Abstract
FtsE and FtsX, which are widely conserved homologs of ABC transporters and interact with each other, have important but unknown functions in bacterial cell division. Coimmunoprecipitation of Escherichia coli cell extracts revealed that a functional FLAG-tagged version of FtsE, the putative ATP-binding component, interacts with FtsZ, the bacterial tubulin homolog required to assemble the cytokinetic Z ring and recruit the components of the divisome. This interaction is independent of FtsX, the predicted membrane component of the ABC transporter, which has been shown previously to interact with FtsE. The interaction also occurred independently of FtsA or ZipA, two other E. coli cell division proteins that interact with FtsZ. In addition, FtsZ copurified with FLAG-FtsE. Surprisingly, the conserved C-terminal tail of FtsZ, which interacts with other cell division proteins, such as FtsA and ZipA, was dispensable for interaction with FtsE. In support of a direct interaction with FtsZ, targeting of a green fluorescent protein (GFP)-FtsE fusion to Z rings required FtsZ, but not FtsA. Although GFP-FtsE failed to target Z rings in the absence of ZipA, its localization was restored in the presence of the ftsA* bypass suppressor, indicating that the requirement for ZipA is indirect. Coexpression of FLAG-FtsE and FtsX under certain conditions resulted in efficient formation of minicells, also consistent with an FtsE-FtsZ interaction and with the idea that FtsE and FtsX regulate the activity of the divisome.
To ensure their survival, bacteria must successfully duplicate their genetic material and partition it equally to each daughter cell just prior to cytokinesis. In Escherichia coli, at least 14 proteins are directly linked to the process of cell division. FtsZ, the prokaryotic homolog of tubulin, is the first known component of the cell division apparatus to localize to the future division site (1, 5). Once localized, FtsZ polymerizes into the Z ring and is proposed to provide a scaffold upon which FtsA, ZipA, FtsE, FtsX, FtsK, FtsQ, FtsL, FtsB, FtsW, FtsI, FtsN, and AmiC are recruited (17, 34, 48). The resulting putative protein complex, or divisome, is required for the synthesis of the division septum and subsequent formation of new cell poles. Removal or inactivation of any of these proteins causes an arrest in cell division, yet cell growth continues, resulting in cell filamentation, lysis, and subsequent death. Although most components of the cell division machinery have likely been identified, how this multisubunit complex is assembled within the membrane and what the precise role of each protein is remain unknown.
In E. coli, cell division proteins localize to the future division site via a linear order of dependency (17). Because downstream cell division proteins fail to localize upon deletion or inactivation of an immediately upstream component, this suggests that each cell division protein interacts with the cell division component localizing immediately upstream. Nevertheless, evidence for a more complex system of interactions is mounting. For example, ZipA is necessary for localization of FtsK and downstream cell division proteins (24, 40), but it can be deleted from cells carrying a gain-of-function mutation in ftsA (14). This result indicates that ZipA does not recruit downstream cell division proteins directly but rather enhances the ability of the Z ring to recruit these proteins. Likewise, the same mutation in ftsA, or increased levels of FtsZ, FtsA, and/or FtsQ, can partially bypass the loss of FtsK; in suppressed strains lacking FtsK, downstream cell division proteins, such as FtsI, are still recruited to the septum, indicating that FtsK is not required for their recruitment (15).
Recent evidence also supports the idea that cell division proteins in E. coli can exist in independent subassemblies. For example, a subassembly of FtsA, FtsI, and FtsN can be forced to localize to the cell poles independently of the rest of the cell division machinery (8). FtsQ can be artificially recruited to the Z ring at an early step, and this results in the recruitment of some late cell division proteins to a subassembly independently of some earlier proteins (18, 19). FtsL and FtsB can be isolated in a subcomplex with FtsQ (6). Finally, bacterial two-hybrid analyses indicate that cell division proteins interact with multiple partners (13, 28), consistent with the idea that the divisome assembles as a web of protein-protein interactions instead of a chain of binary interactions. However, bacterial two-hybrid studies can be problematic because of potential bridging proteins mediating protein-protein interactions. Moreover, when potential interactions among cell division proteins were tested with either of the bacterial two-hybrid systems, one cell division protein not tested was FtsE.
FtsE and FtsX show high homology to ATP-binding cassette (ABC) transporters, with FtsE being the ABC component and FtsX the component in the cytoplasmic membrane (11, 16). FtsE and FtsX localize to the Z ring, and the localization of FtsX is dependent on FtsZ, ZipA, and FtsA but not downstream proteins, FtsK, FtsQ, FtsW, and FtsI (43). Depletion of FtsE results in inhibition of cell division and growth, but this effect is mainly observed in growth medium lacking salt. The cell viability defect could be suppressed by addition of 1% NaCl or other electrolytes, including NaH2PO4, Na2SO4, NH4Cl, and CaCl2 (11, 16). Recently, Reddy demonstrated that high-osmolarity conditions alone suppress the loss of FtsE and further showed that increased expression of ftsQAZ or ftsN, either of which can suppress the loss of FtsK (15, 20), can also compensate for the loss of FtsE (42). Consistent with the idea that FtsE and FtsX are cell division proteins, disruption of FtsE/FtsX functions in other bacteria is also associated with cell division defects (3, 29, 36). Because of the position of FtsE and FtsX relatively early in the recruitment pathway, we became interested in understanding how these proteins might function in cytokinesis and influence the activity of the Z ring. In this study, we provide evidence for how FtsE is targeted to the Z ring.
MATERIALS AND METHODS
Growth conditions.
LB medium (0.5% yeast extract, 1% tryptone, 0.5% NaCl) was used for routine growth experiments. High-salt and no-salt LB media contained 1% or 0% NaCl, respectively, instead of 0.5%. Kanamycin (50 μg/ml), ampicillin (100 μg/ml), or tetracycline (10 μg/ml) was used as necessary. IPTG (isopropyl-β-d-thiogalactopyranoside) at 400 mM was kept as a stock solution in N,N-dimethyl formamide.
Strain construction.
Strains and plasmids are listed in Table 1. To delete ftsX from the chromosome, we constructed PCR primers CTCACCCTGAGCGATGGTCACTTGCATGGAGGCGTGGGCCGCCACGTTGTGTCTC and ACGGAACCCATTGCAGGGAAAGAGTATAACACGCTTTTAGAAAAACTCATCGAGC to amplify the kanamycin resistance cassette from plasmid pUC4K. The PCR product consisted of the 813-bp Kanr cassette with an additional 40 bp upstream and 36 bp downstream, corresponding to the flanking regions of ftsX (underlined in the primer sequence). The linear DNA product was cut with DpnI and transformed into DY330 competent cells as described previously (50), and transformants were selected on high-salt LB containing kanamycin. A phage P1 lysate was made from this strain (WM2650) and used to transduce W3110 to Kanr on high-salt LB to generate strain WM2712. We confirmed that this strain had ftsX deleted, as we were unable to amplify ftsX from the chromosome with PCR primers internal to the gene and could obtain a PCR product corresponding to the size of the kanamycin cassette following amplification of ftsX flanking regions. In addition, cell division was largely rescued in high salt; most cells were highly filamentous in salt-free medium, whereas 50% of the cells were slightly filamentous in high salt.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strain | ||
| TX3772 | MG1655 ΔlacU169 | Laboratory collection |
| Top10 | F−mcrA (mrr-hsdRMS-mcrBC) Φ80lacZΔM15 lacX74 recA1 | |
| ara139 (ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen | |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 | |
| lac (F ΔproAB lacIqZΔM15 Tn10) | Stratagene | |
| W3110 | Wild-type strain | Laboratory collection |
| DY330 | W3110 ΔlacU169 gal490 λcI857 Δ(cro-bioA) | 50 |
| WM746 | WX7/pCX41, leu::Tn10 ftsZo (repA(Ts) ftsZ+) | 46 |
| WM971 | pET11a-ftsZ in BL21(DE3); FtsZ overproducer | H. Erickson |
| WM1099 | WM746 with leu::Tn10 replaced by leu::Tn5 | 33 |
| WM1109 | TX3772 leu::Tn10 | 52 |
| WM1115 | TX3772 leu::Tn10 ftsA12(Ts) | 14 |
| WM1125 | TX3772 ftsZ84(Ts) | 52 |
| WM1281 | CH2 (recA::Tn10, ftsA0)/pDB280 (repA(Ts) ftsA+) | 23 |
| WM1282 | CH5 (recA::Tn10 ΔzipA::kan)/pCH32 (repA(Ts) zipA+ ftsZ+) | 22 |
| WM1657 | TX3772 ΔzipA::kan, ftsA*(R286W) | 14 |
| WM1659 | TX3772 ftsA*(R286W) | 14 |
| WM2497 | MG1655 ftsE::kan (FB21223, Tn5-Sce-I at position 284) | F. Blattner |
| WM2650 | DY330 ΔftsX::kan | This study |
| WM2712 | W3110 ΔftsX::kan | This study |
| WM2933 | W3110 + pWM2270 | This study |
| Plasmids | ||
| pWM176 | Tetr; IncP plasmid containing the tac promoter | 35 |
| pMK4 | Tetr; E. coli wild type ftsZ (ftsZ1-383) in pWM176 | 32 |
| pWM1201 | Tetr; pWM176-ftsZΔC2 (ftsZ1-316) | 33 |
| pDSW209 | Ptrc-gfp (no stop codon) pBR322 derivative (Ampr) | 49 |
| pWM2244 | pflag-MAC cloning vector (Ampr) | Sigma-Aldrich |
| pWM2270 | pflag-ftsE | This study |
| pWM2529 | pflag-ftsE ftsX | This study |
| pflag-Spe-Sse1D8N | pflag-MAC with SpeI site expressing Saccharomyces cerevisiae Sse1 | |
| D8N mutant with N-terminal flag | 44 | |
| pflag-BAP | pflag-MAC expressing E. coli phoA with N-terminal flag | Sigma-Aldrich |
| pWM2800 | pDSW209 with Ptrc-ftsE, (gfp-ftsE, ftsX) | This study |
| pWM2853 | pDSW209 with Ptrc-ftsE (gfp-ftsE) | This study |
| pWM2654 | pWM176 with Ptac-ftsZ1-268 | This study |
| pWM2655 | pWM176 with Ptac-ftsZ1-218 | This study |
Plasmid construction.
