Abstract
Monovalent cation proton antiporter-3 (Mrp) family antiporters are widely distributed and physiologically important in prokaryotes. Unlike other antiporters, they require six or seven hydrophobic gene products for full activity. Standard fluorescence-based assays of Mrp antiport in membrane vesicles from Escherichia coli transformants have not yielded strong enough signals for characterization of antiport kinetics. Here, an optimized assay protocol for vesicles of antiporter-deficient E. coli EP432 transformants produced higher levels of secondary Na+(Li+)/H+ antiport than previously reported. Assays were conducted on Mrps from alkaliphilic Bacillus pseudofirmus OF4 and Bacillus subtilis and the homologous antiporter of Staphylococcus aureus (Mnh), all of which exhibited Na+(Li+)/H+ antiport. A second paralogue of S. aureus (Mnh2) did not. K+, Ca2+, and Mg2+ did not support significant antiport by any of the test antiporters. All three Na+(Li+)/H+ Mrp antiporters had alkaline pH optima and apparent Km values for Na+ that are among the lowest reported for bacterial Na+/H+ antiporters. Using a fluorescent probe of the transmembrane electrical potential (ΔΨ), Mrp Na+/H+ antiport was shown to be ΔΨ consuming, from which it is inferred to be electrogenic. These assays also showed that membranes from E. coli EP432 expressing Mrp antiporters generated higher ΔΨ levels than control membranes, as did membranes from E. coli EP432 expressing plasmid-borne NhaA, the well-characterized electrogenic E. coli antiporter. Assays of respiratory chain components in membranes from Mrp and control E. coli transformants led to a hypothesis explaining how activity of secondary, ΔΨ-consuming antiporters can elicit increased capacity for ΔΨ generation in a bacterial host.
Monovalent cation/H+ antiporters of bacteria have critical roles in alkali tolerance, efflux of toxic monovalent cations, and establishment of a sodium gradient that can drive solute uptake and motility (44, 46). Members of the family of monovalent cation/H+ antiporters referred to here as the Mrp family have diverse designations, including Mrp (19), Mnh (17), Pha (48), Sha (31), and Sno (3), and are widely distributed among physiologically diverse prokaryotes, including numerous pathogenic bacteria (56). Important physiological functions have been attributed to the monovalent cation/H+ activity of Mrp antiporters, since mutational loss or compromise of this activity has been associated with: alkali and Na+ sensitivity in alkaliphilic Bacillus (14) and Anabaena (4); Na+ sensitivity, alkali sensitivity, and a sporulation defect in Bacillus subtilis (19, 20, 31, 32); a growth defect in Staphylococcus aureus (3, 24); and Na+ sensitivity and decreased virulence in Pseudomonas aeruginosa (33). Several bacterial strains have more than one Mrp system. In Sinorhizobium species, where dual systems have been named Pha1 and Pha2, mutational loss of Pha1 results in K+ sensitivity and a nitrogen fixation defect (48) while loss of Pha2 results in Na+ sensitivity (67). The recent observation of increased Mrp expression under specific metabolic conditions, e.g., during growth of the archaeon Methanosarcina acetivorans on acetate (35), suggests that additional settings in which Mrp has important roles will continue to be identified.
In spite of their distribution and importance, Mrp antiporters are still very incompletely characterized, but they are clearly unique among monovalent cation/H+ antiporters. Mrp antiporters constitute their own family, the cation proton antiporter-3 (CPA-3) family (TC 2.A.63), in the sequence-based transporter classification (TC) because of particular sequence features and apparent complexity (6, 54). Mrp antiporter systems are encoded by six or seven gene operons that are highly conserved. All the genes are required for full function of Mrp antiporters in Na+ and alkali resistance (17, 20). Each mrp gene product is a hydrophobic membrane protein that is predicted to span the membrane multiple times. Three of them, MrpA, MrpC, and MrpD, exhibit significant sequence similarity to hydrophobic subunits that are part of the H+-translocating domains of complexes such as respiratory chain complex I and hydrogenases (40, 41, 56). By contrast, all other monovalent cation/proton antiporters are encoded by a single gene with a single gene product that functions as a monomer or homodimer (11, 53). Mrp proteins have been proposed but not yet shown to form a functional complex (17, 56). We have hypothesized that such a complex is a consortium of distinct transporters that includes one or more Na+/H+ antiporters (56, 58), which were proposed by Mathiesen and Hägerhäll to be MrpA and MrpD (40). The putative Mrp complex is also expected to include one or more anion transporters whose particular substrate(s) may be species specific (56, 58). This is suggested by roles of Mrp in bile salt and arsenite resistance in B. subtilis and Agrobacterium tumefaciens, respectively (19, 20, 27). A multifunctional complex could provide synergies such as the presentation of a large protein surface area on the external membrane surface. This could enhance proton capture in support of the monovalent cation/proton antiport function that supports alkali resistance (56, 58).
