. A plus sign indicates that a band was detected by chemiluminescence, and a minus sign indicates that no bands were visible. For positive controls we utilized linearized plasmid DNA. The lane numbers in parentheses indicated the lanes in the gel. pptS, PPT gene (accession no. BAA10326) from Synechocystis sp. strain PCC 6803; ppt3N, gene (accession no. ZP_00110892) encoding a PPT in N. punctiforme ATCC 29133; PPTNs, gene encoding a PPT in N. spumigena NSOR10.
Expression and purification of recombinant proteins.
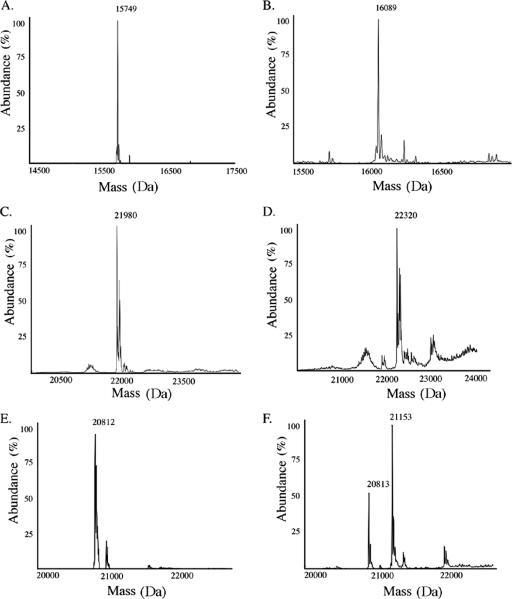
In order to confirm the PPT activity of PPTNs, the enzyme was expressed as a 32.7-kDa, His-tagged, soluble protein. Attempts to produce ACPNs and PCPNs from N. spumigena NSOR10 were not successful. Our attempts to resolve this problem included varying the expression time (2 to 24 h), temperature (18 to 37°C), IPTG induction concentration (0.1 to 1 mM), growth medium (Luria broth and tryptone phosphate), and helper plasmids (pRARE and pLysE; Novagen). As the partial hetM gene in N. spumigena NSOR10 does not encode an ArCP domain, the protein in N. punctiforme ATCC 29133 (designated ArCPNp) that exhibited 90% similarity, encoded by hetM, was selected for expression and in vitro activation analysis. Although soluble HetM expression was observed, analysis of HetM by nanospray ion trap mass spectrometry could not be performed due to low yields. The expression plasmid encoding only the ArCP domain of HetM was therefore utilized, and the 15.6-kDa ArCPNp protein was subsequently expressed with suitable yields. MPCP and ACPNp were expressed as 22.0- and 20.8-kDa proteins, respectively. No phosphopantetheinylation of the cyanobacterial carrier proteins was seen after expression in E. coli (Fig. 3 and Table 2).
FIG. 3.
Mass spectrometry of ArCP, PCP, and ACP phosphopantetheinylation. (A) Mass spectrum of ArCPNp. (B) Mass spectrum of phosphopantetheinylated ArCPNp after incubation with PPTNs. (C) Mass spectrum of MPCP. (D) Mass spectrum of MPCP phosphopantetheinylated after incubation with PPTNs. (E) Mass spectrum of ACPNp. (F) Mass spectrum of phosphopantetheinylated ACPNp after incubation with PPTNs.
TABLE 2.
Phosphopantetheinylation of carrier proteins by PPTNs, detected by addition of a phosphopantetheinyl moiety (340 Da), using mass spectrometry
| Carrier protein | PPT | Expected molecular mass (Da) | Observed molecular mass (Da) | Observed molecular mass of pantetheinylated carrier protein (Da) | % Holo-carrier proteina |
|---|---|---|---|---|---|
| ArCPNp (glycolipid biosynthesis) | PPTNs | 15,751 | 15,749 | 16,089 | 100 |
| MPCP (microcystin biosynthesis, NRPS) | PPTNs | 21,978 | 21,980 | 22,320 | 100 |
| ACPNp (nostopeptolide biosynthesis, PKS) | PPTNs | 20,812 | 20,812 | 21,153 | 69 |
The percentage of holo-carrier protein was estimated by comparing holo-carrier protein abundance and apo-carrier protein abundance in mass spectra as previously described (31).
PPTNs activity.