Plasmid pWM2270 (pflag-ftsE) was constructed by amplifying ftsE from the chromosome of TX3772 using primers FtsE fp EcoR1b (CCGGAATTCCATGATTCGCTTTGAA; restriction site is underlined) and FtsE rv Kpn (CGGGGTACCTTCATGGCCCACGCC). The product was then cloned as a EcoRI-KpnI fragment into pWM2244. Plasmid pWM2529 was constructed by amplifying ftsE and ftsX from the chromosome using primers FtsE fp EcoR1b (CCGGAATTCCATGATTCGCTTTGAA) and FtsX rp Kpn (CGG GGTACCTTCAGGCGTAAAGTG). This 1,728-bp DNA product was then cloned as an EcoRI-KpnI fragment into pWM2244.
To make C-terminal deletions of FtsZ, we used primer FtsZfwd (ACGAAGCTTAGGCGACAGGCACAAATCGGAGAG) and reverse primer ftsZdelta804 (AAAACTGCAGTTAGAAGCCCGCCGTGATGTTAA) or ftsZdelta654 (AAAACTGCAGTTAAGACATTACGGTGCGTACGT). FtsZ PCR products were cloned as HinDIII-PstI fragments into pMK4. To make an N-terminal FtsZ deletion, we amplified FtsZ using primer Zdelta93 (ACGAAGCTTAGGCGACAGGCACAAATCGGAGAGAAACTATGGAGCGCATTGAAGGT) and primer Zrptruncations (CGCTCTAGAACTAGTGGATCCCCCGGGCTGCAGAA) and cloned the PCR product as a HinDIII-PstI fragment into pMK4.
Plasmid pDSW209 (49), containing a weakened trc promoter, was used to express IPTG-inducible gfp fusions to ftsE. The ftsE gene was amplified from TX3772 chromosomal DNA by PCR using primers GFP-ftsEX-F-EcoRI (CCG GAA TTC ATG ATT CGC TTT GAA CAT) and GFP-ftsE-R-HindIII (CGG AAG CTT TTA TTC ATG GCC CAC GCC T). The PCR product was cloned as an EcoRI-HindIII fragment into pDSW209, resulting in pWM2853 (expressing gfp-ftsE). Plasmid pDSW209-ftsE-ftsX was constructed using the same strategy, except that the ftsEX genes from the chromosome were amplified in tandem by PCR using the antisense primer GFP-ftsEX-R-HindIII (CGG AAG CTT TTA TTC AGG CGT AAA GTG) and the sense primer GFP-ftsEX-F-EcoRI as described above. The resulting plasmid, pWM2800, coexpressed gfp-ftsE and ftsX. Top10 or XL1-blue was used as a recipient strain for the initial cloning and characterization of these plasmids.
Immunoprecipitation.
Unless otherwise indicated, bacterial cultures (5 ml) were grown in LB plus antibiotic to an optical density at 600 nm (OD600) of 0.1, and then 0.5 mM IPTG was added to induce expression of pflag-ftsE for 2 to 4 h. Equal numbers of cells, as measured by OD600, were pelleted, resuspended in 1 ml RIPA buffer (10 mM Tris-Hcl, pH 7.5, 150 mM NaCl, 1% NP-40, 1% deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) containing 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1% beta-mercaptoethanol, 50 μl protease inhibitor cocktail (1 tablet Roche complete, mini, EDTA-free, dissolved in 1.5 ml RIPA buffer), and 100 μg/ml lysozyme, and incubated on ice for 45 min. The cells were then sonicated for four 10-s cycles (20% duty cycle; output, 4) using a Branson sonifier 250 microtip apparatus. The suspension was then incubated at 4°C for 4 h with vigorous shaking, followed by centrifugation for 20 min to remove unsolubilized material. The supernatant fraction was then collected and mixed with a 20-μl bed volume of protein A-Sepharose (Sigma-Aldrich) for 1 h to remove nonspecific binding proteins. An aliquot (50 μl) of precleared supernatant fraction was used as a control for total protein levels prior to immunoprecipitation. The remainder of this mixture was collected and incubated with a 50-μl bed volume of protein A-Sepharose and 2 μl of anti-FLAG M2 monoclonal antibody (Sigma) for 2 h or overnight. Beads were washed three times with RIPA buffer, resuspended in loading buffer, and boiled for 15 min. Samples were subsequently analyzed by SDS-polyacrylamide gel electrophoresis, followed by immunoblotting.
FtsZ depletion strains used to perform immunoprecipitation reactions were grown at 42°C for 5 h (at which time 0.5 mM IPTG was added to induce expression of the pWM176-ftsZ derivative) and allowed to grow for an additional 2 h before being harvested. To conduct immunoprecipitation reactions from the ftsA12(Ts) strain WM1115, cells were grown at the permissive temperature of 30°C to early log phase and then shifted to the nonpermissive temperature of 42°C for 1 h before being harvested. For immunoprecipitation from the ZipA depletion strain WM1282, the strain was grown at 42°C for 4 h, followed by IPTG induction (0.5 mM) of flag-ftsE for 1 h before the cells were harvested. For immunoprecipitation from the ftsX deletion strain WM2712, 10-ml cultures were grown in LB plus 1% NaCl medium at 30°C for 4 h. Then, the culture was split, with 5 ml pelleted and resuspended in LB without added NaCl. IPTG (0.5 mM) was added to both high-salt and no-salt cultures, which were grown for one additional hour at 30°C before being harvested.
Protein purification and immunoblotting.
FtsZ was purified from WM971 as described previously (4). For purification of FLAG-FtsE to test for FtsZ copurification, W3110 cells harboring pWM2270 (WM2933) were grown to an OD600 of 0.6 at 37°C and then induced for four more hours with 0.5 mM IPTG. The cell pellet was then incubated with buffer and 0.4 mg/ml lysozyme at 0°C for 1 h, sonicated, and centrifuged at 19,800 × g for 25 min. The membrane fraction was resuspended in TBS (50 mM Tris-HCl, pH 7.4, 150 mM NaCl)-1% NP-40-0.1 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} for 1 h at 0°C, sonicated, kept at 0°C for 1 h, and centrifuged at 264,000 × g for 30 min. The clear supernatant fraction was passed twice through an anti-FLAG M2 (Sigma-Aldrich; A2220) affinity column and washed with 20 column volumes of TBS, and FLAG-FtsE was eluted with 0.1 M glycine (pH 3.5) and neutralized by combining aliquots with 1/10 volume of 1 M Tris-HCl, pH 8.0; the column was also neutralized with the same buffer and stored in TBS-50% glycerol-0.02% NaN3.