To date, little exploration has been initiated on Mrp activities other than monovalent cation/H+ antiport, and this activity of Mrp has only been minimally characterized, even for the most intensively studied B. subtilis Mrp (19, 20, 21, 31, 34). Like the activities of well-characterized prokaryotic antiporters that are single gene products, Mrp-dependent antiport is secondary antiport that is energized by the respiration-generated electrochemical proton gradient, Δp, as demonstrated in whole-cell and membrane vesicle assays (29, 46, 56). In addition, an imposed transmembrane potential is sufficient to energize Na+ efflux by Bacillus Mrp antiporters from whole cells (14, 19, 20); this is consistent with movement of net charge during Mrp antiport, i.e., with an electrogenic antiport. Electrogenicity is an important property for Na+/H+ antiporters that support alkali resistance by generating a pH gradient in which the cytoplasm is more acidic than the external medium (38, 46). However, neither kinetic properties of Mrp antiport nor a definitive in vitro demonstration of electrogenicity has been presented for any Mrp system. The standard fluorescence-based antiporter assays of these properties are usually conducted with everted (inside-out) membrane vesicles from antiporter-deficient Escherichia coli strains expressing the test antiporter (12, 52). Such assays of Mrp systems have thus far yielded signals that were too low for kinetic analyses or assays of transmembrane electrical potential (ΔΨ) consumption (17, 21, 33, 34).
Here, an optimized assay for secondary Mrp antiport in E. coli EP432 (ΔnhaA ΔnhaB) (47) facilitated a more detailed characterization of secondary Mrp antiport than has heretofore been possible. The Mrp systems chosen for study were from three gram-positive bacteria, Bacillus pseudofirmus OF4 Mrp (MrpBpOF4), B. subtilis Mrp (MrpBs), and S. aureus Mnh. The study included a second S. aureus Mrp system (designated Mnh2), of unknown catalytic capacity or function, that is found in staphylococci. All four Mrp systems are “group 1” Mrp systems that are encoded by canonical seven-gene operons (mrpA to mrpG) (56). The substrates for Mnh2 were not identified, but results with the other three Mrp systems show that they catalyze secondary, electrogenic Na+(Li+)/H+ antiport with a very low apparent Km value for Na+. In addition, experiments conducted here demonstrate that E. coli EP432 transformants expressing MrpBpOF4 or Mnh from a plasmid exhibit a higher capacity to generate a transmembrane potential, the ΔΨ (inside negative relative to outside), than a control transformant. Plasmid-borne NhaA, the extensively studied electrogenic Na+(Li+)/H+ antiporter of E. coli (18, 45), produced the same effect on ΔΨ generation. The host response to the presence of an electrogenic antiporter in multicopy is of interest because it mimics situations in which such antiporters are activated in their natural settings. The nature of the host response that results in increased ΔΨ generation in the current experimental system was therefore probed by assays of respiratory chain components in the E. coli EP432 transformant expressing MrpBpOF4 versus a control.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. coli strains used in this study were DH5αMCR (Gibco-BRL), KNabc (ΔnhaA ΔnhaB ΔchaA) (43), and EP432 (ΔnhaA ΔnhaB) (47) (Table 1). The strains were grown routinely in LBK medium, pH 7.5 (12), at 37°C. The plasmids used in this study were pMW118 (Nippon Gene), pGEM-3Zf(+) (Promega), and the recombinant pGEM-3zf(+) derivatives containing MrpBs, MrpBpOF4, Mnh, or Mnh2 (Table 1). The operons cloned in these four recombinant plasmids of pGEM-3Zf(+) contained the B. subtilis mrp operon, the B. pseudofirmus OF4 mrp operon, the S. aureus mnh operon, and the S. aureus mnh2 operon, respectively; the operons were cloned behind their own promoters. They were designated, respectively, MrpBs, MrpBpOF4, Mnh, and Mnh2. For the construction of MrpBs and MrpBpOF4, the starting material was a pair of recombinant plasmids constructed earlier, pMWBSMrp and