Activity was detected by ionization mass spectrometry by addition of 340 Da. This mass was related to the incorporation of the phosphopantetheinyl arm of CoA transferred by the PPT to the carrier protein (Fig. 3). Mass spectra of phosphopantetheinylated carrier proteins were analyzed alongside controls (with no PPT) to verify phosphopantetheinylation by PPTNs.
PPTNs phosphopantetheinyl activity was confirmed by employing the HetM ArCP encoded by the hetMNI gene cluster in N. punctiforme ATCC 29133 (ArCPNp). The conversion of apo-ArCPNp (15.75 kDa) to holo-ArCPNp (16.09 kDa) (Fig. 3) revealed that PPTNs can phosphopantetheinylate the HetM carrier protein which is involved in heterocyst glycolipid synthesis (Table 2).
PPTNs activity in PKS and NRPS secondary metabolism (ACPs and PCPs) was subsequently examined utilizing ACPNp and MPCP (Table 2). Mass spectrometry analyses revealed addition of 340 Da of the phosphopantetheinyl moiety from CoA to each of these carrier proteins. Control reactions were performed without PPTNs (Fig. 3).
DISCUSSION
This study included the first enzymatic analysis of a cyanobacterial PPT. After incubation of PPTNs with apo-ArCPNp, complete conversion to holo-ArCPNp was observed. No phosphopantetheinylation of the HetM ArCPNp was seen after heterologous expression in E. coli. Therefore, the PPTs of E. coli were not capable of phosphopantetheinylating the HetM ArCP from N. punctiforme ATCC 29133. The specificity of E. coli PPTs has been reported previously, and negligible phosphopantetheinylation of the P. aeruginosa PAO1 ArCP has been observed after heterologous expression in E. coli (13).
Although not conclusive, the Southern hybridization results suggest that only one PPT may be encoded in the N. spumigena NSOR10 genome. This suggestion is supported by the presence of only one PPT encoded in the recently available N. spumigena CCY 9414 genome. However, further phosphopatentheinylation assays are necessary to determine whether PPTNs is able to activate the ACP involved in FAS. The possibility that PPTNs is the only PPT encoded in the genome of N. spumigena NSOR10 led to the hypothesis that PPTNs could also phosphopantetheinylate the PKS/NRPS carrier proteins, such as those in the hybrid PKS/NRPS multienzyme responsible for nodularin production in N. spumigena NSOR10 (30). However, the N. spumigena NSOR10 nodularin carrier proteins could not be expressed. Similar results for problematic carrier protein expression have been reported previously and have included the failure to express a P. aeruginosa PAO1 NRPS PCP (13) or the Streptomyces glaucescens NRPS PCP in E. coli (39). Successful expression of a PKS ACP from the N. punctiforme ATCC 29133 nostopeptolide A (nosB) biosynthetic gene cluster and an M. aeruginosa PCC 7806 NRPS PCP from the microcystin (mcyG) biosynthetic gene cluster allowed the analysis of PPTNs phosphopantetheinylation in secondary metabolism. Phosphopantetheinylation of both carrier proteins was observed in a 30-min assay, suggesting that PPTNs could also act as an efficient cofactor for the synthesis of NRPSs and PKSs in N. spumigena NSOR10.
Although the NRPS PCP used here is produced by a unicellular cyanobacterial species, M. aeruginosa PCC 7806, phosphopantetheinylation by a PPT from a filamentous, heterocyst-forming cyanobacterial species was not surprising. This PCP was from a domain in McyG, which is encoded by the M. aeruginosa PCC 7806 microcystin biosynthetic gene cluster. This 300-kDa protein is 70% identical to the corresponding NdaC protein encoded by the nodularin biosynthetic gene cluster in N. spumigena NSOR10. A common evolutionary pathway has been proposed for these two hepatotoxins, with transposition and NRPS module deletion thought to be responsible for the variation in the structures of the two cyclic peptides (30, 35). McyG is putatively involved in the activation of phenylpropanoids, which is the first step in the biosynthesis of the highly modified amino acid Adda (19). The unusual and complex biosynthesis of this amino acid, which occurs only in cyanobacteria, involves a hybrid NRPS/PKS.