Proteins from this experiment and from immunoprecipitations were separated by SDS-polyacrylamide gel electrophoresis and either stained with Coomassie blue or transferred to nitrocellulose membranes for immunoblotting. To detect FtsZ or green fluorescent protein (GFP), blots were probed with affinity-purified polyclonal antibodies against either FtsZ or GFP and goat anti-rabbit antibody conjugated to horseradish peroxidase. To detect FLAG, blots were probed with monoclonal anti-FLAG and goat anti-mouse secondary antibody conjugated to horseradish peroxidase. The membranes were then developed using standard enhanced-chemiluminescence reagents (Amersham or Pierce).
Localization of GFP fusion proteins in live cells.
GFP-FtsE produced from pWM2800 (coproduced with FtsX) was visualized in wild-type cells either with or without IPTG to induce higher levels of expression. In general, uninduced levels of GFP fusion protein were sufficient for visualization; the promoter on this plasmid is sufficiently weak that induction with IPTG only modestly increased the fluorescence intensities of the bands. To examine GFP-FtsE localization in the ftsA12(Ts) strain WM1115 or the ftsZ84(Ts) strain WM1125, cells with the pWM2800 plasmid were grown at 30°C to early exponential phase and then shifted to the nonpermissive temperature of 42°C for 90 to 120 min to inactivate FtsA or FtsZ. To examine GFP-FtsE localization in the ZipA depletion strain WM1282 or the FtsA depletion strain WM1281, cells with the pWM2800 plasmid were grown to early exponential phase and shifted to 42°C for 2.5 to 4 h to dilute out the thermosensitive plasmid carrying zipA or ftsA and thus deplete the cells of the ZipA or FtsA protein. To visualize GFP-FtsE in other strains, such as WM1657, WM1659, or the ftsE null mutant WM2497, cells with pWM2800 or the plasmid producing GFP-FtsE without FtsX (pWM2853) were grown in LB at 30°C to mid-exponential phase. In all cases, the cells were immobilized in LB containing low-melting-point agarose and visualized by fluorescence microscopy with an Olympus BX60 epifluorescence microscope containing a 100× oil immersion objective. Images were captured with QED software and a Photometrics Coolsnap fx charge-coupled device camera.
Localization of FLAG-FtsE by immunofluorescence microscopy.
To observe the localization of FLAG-FtsE, cells were grown to mid-log phase and fixed with methanol as described previously (51). Each sample was processed using anti-FLAG M2 monoclonal antibody (1:200) and goat anti-mouse secondary antibody conjugated to rhodamine red (1:500; Molecular Probes) with an incubation period of 1 h.
RESULTS
A flag-ftsEX fusion construct is functional.
To elucidate the function of FtsE and to facilitate purification, the entire ftsE gene, coding for a 24-kDa protein, was cloned downstream of the tac promoter in a pflag-MAC cloning vector. This newly created vector, referred to as pWM2270, produced FtsE with an amino-terminal FLAG tag. Most cells producing the FLAG-FtsE fusion protein had a phenotype similar to that of wild-type cells, although a small percentage of cells were 1.5 to 3 times longer than normal. Immunofluorescence microscopy experiments with antibody directed against the FLAG epitope revealed that the majority of cells examined contained intense fluorescent staining at midcell (Fig. 1A), suggesting that the FLAG-FtsE fusion protein targeted the Z ring correctly. However, there was also a high background of extraneous fluorescence that made it difficult to evaluate the dependency of FLAG-FtsE localization on other cell division proteins in filamentous cells; this was addressed with a GFP fusion (see below).
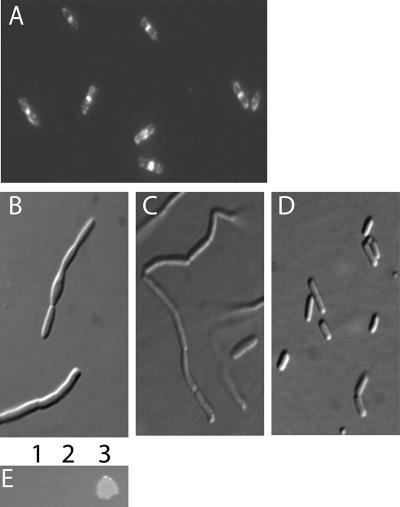
FIG. 1.
A flag-ftsE fusion is functional when coexpressed with ftsX. (A) W3110 cells expressing flag-ftsE from pWM2270 that were fixed and immunostained with anti-FLAG antibody. (B to D) Micrographs (differential interference contrast) of the ftsE::kan strain (WM2497) grown in LB no-salt growth medium with no plasmid (B), pflag-ftsE plasmid pWM2270 (C), or plasmid pWM2529 expressing both flag-ftsE and ftsX (D). IPTG (100 μM) was added to cells in panels A to C to induce expression of flag-ftsE from pWM2270, but basal levels of expression were sufficient to allow complementation by pWM2529 in panel D. (E) Colony growth overnight on LB no-salt agar of the ftsE::kan strain (WM2497) containing either no plasmid (lane 1), pflag vector pWM2244 (lane 2), or pflag-ftsE plasmid pWM2270 (lane 3).
Although FtsE is required for bacterial cell division, the requirement can be largely suppressed when cells are grown in high salt (1% NaCl). In salt-free medium (0% NaCl), however, cell division was blocked as growth continued, resulting in cell filamentation and loss of viability, as observed previously (43). To further characterize the functionality of the FLAG-FtsE fusion construct, we investigated whether this fusion was sufficient to complement an ftsE allele disrupted by insertion of a kanamycin cassette (WM2497; ftsE::kan) under no-salt growth conditions. Analysis of cell growth on salt-free plates incubated overnight revealed that the ftsE::kan strain expressing FLAG-FtsE was capable of forming colonies (Fig. 1E, spot 3), whereas cells containing no plasmid, or empty vector alone, failed to grow (Fig. 1E, spots 1 and 2). Although expression of FLAG-FtsE permitted colony growth, the cells in the colonies were filamentous (data not shown), similar morphologically to the ftsE::kan strain (Fig. 1B) or ftsE::kan/FLAG-FtsE cells grown in no-salt LB with expression of FLAG-FtsE induced with 100 μM IPTG (Fig. 1C). This suggested that our FLAG-FtsE fusion, even when expressed at high levels, was not completely able to complement the ftsE::kan strain.
We reasoned that this might be caused by polarity of the Tn5 insertion within ftsE on the downstream ftsX gene and therefore cloned ftsX downstream of the flag-ftsE construct in the plasmid. This plasmid carrying flag-ftsE-ftsX (pWM2529) was sufficient to complement the ftsE::kan strain completely, allowing growth of colonies and mostly normal division of cells without IPTG induction (Fig. 1D). Therefore, we can conclude that our FLAG-FtsE fusion protein is fully functional when cosynthesized with FtsX.
FtsE interacts with FtsZ.
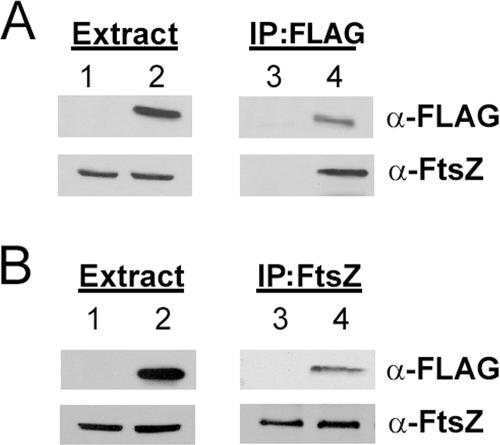
To decipher the role of FtsE in bacterial cell division, and to rectify the specific gap in knowledge about which proteins interact with FtsE, we used coimmunoprecipitation to investigate possible interaction partners. Using antibodies directed against the FLAG epitope, we were able to immunoprecipitate FLAG-FtsE (Fig. 2A, lane 4, top). Because FtsZ is abundant in the cell and we have a good polyclonal antibody against it, we wanted to see if FtsE could pull down a complex that contained FtsZ. Probing with anti-FtsZ, we found that FLAG-FtsE efficiently pulled down FtsZ (Fig. 2A, lane 4, bottom). In the converse experiment, we could immunoprecipitate FtsZ with anti-FtsZ (Fig. 2B, lanes 3 and 4, bottom) and were able to detect FLAG-FtsE in the precipitated mixture (Fig. 2B, lane 4, top).