pMWOFMrp, respectively, containing the full operons with their promoters in the low-copy plasmid pMW118 (21). For isolation of MrpBs, pMWBSMrp was digested with EcoRI and XbaI, and then the purified product, containing the mrp operon and its promoter region, was ligated into EcoRI- and XbaI-digested pGEM-3Zf(+) using Quick ligase (New England Biolabs). For isolation of MrpBpOF4, pMWOFMrp was digested with EcoRI and XmaI, and then the purified product, containing the mrp operon and its putative promoter region, was ligated into EcoRI- and XmaI-digested pGEM-3Zf(+) using Quick ligase. For construction of a recombinant plasmid bearing S. aureus Mnh, PCR was performed on S. aureus RF4220 chromosomal DNA with primers SaMnh1-F-NheI and SaMnh1-R-XhoI. SaMnh1-F-NheI (5′-GCT AGC TTG TTA CAT ATT GCG GTG-3′) corresponded to the sequence of the database entry GenBank accession no. DQ659238 and additional nucleotides containing an NheI site at the 5′ end of the sequence. SaMnh1-R-XhoI (5′-CTC GAG TTT TGT GTC TTT TAA GTC TTC CG-3′) corresponded to the complementary sequence of the database entry GenBank accession no. DQ659238 and additional nucleotides containing an XhoI site at the 5′ end of the sequence. The PCR product was ligated into SmaI-digested pMW118. After sequence confirmation, the recombinant plasmid was digested with EcoRI and HindIII and ligated into EcoRI- and HindIII-digested pGEM-3Zf(+). For construction of pGEMSaMnh2, PCR was performed on S. aureus RF4220 chromosomal DNA with primers SaMnh2-F-XbaI and SaMnh2-R-XhoI. SaMnh2-F-XbaI (5′-TCT AGA AAC AAA GGA GGC TAA TAA TGA GTT TGG-3′) corresponded to the sequence of the database entry GenBank accession no. DQ659239 and additional nucleotides containing an XbaI site at the 5′ end of the sequence. SaMnh2-R-XhoI (5′-CTC GAG ATC GTT TTG ATA CCA TTT CTT ACG-3′) corresponded to the complementary sequence of the database entry GenBank accession no. DQ659239 and additional nucleotides containing an XhoI site at the 5′ end of the sequence. The PCR product was ligated into SmaI-digested pMW118. After sequence confirmation, the recombinant plasmid was digested with EcoRI and HindIII and ligated in EcoRI- and HindIII-digested pGEM-3Zf(+). For all the plasmid selections, blue-white screening in E. coli DH5α was employed and complete DNA sequencing was used to confirm that the plasmids ultimately used were free of errors. The plasmids were transformed into E. coli KNabc and EP432 for use in the experiments. Plasmid preparations made from those parallel transformants indicated that the transformant with pGEMBsMrp consistently had a lower level of plasmid than transformants with the other three mrp-bearing plasmids. Therefore, direct quantitative comparisons of the total activity or complementation capacities of MrpBs with those of the other systems could not be made.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| EP432 | melB(LiDa) nhaA1::kan ΔnhaB1::cam ΔlacZY thr-1 | 47 |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Gibco-BRL |
| KNabc | TG1 (ΔnhaA ΔnhaB ΔchaA) | 43 |
| Plasmids | ||
| pMW118 | Cloning vector; Ampr | NipponGene, Toyama, Japan |
| pMWBSMrp | pMW118(+) derivative; contains the full mrp operon from B. subtilis | 21 |
| pMWOF4Mrp | pMW118(+) derivative; contains the full mrp operon from B. pseudofirmus OF4 | 21 |
| pMWSaMnh | pMW118(+) derivative; contains the full mnh operon from S. aureus | This study |
| pMWSaMnh2 | pMW118(+) derivative; contains the full mnh2 operon from S. aureus | This study |
| pGEM-3zf | Cloning vector; Ampr | Promega |
| pGEMBs-Mrp | pGEM-3Zf(+) derivative; contains the full mrp operon from B. subtilis | This study |
| pGEMBpOF4-Mrp | pGEM-3Zf(+) derivative; contains the full mrp operon from B. pseudofirmus OF4 | This study |
| pGEMMnh | pGEM-3Zf(+) derivative; contains the full mnh operon from S. aureus | This study |
| pGEMMnh2 | pGEM-3Zf(+) derivative; contains the full mnh2 operon from S. aureus | This study |
| pBR322 | Cloning vector; Ampr | New England Biolabs |
| pGM36 | pBR322 derivative; contains the full nhaA gene from E. coli | 12 |
LiD, Li+ dependent.