ACPNp was also phosphopantetheinylated, indicating that PPTNs can activate carrier proteins from biosynthetic pathways that are not present in N. spumigena NSOR10, such as those involved in nostopeptolide biosynthesis. The incomplete conversion (approximately 65% conversion to holo-ACPNp) indicated that there was reduced specificity for noncognate carrier proteins. The terminal 100 amino acids (incorporating the ACP) of the putative NosB protein from N. punctiforme ATCC 29133 exhibit 60% similarity to the corresponding amino acids of the NdaC PKS ACP.
A similar scenario of a singular PPT participating in multiple metabolic pathways is typified by PcpS in P. aeruginosa PAO1 (13). This PPT was catalytically characterized to show that it is most efficient with the ACPs of FAS. It was suggested that a small reduction in the efficiency of this PPT would lead to the failure of PcpS to phosphopantetheinylate carrier proteins involved in secondary metabolism. In cyanobacteria, where the majority of species harbor a single PPT, genetic mutations could potentially prevent toxin production by inhibiting the ability of the PPT to activate carrier proteins in secondary metabolism biosynthetic pathways.
Cyanobacterial species associated with slow growth and complicated or unattainable culture requirements often include strains that harbor the gene clusters for the synthesis of pharmaceutically interesting compounds (14, 17, 37). Molecular methods may yield the genetic information (5, 30), but they have not allowed isolation and production of novel compounds without recombinant expression (2). The PPT Sfp has been used to overcome problems associated with carrier protein modification in E. coli heterologous hosts (23, 32); however, modification is often incomplete (15). Alternative expression hosts, including Pseudomonas sp. (16). and Streptomyces spp. (22, 36), are being investigated for heterologous expression and biochemical characterization of secondary metabolite gene clusters from cyanobacteria.
PPTNs is capable of phosphopantetheinylating a broad range of carrier protein substrates, which, based on the evidence presented here, may be a common characteristic of all cyanobacterial PPTs. If so, the occurrence of multiple PPTs in heterocyst-forming species, such as the three PPTs of N. punctiforme ATCC 29133, would be superfluous to the phosphopantetheinylation needs of the organism. An alternative scenario is that this broad range of activity is present only in cyanobacterial species with multiple phosphopantetheinylation pathways (FAS, NRPS, and PKS) that harbor a single Sfp-like PPT. If this is true, the multiple PPTs encoded by the genome of N. punctiforme ATCC 29133 would be expected to show dedication to a single phosphopantetheinylation pathway.
The ability of PPTNs to activate noncognate secondary metabolite carrier proteins has great potential for application in biotechnology. Phosphopantetheinylation of noncognate carrier proteins from alternative pathways, such as those involved in microcystin or nostopeptolide biosynthesis, has demonstrated the potential of using this PPT to harness the cyanobacterial biosynthetic pathways in heterologous hosts.
Acknowledgments
The Australian Research Council and Diagnostic Technology P/L financially supported this work. J.N.C. was also supported by an Adrian Lee Travel Scholarship, University of New South Wales.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Becker, J. E., R. E. Moore, and B. S. Moore. 2004. Cloning, sequencing, and biochemical characterization of the nostocyclopeptide biosynthetic gene cluster: molecular basis for imine macrocyclization. Gene 325:35-42. [DOI] [PubMed] [Google Scholar]
- 2.Brady, S. F., C. J. Chao, J. Handelsman, and J. Clardy. 2001. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 3:1981-1984. [DOI] [PubMed] [Google Scholar]
- 3.Burja, A. M., B. Banaigs, E. Abou-Mansour, J. G. Burgess, and P. C. Wright. 2001. Marine cyanobacteria—a prolific source of natural products. Tetrahedron 57:9347-9377. [Google Scholar]