FIG. 2.
Identification of an interaction between FtsE and FtsZ. (A) Immunoblots from a typical immunoprecipitation experiment performed on RIPA-solubilized total cell protein with anti-FLAG. (B) Immunoblots from a typical immunoprecipitation experiment performed on RIPA-solubilized total cell protein with anti-FtsZ. Antibodies used to probe the blots are outlined on the right. In all panels, lanes 1 and 3 contain cell extracts from Top10 cells expressing a pflag vector (pWM2244) control, whereas lanes 2 and 4 contain cell extracts from Top10 cells expressing pflag-ftsE (pWM2270).
FtsZ was immunoprecipitated equally efficiently in cells expressing either the pflag vector control (Fig. 2B, lane 3, bottom) or pflag-ftsE (Fig. 2B, lane 4, bottom), but a band corresponding to FLAG-FtsE was detected only in the reactions that included FLAG-FtsE (Fig. 2B, compare lanes 3 and 4, top). At the time the immunoprecipitation reactions were performed, the concentrations of FtsZ protein were similar in all extracts tested (Fig. 2A and B, lanes 1 and 2, bottom). The anti-FLAG antibody was specific in detecting the FLAG-FtsE protein (Fig. 2A and B, lanes 1 and 2, top).
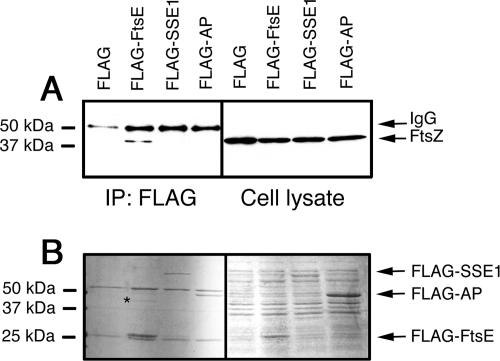
To confirm that FtsZ interacted with the FtsE portion of FLAG-FtsE and not with the FLAG tag, we performed parallel immunoprecipitations with FLAG-FtsE and other FLAG-tagged proteins. These included FLAG-SSE1, a yeast chaperone protein, and FLAG-AP (E. coli alkaline phosphatase). Both were efficiently expressed in E. coli cells from a derivative of the same pflag-MAC vector (Fig. 3B, right). Blots of the immunoprecipitated proteins from the corresponding cell extracts revealed that FtsZ was pulled down only by FLAG-FtsE, indicating that FtsZ interacts with FLAG-FtsE but not FLAG-SSE1 or FLAG-AP (Fig. 3A, left). FtsZ levels were similar in all the extracts used (Fig. 3A, right).
FIG. 3.
FtsZ is pulled down by FLAG-FtsE but not by other FLAG-tagged proteins. (A) Immunoblots of RIPA-solubilized total cell protein (right) or after immunoprecipitation with anti-FLAG (left) probed with anti-FtsZ antibodies. The positions of protein size markers, FtsZ, and the IgG heavy chain are shown. The sources of extracts were strains overproducing either FLAG alone, FLAG-FtsE, FLAG-SSE1, or FLAG-AP (see Materials and Methods). (B) Coomassie-stained SDS-polyacrylamide gels corresponding to the blots in panel A; size markers and the positions of the overproduced FLAG fusion protein bands are shown. A band in the FLAG-FtsE lane presumed to be FtsZ is indicated by the asterisk.
Coomassie blue staining of the corresponding SDS gels of immunoprecipitated extracts showed that the predominant bands were the FLAG-tagged proteins and the immunoglobulin G (IgG) heavy and light chains (Fig. 3B, left). Interestingly, several faint bands between 40 and 50 kDa were detectable in the FLAG-FtsE lane but absent in the others, suggesting that they were specifically pulled down by FtsE. One band at ∼40 kDa (Fig. 3B) was suggestive of FtsZ. Probing with anti-ZipA antibodies revealed a faint signal just below the IgG heavy chain in the FLAG-FtsE lane that might be ZipA (data not shown). Two other faint bands larger than 80 kDa were also visible only in the FLAG-FtsE pull-down, but their identities are unknown.
FtsZ-FtsE interaction is independent of FtsA or ZipA.
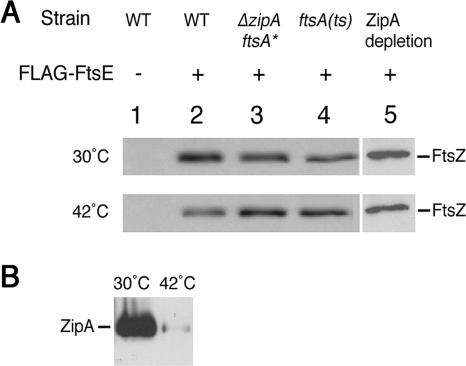
According to the linear-dependency order of recruitment, the only two proteins localizing to the septum following FtsZ, but prior to FtsE, are ZipA and FtsA (43). Thus, we investigated if we could delete ZipA or inactivate FtsA and still retain an FtsE-FtsZ complex. To inactivate FtsA, we utilized a strain containing an ftsA12(Ts) allele and performed immunoprecipitation reactions on FLAG-FtsE and FtsZ from cells grown at both permissive and nonpermissive temperatures. To remove ZipA, we utilized a strain harboring a gain-of-function mutation in FtsA, R286W (also known as FtsA*), which allowed the viability of a ΔzipA null allele (14).
When ZipA was absent (Fig. 4, lane 3 top and bottom) or FtsA was thermoinactivated (Fig. 4, lane 4, bottom), immunoprecipitation of FLAG-FtsE with anti-FLAG still resulted in the efficient detection of FtsZ. The intensities of the FtsZ bands on the immunoblot were roughly equivalent to those from the permissive temperature for ftsA12(Ts) (Fig. 4, lane 4, top) or from wild-type cells (lane 2, top and bottom). As expected, no FtsZ was detected when only the pflag vector was used in the immunoprecipitation reaction (lane 1), indicating that the FtsZ pull-down was FtsE-specific. These results suggest that FtsE and FtsZ can interact in the absence of functional FtsA or in the absence of ZipA when compensated for by the FtsA R286W bypass mutant.
FIG. 4.
The FtsE-FtsZ interaction is independent of FtsA or ZipA. (A) Immunoblots of immunoprecipitation experiments performed on RIPA-solubilized total cell protein by using anti-FLAG and subsequently probed with anti-FtsZ antibodies. Cells were grown at permissive (top) or nonpermissive (bottom) temperatures. Solubilized total proteins were from cells expressing either the pflag vector pWM2244 (lane 1) or pflag-ftsE plasmid pWM2270 (lanes 2 to 5). Cell lysates were from the TX3772 wild-type (WT) parent strain (lanes 1 and 2), a strain (WM1657) that lacks zipA but contains the ftsA*(R286W) suppressor mutation and therefore lacks ZipA at both 30°C and 42°C (lane 3), cells containing an ftsA12(Ts) allele (WM1115) that is functional at 30°C but thermoinactivated at 42°C (lane 4), or a ZipA depletion strain that contains ZipA at 30°C but is depleted of ZipA after 4 h at 42°C (lane 5). Lanes 1 to 4 are all from one blot, and lane 5 is from a separate blot. (B) Extent of ZipA depletion for the experiment in lane 5 of panel A; extracts from equivalent densities of cells grown at 30°C or 42°C for 4 h were probed with anti-ZipA antibodies, and the resulting ZipA protein band, which migrates at ∼50 kDa, is highlighted.