Preparation of everted membrane vesicles.
Everted membrane vesicles were prepared by breaking cells with a French Pressure cell as described by others (1, 52); the buffer used in the preparations was 10 mM bis-[tris(hydroxymethyl)methylamino]-propane (BTP) (pH 7.5), 10% glycerol, a protease inhibitor tablet (Roche), and 1 mM phenylmethylsulfonyl fluoride for the assays of ΔΨ generation. For the antiport assays, the buffer used to prepare everted membrane vesicles was 10 mM Tris-HCl (pH 7.5) containing 140 mM choline chloride, 0.5 mM dithiothreitol, 10% glycerol, a protease inhibitor tablet (Roche), and 1 mM phenylmethylsulfonyl fluoride. For storage, glycerol (to 10%, vol/vol) was added to all everted membrane vesicles, and the vesicles were shock frozen in liquid nitrogen and then stored at −80°C. Protein content was measured by the method of Lowry et al., using lysozyme as the standard (37).
Assays of ΔpH-dependent antiport activity.
Antiport assays were conducted in 10 mM BTP-sulfate, 140 mM choline-Cl, 5 mM MgSO4, and 1 μM acridine orange at pH 7.0 to 9.0. Measurements were conducted using a Perkin-Elmer LS50B luminescence spectrometer with excitation at 420 nm (10-mm slit) and emission at 500 nm (10-mm slit). Respiration was initiated by the addition of Tris-succinate to a final concentration of 2.5 mM. The high chloride content ensured that the Δp established by the addition of the electron donor was entirely in the form of a ΔpH, acid inside the everted vesicles. Establishment of this ΔpH was monitored by quenching of acridine orange fluorescence. Dequenching of fluorescence, which results from subsequent cation addition, reflected cation-dependent proton movement out of the everted vesicles (12). Addition of 10 mM ammonium chloride was used to dissipate the remaining Δp to bring the fluorescence back to baseline. The concentration of cation yielding the half-maximal dequenching has been validated as a good estimate of the apparent Km of monovalent cation/H+ antiporters (51, 61).
Fluorescence-based assays of ΔΨ generation and antiporter-dependent consumption.
ΔΨ-dependent fluorescence of oxonol VI was used to measure the generation of a positive inside potential with the addition of 1 mM NADH. Evaluation of electrogenicity was conducted by adding 10 mM NaCl to energized membranes and observing a reversal of the quench. The reversal of quenching represents antiport-dependent consumption of the ΔΨ, which reflects the activity of an electrogenic antiport that can be energized by, and thus partially dissipate, ΔΨ (46). Addition of 10 μM carbonyl cyanide m-chlorophenylhydrazone, a protonophore that can abolish the ΔΨ, was used to bring the fluorescence back to baseline. The assay mixture contained 10 mM BTP, 5 mM MgSO4, 200 mM K2SO4, 1 μM nigericin (pH 7.5), and 1 μM oxonol VI. Measurements were conducted on a Perkin-Elmer LS50B luminescence spectrometer. The excitation wavelength was set at 580 nm with a 10-mm slit, and emission was at 631 nm with a 10-mm slit (64). The final concentration of vesicle protein was 200 μg/ml.