- 4.Burns, B. P., M. L. Saker, M. C. Moffitt, and B. A. Neilan. 2004. Molecular detection of genes responsible for cyanobacterial toxin production in the genera Microcystis, Nodularia and Cylindrospermopsis. Methods Mol. Biol. 268:213-222. [DOI] [PubMed] [Google Scholar]
- 5.Burns, B. P., A. Seifert, F. Goh, A. D. Jungblut, A. Serhat, and B. A. Neilan. 2005. Genetic potential for secondary metabolite production in stromatolite communities. FEMS Microbiol. Lett. 243:293-301. [DOI] [PubMed] [Google Scholar]
- 6.Callahan, S. M., and W. J. Buikema. 2001. The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol. Microbiol. 40:941-950. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, E. L., M. F. Cohen, and J. C. Meeks. 1997. A polyketide-synthase-like gene is involved in the synthesis of heterocyst glycolipids in Nostoc punctiforme strain ATCC 29133. Arch. Microbiol. V. 167:251-258. [DOI] [PubMed] [Google Scholar]
- 8.Carmichael, W. W., J. T. Eschedor, G. M. Patterson, and R. E. Moore. 1988. Toxicity and partial structure of a hepatotoxic peptide produced by the cyanobacterium Nodularia spumigena Mertens emend. L575 from New Zealand. Appl. Environ. Microbiol. 54:2257-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Z., P. Flatt, W. H. Gerwick, V.-A. Nguyen, C. L. Willis, and D. H. Sherman. 2002. The barbamide biosynthetic gene cluster: a novel marine cyanobacterial system of mixed polyketide synthase (PKS)-non-ribosomal peptide synthetase (NRPS) origin involving an unusual trichloroleucyl starter unit. Gene 296:235-247. [DOI] [PubMed] [Google Scholar]
- 10.Christiansen, G., J. Fastner, M. Erhard, T. Borner, and E. Dittmann. 2003. Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J. Bacteriol. 185:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Copp, J. N., and B. A. Neilan. 2006. The phosphopantetheinyl transferase superfamily: phylogenetic analysis and functional implications in cyanobacteria. Appl. Environ. Microbiol. 72:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 13.Finking, R., J. Solsbacher, D. Konz, M. Schobert, A. Schafer, D. Jahn, and M. A. Marahiel. 2002. Characterization of a new type of phosphopantetheinyl transferase for fatty acid and siderophore synthesis in Pseudomonas aeruginosa. J. Biol. Chem. 277:50293-50302. [DOI] [PubMed] [Google Scholar]
- 14.Fortman, J. L., and D. H. Sherman. 2005. Utilizing the power of microbial genetics to bridge the gap between the promise and the application of marine natural products. Chembiochem 6:960-978. [DOI] [PubMed] [Google Scholar]
- 15.Gokhale, R. S., S. Y. Tsuji, D. E. Cane, and C. Khosla. 1999. Dissecting and exploiting intermodular communication in polyketide synthases. Science 284:482-485. [DOI] [PubMed] [Google Scholar]
- 16.Gross, F., D. Gottschalk, and R. Müller. 2005. Posttranslational modification of myxobacterial carrier protein domains in Pseudomonas sp. by an intrinsic phosphopantetheinyl transferase. Appl. Microbiol. Biotechnol. 68:66-74. [DOI] [PubMed] [Google Scholar]
- 17.Handelsman, J., M. R. Rondon, S. F. Brady, J. Clardy, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245-R249. [DOI] [PubMed] [Google Scholar]
- 18.Heresztyn, T., and B. C. Nicholson. 1997. Nodularin concentrations in Lakes Alexandrina and Albert, South Australia, during a bloom of the cyanobacterium (blue-green alga) Nodularia spumigena and degradation of the toxin. Environ. Toxicol. Water 12:273-282. [Google Scholar]
- 19.Hicks, L. M., M. C. Moffitt, L. L. Beer, B. S. Moore, and N. L. Kelleher. 2006. Structural characterization of in vitro and in vivo intermediates on the loading module of microcystin synthetase. ACS Chem. Biol. 1:93-102. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann, D., J. M. Hevel, R. E. Moore, and B. S. Moore. 2003. Sequence analysis and biochemical characterization of the nostopeptolide A biosynthetic gene cluster from Nostoc sp. GSV224. Gene 311:171-180. [DOI] [PubMed] [Google Scholar]
- 21.Hunsucker, S. W., K. Klage, S. M. Slaughter, M. Potts, and R. F. Helm. 2004. A preliminary investigation of the Nostoc punctiforme proteome. Biochem. Biophys. Res. Commun. 317:1121-1127. [DOI] [PubMed] [Google Scholar]