To further confirm that the FtsZ-FtsE interaction was independent of ZipA, we tested for the ability of FLAG-FtsE to pull down FtsZ from extracts of a ZipA depletion strain lacking the ftsA* suppressor. After ZipA depletion in this strain at 42°C, cells cease dividing and form long filaments, similar to those carrying the ftsA12(Ts) allele. As with the ΔzipA ftsA* strain, FtsZ was pulled down by FLAG-FtsE (Fig. 4A, lane 5) after significant depletion of ZipA (Fig. 4B). Therefore, inactivation of the two known intermediary proteins between FtsE and FtsZ in the linear protein recruitment pathway failed to disrupt formation of an FtsE-FtsZ complex by coimmunoprecipitation, further supporting the idea that FtsZ and FtsE interact.
FtsZ copurifies with FLAG-FtsE.
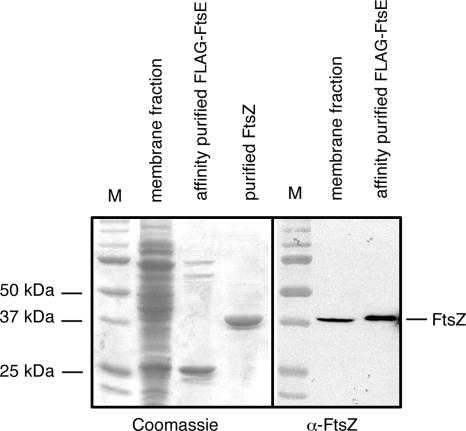
To provide further evidence for an interaction between FtsE and FtsZ, we tested whether FtsZ could copurify with FLAG-FtsE. FLAG-FtsE was overproduced, and the soluble and membrane fractions were then subjected to affinity purification with M2-agarose beads containing anti-FLAG. Whereas the soluble fraction became purer, we were more successful in obtaining largely pure FLAG-FtsE from the membrane fraction (Fig. 5, left), although two bands of larger molecular mass remained visible in the affinity-purified preparation. It is possible that one of these, running at ∼55 kDa, was GroEL, an abundant protein that was shown previously to specifically interact with FtsE (7).
FIG. 5.
FtsZ copurifies with FLAG-FtsE. FLAG-FtsE was overproduced from WM2933 and purified as described in Materials and Methods. The Coomassie-stained SDS-polyacrylamide gel of the crude membrane preparation and affinity-purified FLAG-FtsE is shown in the left panel (middle lanes), along with molecular mass markers (M) and purified FtsZ (outer lanes). An immunoblot made from the same preparations and probed with anti-FtsZ is shown in the right panel, along with the same mass markers, showing that FtsZ copurifies with FLAG-FtsE.
This partially purified FLAG-FtsE preparation was then used to probe for FtsZ on immunoblots. As shown in Fig. 5 (right), FtsZ was present at roughly equivalent levels in the crude membrane fraction of overproduced FLAG-FtsE, where it represented a very small fraction of the total protein, and the purified FLAG-FtsE preparation. Other purified FLAG-FtsE preparations also contained FtsZ (data not shown). These results strongly suggest that FtsZ copurifies with overproduced FLAG-FtsE and support the idea that FtsZ and FtsE interact.
The conserved C-terminal tail of FtsZ is not required for the interaction with FtsE.
FtsZ interacts with FtsA and ZipA via 12 amino acids within its conserved C-terminal tail (23, 26, 30, 33). Because this C-terminal tail of FtsZ is essential in mediating its interaction between ZipA and FtsA, we hypothesized that the domain might be important for FtsE binding.
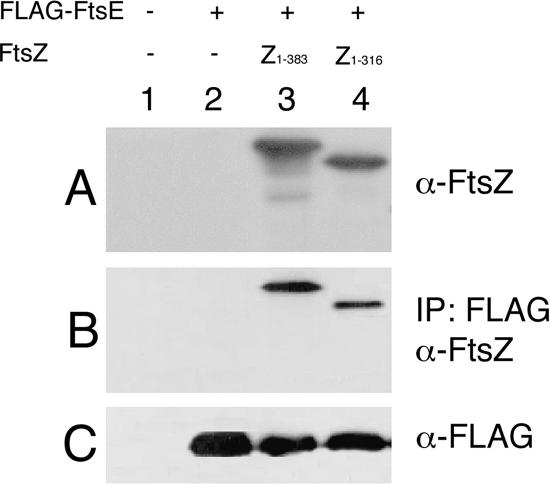
To test this idea, we performed immunoprecipitation reactions with anti-FLAG, using FLAG-FtsE and several C-terminal truncations of FtsZ. Because FtsZ is essential for viability, we utilized an FtsZ depletion strain that contains a null allele of the chromosomal ftsZ, a thermosensitive plasmid expressing ftsZ for depletion of wild-type FtsZ upon temperature shift, and an additional IncP-based low-copy-number plasmid that expresses various deletions of ftsZ from the tac promoter (33). After growth at 42°C and induction with IPTG, the plasmid with the wild-type ftsZ was removed, depleting the wild-type protein and leaving the truncated FtsZ derivatives as the sole FtsZ proteins in the cells. Extracts from these cells were then used in coimmunoprecipitations with FLAG-FtsE, and the abilities of various FtsZ truncation derivatives to be pulled down with FLAG-FtsE were examined.
Immunoblot analysis of FtsZ protein levels within the cell confirmed that at the time the immunoprecipitation reactions were performed, the only detectable form of FtsZ within the cell was the full-length FtsZ (Fig. 6A, lane 3) or the C-terminal FtsZ truncation (Fig. 6A, lane 4). These levels of FtsZ were comparable to those of native FtsZ in wild-type cells (data not shown). Cell extracts of control strains containing an empty IncP vector with (Fig. 6A, lane 2) or without (lane 1) expression of flag-ftsE from the pflag vector did not contain detectable levels of FtsZ at the time they were used for the immunoprecipitation experiments.
FIG. 6.
The FtsZ C-terminal tail is not required for the FtsE-FtsZ interaction. Immunoprecipitation experiments were performed with anti-FLAG antibody on RIPA-solubilized total protein from cells depleted of native FtsZ but expressing various forms of FtsZ from a plasmid. Immunoblots from these experiments are shown. (A) Extracts of cell lysates prior to immunoprecipitation probed with anti-FtsZ antibody. (B) Immunoprecipitation reactions probed with anti-FtsZ antibody. (C) Whole-cell lysates immunoprecipitated and probed with anti-FLAG antibody. Lane 1 contains extracts from cells that do not express FtsZ or the pFLAG empty vector. Extracts in lane 2 are from cells producing FLAG-FtsE but not FtsZ. Lanes 3 and 4 contain extracts from cells synthesizing wild-type FtsZ or FtsZ1-316, respectively. WM1099 was the strain background used for all of the FtsZ depletion extracts.
Surprisingly, the truncated FtsZ derivative (FtsZ1-316) missing its entire C-terminal 67 amino acids was reproducibly pulled down by FLAG-FtsE (Fig. 6B, lane 4), although the amount coimmunoprecipitated was reproducibly less than that for full-length FtsZ, based on band intensities (Fig. 6B, compare lanes 3 and 4). Because deletion of just 12 residues from the C terminus of FtsZ is normally sufficient to abolish its interaction with ZipA and FtsA, it was striking that we could remove this domain, as well as much of the flexible linker domain (residues 317 to 371), and still detect a functional interaction between FtsE and FtsZ.
This result prompted us to investigate whether more extensive C-terminal truncations of FtsZ were also able to interact with FtsE. FtsZ1-268 and FtsZ1-218 lack not only the C-terminal tail and flexible linker regions, but also large portions of the conserved C-terminal globular domain (37). These FtsZ derivatives were synthesized in the depletion strain but were probably insoluble, as they were not detectable in the lysate just prior to immunoprecipitation (data not shown) and consequently were not pulled down by FLAG-FtsE (data not shown). Therefore, the only firm conclusion we can draw is that the C-terminal tail and flexible linker regions of FtsZ are not required for its interaction with FtsE.
The FtsE-FtsZ interaction is independent of FtsX.
FtsX is the membrane component of the hypothetical FtsEX ABC transporter. Subcellular fractionation studies have shown that FtsE is incorporated into the membrane upon overexpression of FtsX. In addition, FtsE has been shown to pull down FtsX upon overexpression and copurify with a histidine-tagged FtsX (11). Because FtsE interacts with FtsX and is incorporated into the membrane upon overexpression of FtsX, we wanted to investigate whether FtsX was necessary for the FtsE-FtsZ interaction.