Assays of respiratory chain activities in everted membrane vesicles.
Succinate dehydrogenase assays were carried out according to the method of Hatefi and Stiggall (15) by phenazine methosulfate-mediated bleaching of the electron acceptor 2,6-dichloroinophenol (DCPIP), using a Shimadzu UV-1601 UV-visible spectrophotometer. This approach was also used for assays of malate:quinone oxidoreductase as recently described (57). Assays were conducted at room temperature using 100 μg of vesicle protein in 1 ml of 10 mM BTP-sulfate and 5 mM MgSO4 (pH 7.5). NADH oxidase assays were carried out using the standard method of monitoring A340 over time in the presence of 1 mM NADH (66). The NADH-K3Fe(CN)6 reductase activity was measured at 420 nm with added 10 mM KCN, 1 mM NADH, and 1 mM K3Fe(CN)6 as described previously (25). The extinction coefficients used for activity calculations were ɛ340 = 6.22 mM−1 cm−1 for NADH and deamino-NADH (d-NADH) (reduced nicotinamide hypoxanthine dinucleotide), ɛ420 = 1.00 mM−1 cm−1 for K3Fe(CN)6, and ɛ600 = 2 mM−1 cm−1 for DCPIP (2). The NADH analogue d-NADH is a substrate for the proton-pumping Nuo complex but not for the non-proton-pumping Ndh NADH dehydrogenase of E. coli (42).
Cytochrome spectra.
Absorption spectra were recorded at room temperature with a Perkin-Elmer 550 dual-beam spectrophotometer. Scans were performed at 2 nm/s with a slit width of 2 nm. Samples containing 2 mg/ml of membrane protein in 50 mM Tricine-NaOH (pH 8)-0.1% lauryl maltoside were evaluated. A baseline correction was obtained, and then reduced-minus-air-oxidized difference spectra were obtained by addition of a small amount of sodium dithionite and incubation for 1 min. The following wavelength pairs and millimolar extinction coefficients were used: cytochrome b, ΔA560-575 and Δɛ = 17.5; cytochrome d, ΔA622-650 and Δɛ = 18.8 (16, 28, 49).
RNA preparation and real-time RT-PCR.
Cells were grown overnight, and total RNA was isolated after cell lysis with glass beads (0.5 mm) using the RNeasy minikit (QIAGEN). RNA (1 μg) was used for reverse transcription in a reaction mixture of 20 μl. Real-time PCR was performed in a LightCycler (Roche) with a QuantiTect SYBR green reverse transcriptase (RT-PCR) kit (QIAGEN). Primers were designed using the LC probe design software and selected based on an amplicon size of 100 bp and a melting temperature at 55°C. The two genes studied were the succinate dehydrogenase gene (sdh) and the 16S rRNA gene (rrsA). The primer sequences used were the following: rrsA forward, 5′-AGAGATGAGAATGTGC-3′; rrsA reverse, 5′-CACTTTATGAGGTCCG-3′; sdh forward, 5′-TGCTGAACAAACATGG-3′; and sdh reverse, 5′-GTGACAGGTTGGGATA-3′. Reverse transcription was carried out at 50°C for 30 min, and then amplification conditions were as follows: an initial denaturation step for 15 min at 94°C and then 45 cycles of denaturation at 94°C (15 s), annealing at 55°C (25 s), and elongation at 72°C (10 sec). Melting curve analysis was performed to check for a single amplicon and the absence of primer-dimers. LightCycler analysis software was used for determining crossing points. Data were analyzed by the 2−ΔΔCT method and are presented as fold induction/repression of sdh, normalized to rrsA levels (36).
RESULTS AND DISCUSSION
MrpBs, MrpBpOF4, and Mnh complement the Na+ and alkali sensitivities of E. coli EP432.