- 22.Kalaitzis, J. A., and B. S. Moore. 2004. Heterologous biosynthesis of truncated hexaketides derived from the actinorhodin polyketide synthase. J. Nat. Prod. 67:1419-1422. [DOI] [PubMed] [Google Scholar]
- 23.Kealey, J. T., L. Liu, D. V. Santi, M. C. Betlach, and P. J. Barr. 1998. Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc. Natl. Acad. Sci. USA 95:505-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 25.Li, A., and J. Piel. 2002. A gene cluster from a marine Streptomyces encoding the biosynthesis of the aromatic spiroketal polyketide griseorhodin A. Chem. Biol. 9:1017-1026. [DOI] [PubMed] [Google Scholar]
- 26.Luesch, H., G. G. Harrigan, G. Goetz, and F. D. Horgen. 2002. The cyanobacterial origin of potent anticancer agents originally isolated from sea hares. Curr. Med. Chem. 9:1791-1806. [DOI] [PubMed] [Google Scholar]
- 27.Luesch, H., R. E. Moore, V. J. Paul, S. L. Mooberry, and T. H. Corbett. 2001. Isolation of dolastatin 10 from the marine cyanobacterium Symploca species VP642 and total stereochemistry and biological evaluation of its analogue symplostatin 1. J. Nat. Prod. 64:907-910. [DOI] [PubMed] [Google Scholar]
- 28.MacGregor, B. J., B. Van Mooy, B. J. Baker, M. Mellon, P. H. Moisander, H. W. Paerl, J. Zehr, D. Hollander, and D. A. Stahl. 2001. Microbiological, molecular biological and stable isotopic evidence for nitrogen fixation in the open waters of Lake Michigan. Environ. Microbiol. 3:205-219. [DOI] [PubMed] [Google Scholar]
- 29.Meeks, J. C., E. L. Campbell, M. L. Summers, and F. C. Wong. 2002. Cellular differentiation in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. V178:395-403. [DOI] [PubMed] [Google Scholar]
- 30.Moffitt, M. C., and B. A. Neilan. 2004. Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Appl. Environ. Microbiol. 70:6353-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mofid, M. R., R. Finking, and M. A. Marahiel. 2002. Recognition of hybrid peptidyl carrier proteins/acyl carrier proteins in nonribosomal peptide synthetase modules by the 4′-phophopantetheinyl transferases AcpS and Sfp. J. Biol. Chem. 277:17023-17031. [DOI] [PubMed] [Google Scholar]
- 32.Mootz, H. D., K. Schorgendorfer, and M. A. Marahiel. 2002. Functional characterization of 4′-phosphopantetheinyl transferase genes of bacterial and fungal origin by complementation of Saccharomyces cerevisiae lys5. FEMS Microbiol. Lett. 213:51-57. [DOI] [PubMed] [Google Scholar]
- 33.Provasoli, L., J. J. McLaughlin, and M. R. Droop. 1957. The development of artificial media for marine algae. Arch. Mikrobiol. 25:392-428. [DOI] [PubMed] [Google Scholar]
- 34.Quadri, L. E. N., P. H. Weinreb, M. Lei, M. M. Nakano, P. Zuber, and C. T. Walsh. 1998. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37:1585-1595. [DOI] [PubMed] [Google Scholar]
- 35.Rantala, A., D. P. Fewer, M. Hisbergues, L. Rouhiainen, J. Vaitomaa, T. Borner, and K. Sivonen. 2004. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. USA 101:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez, E., S. Ward, H. Fu, W. P. Revill, R. McDaniel, and L. Katz. 2004. Engineered biosynthesis of 16-membered macrolides that require methoxymalonyl-ACP precursors in Streptomyces fradiae. Appl. Microbiol. Biotechnol. 66:85-91. [DOI] [PubMed] [Google Scholar]
- 37.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouhiainen, L., T. Vakkilainen, B. L. Siemer, W. Buikema, R. Haselkorn, and K. Sivonen. 2004. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Appl. Environ. Microbiol. 70:686-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez, C., L. Du, D. J. Edwards, M. D. Toney, and B. Shen. 2001. Cloning and characterization of a phosphopantetheinyl transferase from Streptomyces verticillus ATCC15003, the producer of the hybrid peptide-polyketide antitumor drug bleomycin. Chem. Biol. 8:725-738. [DOI] [PubMed] [Google Scholar]
- 40.Sieber, S. A., C. T. Walsh, and M. A. Marahiel. 2003. Loading peptidyl-coenzyme A onto peptidyl carrier proteins: a novel approach in characterizing macrocyclization by thioesterase domains. J. Am. Chem. Soc. 125:10862-10866. [DOI] [PubMed] [Google Scholar]
- 41.Tillett, D., E. Dittmann, M. Erhard, H. von Dohren, T. Borner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]