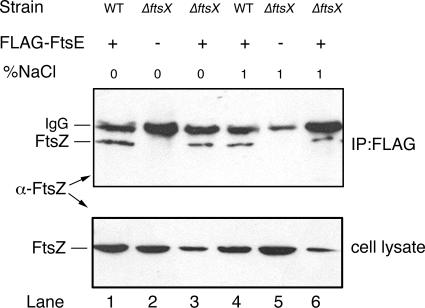
To address this question, we first constructed a ΔftsX::kan strain, which could survive when grown on high salt. We then used extracts from ΔftsX::kan cells with or without pflag-ftsE for immunoprecipitation reactions to determine if FLAG-FtsE could pull down FtsZ in a ΔftsX::kan background. Cells were either grown in 1% NaCl to suppress the division defect of the ΔftsX::kan allele or switched from 1% to 0% for 1 h to create nonpermissive conditions for cell division. Immunoblots probed for FtsZ revealed that FLAG-FtsE could pull down FtsZ in extracts lacking FtsX (Fig. 7, top, lanes 3 and 6) at levels similar to the wild-type background (top, lanes 1 and 4). The ability of FtsZ to be pulled down by FLAG-FtsE in all cases was independent of the presence or absence of NaCl in the growth medium. Control immunoprecipitations from extracts lacking FLAG-FtsE did not pull down FtsZ (Fig. 7, top, lanes 2 and 5). FtsZ levels were similar in ftsX+ strains expressing FLAG-FtsE and ftsX mutant strains not expressing FLAG-FtsE (Fig. 7A, lanes 1, 2, 4, and 5). The band intensities of FtsZ were lower in extracts from ftsX mutant strains expressing FLAG-FtsE, possibly because of toxicity; nevertheless, FtsZ was still pulled down by FLAG-FtsE in these extracts (Fig. 7A, lanes 3 and 6). These results suggested that the FtsE-FtsZ interaction does not require FtsX, is not affected significantly by FtsX, and is independent of added NaCl.
FIG. 7.
The FtsE-FtsZ interaction is not dependent on FtsX. Shown are immunoblots, probed with anti-FtsZ, of immunoprecipitations (IP) with anti-FLAG antibody (top) or RIPA-solubilized total cell protein (bottom). Extracts were prepared from cells containing either the pflag empty vector (lanes 2 and 5) or pflag-ftsE (lanes 1, 3, 4, and 6) grown in LB with either 0% or 1% NaCl added, as described in Materials and Methods. The extracts used in lanes 2 and 3 and lanes 5 and 6 are from cells with a ΔftsX::kan insertion (WM2712), whereas cell extracts used in lanes 1 and 4 are from the wild-type (WT) parent strain W3110. The positions of FtsZ (∼40 kDa) or the IgG heavy chain (∼50 kDa) are shown. Lower levels of FtsZ in lanes 3 and 6 may have resulted from toxicity of FLAG-FtsE overproduction in the absence of chromosomal ftsX.
Targeting of GFP-FtsE to Z rings and its dependence on ZipA and FtsA.
To provide in vivo evidence for FtsE-FtsZ interaction, we asked whether FtsE could localize to Z rings independently of other cell division proteins. Other division proteins that interact directly with FtsZ, such as FtsA, ZipA, and ZapA of E. coli, do not depend on any known divisome proteins other than FtsZ for their localization to Z rings (21, 23, 31). As mentioned above, detection of FLAG-FtsE by immunofluorescence was complicated by the high background fluorescence often observed, particularly in filamentous cells. Therefore, we constructed a GFP-FtsE fusion to detect FtsE localization in live cells. However, in wild-type cells, GFP-FtsE fluorescence was usually uniformly distributed throughout the cell, and midcell bands were difficult to detect. This poor localization of GFP-FtsE to Z rings was also noted in a previous study (43).
Because FtsE and FtsX interact and the ftsEX gene cluster was more effective at complementing the ftsE null allele than ftsE alone, as shown above, we reasoned that including the ftsX gene downstream of the gfp-ftsE construct might provide the proper ratio of FtsE to FtsX to potentially stabilize the GFP-FtsE fusion. This indeed seemed to be the case, as expression of gfp-ftsE-ftsX from pDSW209-ftsE-ftsX gave rise to fluorescent bands at midcell in most cells in the population in a variety of strains that were proficient for cell division (Fig. 8A, C, E, and H). Moreover, the GFP-FtsE fusion protein made from pDSW209-ftsE-ftsX was functional, because it permitted colony formation of the ftsE::kan strain WM2497 on LB plates lacking added NaCl and produced fluorescent bands at division sites in these cells (data not shown). The failure of GFP-FtsE alone to localize was not simply because of protein degradation, as the low levels of the fusion protein were similar to those made in the presence of coproduced FtsX, as determined by immunoblotting with anti-GFP (data not shown). GFP-FtsE also did not cause destabilization of Z rings, because wild-type cells producing GFP-FtsE alone without extra FtsX divided mostly normally (data not shown).
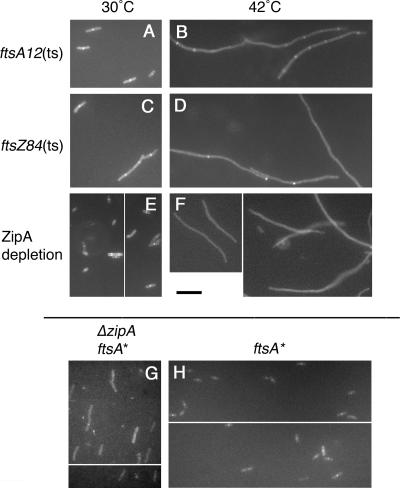
FIG. 8.
Requirements for localization of GFP-FtsE to Z rings. Shown are fluorescence micrographs of representative fields of cells producing GFP-FtsE and FtsX from plasmid pWM2800 in the ftsA12(Ts) strain WM1115 at 30°C (A) or after 90 min at 42°C (B), in the ftsZ84(Ts) strain WM1125 at 30°C (C) or after 90 min at 42°C (D), in the ZipA depletion strain WM1282 at 30°C (E) or after 5 h at 42°C (F), and in an ftsA* strain either lacking (G; WM1657) or containing (H; WM1659) the zipA gene, both at 30°C. Panels E, F, G, and H are subdivided to show two representative fields of cells. Scale bar, 10 μm.
We then tested whether localization of GFP-FtsE (again, with ftsX expressed in cis) to potential division sites required FtsZ, ZipA, or FtsA, with the idea that GFP-FtsE should be able to localize to Z rings in the absence of ZipA or FtsA if FtsE interacted directly with FtsZ. As expected, GFP-FtsE exhibited diffuse localization in ftsZ84(Ts) cells at 42°C, a temperature at which Z rings are absent in this mutant, indicating that FtsE localization to division sites requires the presence of Z rings (Fig. 8D). GFP-FtsE also failed to localize to Z rings in filaments depleted of ZipA (Fig. 8F) but localized proficiently to potential division sites in ftsA12(Ts) filaments at 42°C (Fig. 8B) or in filaments depleted of FtsA in strain WM1281 (data not shown). These results indicate that localization of FtsE was dependent on ZipA but independent of FtsA.
The dependence of FtsE localization on ZipA would seem at first to contradict the idea that FtsE and FtsZ interact independently of ZipA. However, our previous isolation of the ftsA* (R286W) mutant that could bypass the requirement for ZipA for cell division prompted a reassessment of how proteins are recruited to the divisome: proteins dependent on ZipA for recruitment, such as FtsK, FtsQ, or FtsN, are not actually recruited directly by ZipA but instead most likely depend on an indirect effect of ZipA on the Z ring that can be bypassed by an altered FtsA. As a result, we explored the possibility that the requirement for ZipA in FtsE recruitment was also indirect. GFP-FtsE (with coproduced FtsX) was localized in WM1657, which contains ftsA* and a deletion of zipA. As predicted, GFP-FtsE could localize to midcell in the slightly elongated cells of this strain (Fig. 8G), although the midcell bands were sometimes more difficult to visualize than those in the zipA+ parent strain, WM1659 (Fig. 8H). These results support the idea that targeting of GFP-FtsE to the Z ring does not require ZipA itself, but rather a state of the Z ring enhanced by ZipA.