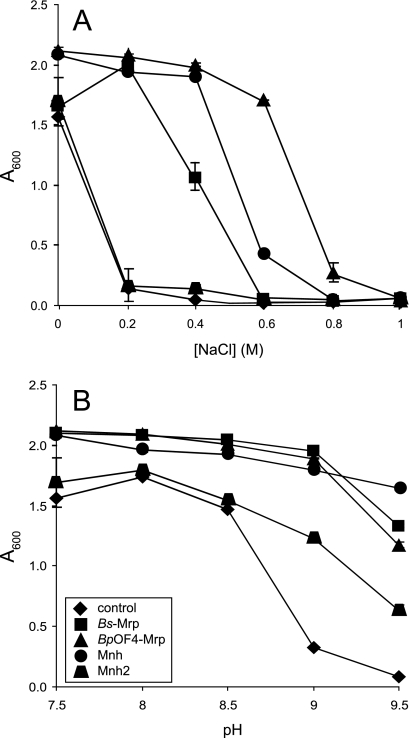
The Na+-sensitive E. coli strain EP432, harboring the plasmid control, is highly sensitive to growth inhibition by NaCl concentrations of 200 mM and above (Fig. 1A). The E. coli EP432-Mnh transformant exhibited marked Na+ resistance at up to 400 mM, with the MrpBpOF4 transformant supporting resistance at up to 600 mM. The Mnh2 transformant did not confer Na+ resistance (Fig. 1A). The B. subtilis Mrp complemented to a lesser extent, which perhaps is related to the lower levels of plasmid in this transformant (see Materials and Methods). All four Mrp systems, including the Mnh2 transformant, supported a high growth yield compared to the control transformant at pHs of up to 9.5 in the absence of added Na+ (Fig. 1B).
FIG. 1.
Effect of NaCl concentration and pH on the growth of E. coli EP432 transformants of Mrp-Mnh antiporters. (A) Transformants with empty vector [pGEM-3Zf(+)], or expressing the Mrp operons from B. subtilis, B. pseudofirmus OF4, or S. aureus were grown on LBK medium, pH 7.5, containing added NaCl at the indicated concentrations. (B) LBK with no added NaCl was adjusted to the pH values indicated. The LBK contained approximately 12 mM contaminating Na+ (62). Cells were grown overnight for 16 h at 37°C, with shaking, after which the A600 of the cultures was measured. The error bars indicate standard deviations from duplicate cultures in three independent experiments.
MrpBs, MrpBpOF4, and Mnh catalyze secondary Na+(Li+)/H+ antiport with a low apparent Km for Na+ and an alkaline pH optimum.
Previous in vitro studies of Mrp-dependent antiport activity via the widely used fluorescence assay (12) employed everted vesicles from the triple antiporter mutant E. coli KNabc (43) transformed with a low-copy-number plasmid expressing the test operons. In this assay, energization of everted membrane vesicles by addition of an electron donor results in development of a ΔpH, acid in, in the everted vesicles. The ΔpH is monitored by quenching of acridine orange fluorescence until steady state is reached. Antiport is then assessed by the percent dequenching resulting from addition of the monovalent cation substrate for antiport. In the earlier Mrp assays conducted with vesicles of E. coli KNabc by our group and others, the percent dequenching that reflected antiport activity was ≤6% dequenching for MrpBs/Sha (21, 34), MrpBpOF4 (21), and Mnh (17). Attempts to achieve significantly higher signals for Mrp-dependent antiport in this triple mutant were made using different vectors and induction protocols, but these were without success because high expression of the Mrp antiporters led to poor growth or poor vesicle quality (data not shown). Further attempts to empirically optimize the fluorescence assay for Mrp systems were therefore mounted in the less impaired double mutant E. coli EP432. This strain differs from E. coli KNabc in retaining a functional chaA gene that encodes an Na+(Ca2+)(K+)/H+ antiporter (23, 50). The background activity in membranes from this strain either can be abolished by the addition of KCl to the assay buffer for antiporters that do not use K+ as a substrate or is small enough to be subtracted from the dequenching observed in assays with other substrates (Table 2); in the current studies, any small background activity was subtracted. In addition to using E. coli EP432, the new antiport assay protocol used the pGEM-3Zf(+) vector instead of the lower-copy-number vector pMW118 and used BTP in the assay buffer instead of Tris, which had been included in our earlier assays (21). The new protocol yielded larger signals for Mrp-dependent activity, i.e., percent dequenching of as high as 40 to 50% (Table 2).