Minicells induced by expression of flag-ftsE-ftsX.
Overproduction of FtsE and FtsX together, as well as FtsX alone, causes cell filamentation and death (11). Cell division is also inhibited when some other cell division proteins, such as FtsZ or ZipA, are overproduced to high levels, suggesting that stoichiometries of different components of the divisome are crucial for proper function (10, 12, 22). However, overproduction of FtsZ and FtsA severalfold over native levels resulted in the formation of minicells, which result from cytokinesis at cell poles, as well as midcell, indicating that cell division was being stimulated (2, 47). We were interested to explore whether production of lower levels of FtsE and/or FtsX might give rise to more subtle and perhaps more interesting cell division phenotypes in cells containing chromosomal ftsEX genes. Top10 cells were transformed with pWM2529, containing an IPTG-dependent flag-ftsE fusion as well as ftsX that was able to complement the ftsE::kan strain (Fig. 1D). Growth of the transformants in Top10, which also contains a chromosomal copy of ftsEX, required addition of 1% glucose to inhibit expression of the plasmid-borne flag-ftsE-ftsX. Colonies were then inoculated into LB broth with or without glucose, and the cell morphology was examined after a few hours of growth.
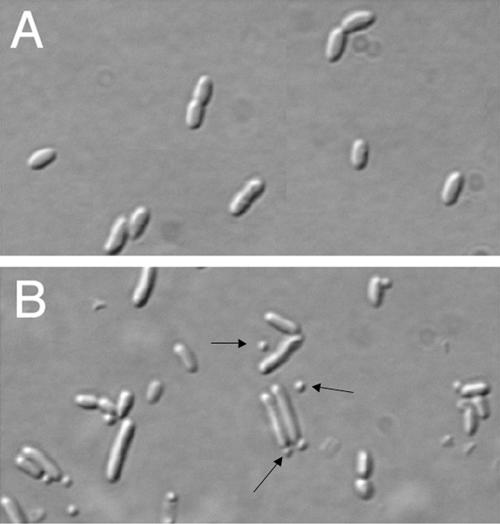
Cells grown in 1% glucose had a normal morphology, with only 0.3% of the cell population displaying a minicell phenotype (Fig. 9A) . However, cells grown without glucose, which presumably allowed higher expression levels of flag-ftsE-ftsX, failed to grow on agar plates, although they were able to grow in broth, where they exhibited a high proportion (∼20%) of minicells (Fig. 9B). It is not clear why cells without glucose were unable to form colonies on plates yet formed minicells without significant filamentation in broth, but this phenotype is reminiscent of moderate overproduction of FtsZ and FtsA and suggests that higher levels of FtsE and FtsX may make FtsZ more resistant to the MinC division inhibitor (38). A smaller proportion of minicells was also observed when flag-ftsE-ftsX from pWM2529 was expressed without IPTG in the ftsE::kan strain (data not shown). However, this effect was not observed in several other strain backgrounds, including TX3772, suggesting that a rather narrow window of FtsE-FtsX protein overproduction and/or strain background may be required to cause minicell formation.
FIG. 9.
Minicells in a strain expressing FLAG-FtsE-FtsX. Shown are composite differential interference contrast micrographs of Top10 cells containing the pflag-ftsE ftsX plasmid pWM2529 with (A) or without (B) glucose to control expression. The arrows highlight minicells.
DISCUSSION
Most of the proteins of the E. coli divisome have likely been identified, and several previous studies have identified or inferred a number of interactions among cell division proteins. However, the interactions between FtsE, which was recently demonstrated to be a component of the divisome, and other proteins of the divisome were not investigated in those studies.
In the present work, we identified an interaction between FtsE and FtsZ by coimmunoprecipitation of FLAG-FtsE and FtsZ from E. coli cell extracts. We also showed that affinity-purified FLAG-FtsE is enriched for FtsZ and that the interaction is not mediated via the FLAG epitope. These results suggest that FtsZ-FtsE interaction may be direct. This idea is supported by the ability of FLAG-FtsE to pull down FtsZ in the absence of functional FtsA, ZipA (with or without FtsA R286W), or FtsX (with or without suppressing levels of salt), which are the nearest neighbors of FtsE in the Z-ring recruitment pathway. The ability of FtsE to localize to Z rings in the absence of FtsA is entirely consistent with a direct interaction with FtsZ, as ZipA and ZapA, which interact directly with FtsZ, also localize to Z rings independently of FtsA. All other known E. coli divisome proteins require FtsA for their recruitment to the Z ring.
Nevertheless, if FtsE interacts directly with FtsZ, why would GFP-FtsE fail to localize to Z rings in ZipA-depleted filaments? ZipA is not well conserved among bacteria, and FtsA R286W can bypass the requirement for ZipA (14). However, FtsA R286W can also suppress other divisome defects, and the molecular mechanism of suppression by R286W is not known. Therefore, it is possible that R286W does not specifically replace ZipA but instead generally stabilizes divisome components, compensating for the weakening of the divisome when a component, such as ZipA, is inactivated. We propose that ZipA probably does not directly recruit cell division components that depend on ZipA for proper targeting to the Z ring, including FtsEX, but instead indirectly enhances the Z ring's ability to recruit these proteins. The mechanism of this enhancement is unknown but may be more efficient anchoring of the Z ring to the membrane (39) and/or increased bundling of FtsZ protofilaments (25, 41).
There are other potential explanations for the failure to detect GFP-FtsE rings in ZipA-depleted filaments. One is that FtsZ did not localize to rings in these filaments, which would prevent GFP-FtsE localization to division sites. However, while the frequency of rings was lower in ZipA-depleted strains producing GFP-FtsEX, many rings remained, ruling out this possibility (data not shown). Another possibility is that the GFP-FtsE fusion was unstable under these conditions. Immunoblotting indicated that the fusion was expressed at low but detectable levels after the several hours at 42°C needed to deplete ZipA compared to the time at 30°C, and there was no detectable increase in degradation of the fusion protein at 42°C (data not shown). It also should be noted that GFP-FtsE localized proficiently at 42°C in the ftsA(Ts) and FtsA depletion strains, so it is likely that protein degradation is not a major issue. We cannot, however, rule out possible subtle effects of the GFP tag on the conformation of FtsE, which would lead to the apparent dependence on ZipA.
The presence of FtsX, which interacts with FtsE, potentially complicates any interpretations of localization requirements. For example, FtsX may also interact directly with FtsA and FtsQ (28). In support of this, previous results showed that localization of GFP-FtsX to Z rings required both ZipA and FtsA (43), in contrast to the clearly FtsA-independent localization of FtsE we observed here. Because our GFP-FtsE fusion without coproduced FtsX did not localize well and because of high nonspecific fluorescence with the FLAG-tagged FtsE, we were unable to determine conclusively whether localization of FtsE depends on FtsX. The ability of FLAG-FtsE to coimmunoprecipitate FtsZ in the absence of FtsX, however, suggests that FtsE probably does not need FtsX for its targeting to the Z ring. This idea is consistent with the ability of GFP-FtsE to localize to Z rings independently of FtsA, in contrast to GFP-FtsX. However, it is also possible that FtsX helps target FtsE to Z rings via its association with the membrane. Such membrane targeting is important for the proper midcell localization of other FtsZ-interacting proteins, such as FtsA or the C-terminal domain of MinC (27, 39). The requirement for coproduced FtsX for localization of GFP-FtsE to Z rings suggests that the ratio between FtsE and FtsX is critical for their proper function. Another possibility, which cannot be ruled out at present, is that the GFP tag causes FtsE to fail to localize under conditions (such as lack of ZipA or coproduced FtsX) that are normally permissive for its localization.
The C-terminal tail of FtsZ is required for its interaction with FtsA and ZipA and anchoring to the cytoplasmic membrane (25, 33, 45). However, this domain of FtsZ is not required for interaction with FLAG-FtsE by coimmunoprecipitation. Although the SOS-inducible division inhibitor SulA binds to the conserved C-terminal globular domain of FtsZ (9), this is the first evidence of a domain of FtsZ apart from its C-terminal tail interacting with an essential divisome protein. Clearly, considerably more work will be needed to define precisely which determinants of FtsZ are necessary for its interaction with FtsE.
The role of FtsEX in E. coli cell division is unknown. The fact that FtsEX function is not essential for cell division at high osmotic strength suggests that other factors can compensate under some conditions. Indeed, it was recently found that expression of extra ftsQAZ or sufI can bypass the requirement for FtsEX (42). This is consistent with other reports of multiple overlapping functions of some other essential cell division proteins of E. coli (14, 15, 40). Whereas the localization of a number of cell division proteins is dependent on FtsEX (43), this dependence is probably a reflection of its effect on the Z ring and not direct recruitment by FtsEX, similar to the ZipA effect discussed above, because FtsEX is dispensable under certain conditions. Therefore, FtsEX, like ZipA, probably functions to maintain the integrity of the Z ring during its assembly and constriction. This idea is supported by the appearance of filamentous ΔftsE::kan cells grown in no salt, which have clear indentations that may be aborted attempts at septation. The efficient production of minicells when FtsEX is overproduced under certain conditions suggests that FtsEX has a stimulatory effect on Z-ring activity. It remains to be determined precisely how FtsEX influences the Z ring, what other proteins are contacted, and how the putative transporter function of FtsEX acts in this process.
Acknowledgments
We thank Eric Cascales, Kevin Morano, and members of his laboratory for helpful advice. We are grateful to Fred Blattner for providing the ftsE::kan strain, Harold Erickson for the FtsZ overproducer (renamed WM971), and Patrick Gibney of the Morano laboratory for the SSE1 and BAP control plasmids.
This work was supported by a grant from the National Institutes of Health (R01-GM61074) to W.M. and a UT-TORCH Training Grant (5 T32 DE015355-04) to Y.W.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Addinall, S. G., E. Bi, and J. Lutkenhaus. 1996. FtsZ ring formation in fts mutants. J. Bacteriol. 178:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begg, K., Y. Nikolaichik, N. Crossland, and W. D. Donachie. 1998. Roles of FtsA and FtsZ in activation of division sites. J. Bacteriol. 180:881-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernatchez, S., F. M. Francis, H. Salimnia, T. J. Beveridge, H. Li, and J. A. Dillon. 2000. Genomic, transcriptional and phenotypic analysis of ftsE and ftsX of Neisseria gonorrhoeae. DNA Res. 7:75-81. [DOI] [PubMed] [Google Scholar]
- 4.Beuria, T. K., S. S. Krishnakumar, S. Sahar, N. Singh, K. Gupta, M. Meshram, and D. Panda. 2003. Glutamate-induced assembly of bacterial cell division protein FtsZ. J. Biol. Chem. 278:3735-3741. [DOI] [PubMed] [Google Scholar]
- 5.Bi, E., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 6.Buddelmeijer, N., and J. Beckwith. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 52:1315-1327. [DOI] [PubMed] [Google Scholar]
- 7.Butland, G., J. M. Peregrin-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emili. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531-537. [DOI] [PubMed] [Google Scholar]
- 8.Corbin, B. D., B. Geissler, M. Sadasivam, and W. Margolin. 2004. A Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J. Bacteriol. 186:7736-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordell, S. C., E. J. Robinson, and J. Löwe. 2003. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc. Natl. Acad. Sci. USA 100:7889-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai, K., and J. Lutkenhaus. 1992. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J. Bacteriol. 174:6145-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Leeuw, E., B. Graham, G. J. Phillips, C. M. ten Hagen-Jongman, B. Oudega, and J. Luirink. 1999. Molecular characterization of Escherichia coli FtsE and FtsX. Mol. Microbiol. 31:983-993. [DOI] [PubMed] [Google Scholar]
- 12.Dewar, S. J., K. J. Begg, and W. D. Donachie. 1992. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J. Bacteriol. 174:6314-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Lallo, G., M. Fagioli, D. Barionovi, P. Ghelardini, and L. Paolozzi. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353-3359. [DOI] [PubMed] [Google Scholar]
- 14.Geissler, B., D. Elraheb, and W. Margolin. 2003. A gain of function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. USA 100:4197-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geissler, B., and W. Margolin. 2005. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol. Microbiol. 58:596-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill, D. R., and G. P. Salmond. 1987. The Escherichia coli cell division proteins FtsY, FtsE and FtsX are inner membrane-associated. Mol. Gen. Genet. 210:504-508. [DOI] [PubMed] [Google Scholar]
- 17.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15:514-526. [DOI] [PubMed] [Google Scholar]
- 18.Goehring, N. W., M. D. Gonzalez, and J. Beckwith. 2006. Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol. Microbiol. 61:33-45. [DOI] [PubMed] [Google Scholar]
- 19.Goehring, N. W., F. Gueiros-Filho, and J. Beckwith. 2005. Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli. Genes Dev. 19:127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goehring, N. W., C. Robichon, and J. Beckwith. 2007. A role for the non-essential N terminus of FtsN in divisome assembly. J. Bacteriol. 189:646-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hale, C. A., and P. A. de Boer. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175-185. [DOI] [PubMed] [Google Scholar]
- 23.Hale, C. A., and P. A. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hale, C. A., and P. A. de Boer. 2002. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J. Bacteriol. 184:2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hale, C. A., A. C. Rhee, and P. A. de Boer. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haney, S. A., E. Glasfeld, C. Hale, D. Keeney, Z. He, and P. de Boer. 2001. Genetic analysis of the Escherichia coli FtsZ-ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 276:11980-11987. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, J. E., L. L. Lackner, and P. A. de Boer. 2002. Targeting of (d)MinC/MinD and (d)MinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J. Bacteriol. 184:2951-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kempf, M. J., and M. J. McBride. 2000. Transposon insertions in the Flavobacterium johnsoniae ftsX gene disrupt gliding motility and cell division. J. Bacteriol. 182:1671-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, Z., A. Mukherjee, and J. Lutkenhaus. 1999. Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol. 31:1853-1861. [DOI] [PubMed] [Google Scholar]
- 31.Low, H. H., M. C. Moncrieffe, and J. Löwe. 2004. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol. 341:839-852. [DOI] [PubMed] [Google Scholar]
- 32.Ma, X., D. W. Ehrhardt, and W. Margolin. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA 93:12998-13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma, X., and W. Margolin. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margolin, W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell. Biol. 6:862-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margolin, W., and S. R. Long. 1994. Rhizobium meliloti contains a novel second copy of the cell division gene ftsZ. J. Bacteriol. 176:2033-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merino, S., M. Altarriba, R. Gavin, L. Izquierdo, and J. M. Tomas. 2001. The cell division genes (ftsE and X) of Aeromonas hydrophila and their relationship with opsonophagocytosis. FEMS Microbiol. Lett. 198:183-188. [DOI] [PubMed] [Google Scholar]
- 37.Oliva, M. A., S. C. Cordell, and J. Löwe. 2004. Structural insights into FtsZ protofilament formation. Nat. Struct. Mol. Biol. 11:1243-1250. [DOI] [PubMed] [Google Scholar]
- 38.Pichoff, S., and J. Lutkenhaus. 2001. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183:6630-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pichoff, S., and J. Lutkenhaus. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55:1722-1734. [DOI] [PubMed] [Google Scholar]
- 40.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raychaudhuri, D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18:2372-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy, M. 2007. Role of FtsEX in cell division of Escherichia coli: viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J. Bacteriol. 189:98-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt, K. L., N. D. Peterson, R. J. Kustusch, M. C. Wissel, B. Graham, G. J. Phillips, and D. S. Weiss. 2004. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaner, L., A. Trott, J. L. Goeckeler, J. L. Brodsky, and K. A. Morano. 2004. The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J. Biol. Chem. 279:2199-2201. [DOI] [PubMed] [Google Scholar]
- 45.Wang, X., J. Huang, A. Mukherjee, C. Cao, and J. Lutkenhaus. 1997. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, X. D., P. A. de Boer, and L. I. Rothfield. 1991. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 10:3363-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward, J. E., and J. Lutkenhaus. 1985. Overproduction of FtsZ induces minicells in E. coli. Cell 42:941-949. [DOI] [PubMed] [Google Scholar]
- 48.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 49.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, X.-C., and W. Margolin. 1999. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32:315-326. [DOI] [PubMed] [Google Scholar]
- 52.Yu, X. C., and W. Margolin. 2000. Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J. Bacteriol. 182:6203-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]