TABLE 2.
Assays of monovalent cation/proton antiport activity at pH 8.5 in everted membrane vesicles of Mrp-expressing E. coli EP432 transformants
| Transformant | % Dequenching observed upon addition ofa:
|
||
|---|---|---|---|
| Na+ | Li+ | K+ | |
| Control | 2.6 ± 0.7 | 1.1 ± 0.0 | 2.6 ± 1.2 |
| MrpBs | 17.3 ± 2.6 | 20.3 ± 0.9 | 4.3 ± 0.5 |
| MrpBpOF4 | 37.2 ± 0.8 | 31.5 ± 1.4 | 2.6 ± 1.1 |
| Mnh | 50.7 ± 0.6 | 46.7 ± 1.3 | 1.8 ± 0.5 |
| Mnh2 | 0 | 0 | 0 |
Vesicles from transformants expressing empty vector [pGEM-3zf(+)] and the Mrp operons from B. subtilis, B. pseudofirmus OF4, and S. aureus (Mnh and Mnh2) were assayed in 2 ml containing 50 mM BTP-sulfate buffer, 140 mM choline chloride, 5 mM MgCl2, 1 μM acridine orange, and 66 μg of vesicle protein. Respiration was initiated by the addition of Tris-succinate to a final concentration of 2.5 mM. After steady-state fluorescence quenching was reached, NaCl, LiCl, or KCl was added to a final concentration of 2.5 mM. The values presented for the subsequent percent dequenching are from duplicate assays from three independent experiments. The percentages represent the average values of the calculated percent dequenching and are shown with the standard deviation of the values.
Assays were first performed to characterize the cation specificity of secondary Mrp-dependent antiport. Initial experiments were conducted at pH 8.5 in anticipation of an alkaline optimum for these antiporters. MrpBpOF4 and Mnh exhibited antiport in the presence of test cations Na+ and Li+ (2.5 mM) at pH 8.5 (Table 2). MrpBs also exhibited significant Na+- and Li+-dependent antiport, although the levels were lower than those observed with MrpBpOF4 and Mnh. Significant K+/H+ antiport was not observed with any of the three antiporters using a range of K+ concentrations of from 2 to 400 mM (Table 2 and data not shown). The possibility of modest Mrp-dependent K+/H+ antiport activity had been raised by physiological experiments on B. subtilis Mrp (19), and the assay data here similarly show a hint but certainly not highly significant levels of such activity, even though some other Mrp systems (e.g., the Pha1 antiporter of S. meliloti) are active K+/H+ antiporters (48). Neither Mg2+- nor Ca2+-dependent antiport was observed for any of the four antiporters in assays conducted in buffers without added Mg2+ (not shown).
Mnh2 from S. aureus, for which no earlier studies had been reported, exhibited no activity with any test cation at 2.5 mM at pH 8.5 (Table 2). Mnh2 also did not exhibit antiport activity at pH 8.5 with any of the monovalent cations when added at concentrations of up to 200 mM at a range of pH values of from 7.0 to 9.0. Finally, assays of Mnh2 were conducted in the absence of Mg2+ to test the possibility that this system has cation/proton activity that is inhibited by Mg2+. These assays were also negative (not shown). It was interesting that the low background level observed in control vesicles was suppressed in the Mnh2-expressing vesicles. This is consistent with an Mnh2 antiporter whose activity during growth repressed the modest background antiport in the heterologous host. Mnh2 may use an unusual efflux substrate and perhaps may require a cosubstrate on the external side to couple with the H+ uptake that is inferred for Mnh2 by its capacity to complement alkali sensitivity in E. coli EP432 (e.g., a citric-malic or malic-lactate type of exchange). The distinct subset of second Mrp-type systems of which Mnh2 is a member is found only in Staphylococcus species (56), so Mnh2 may play a role that is specific to the ecology-metabolism interplay of this pathogen.
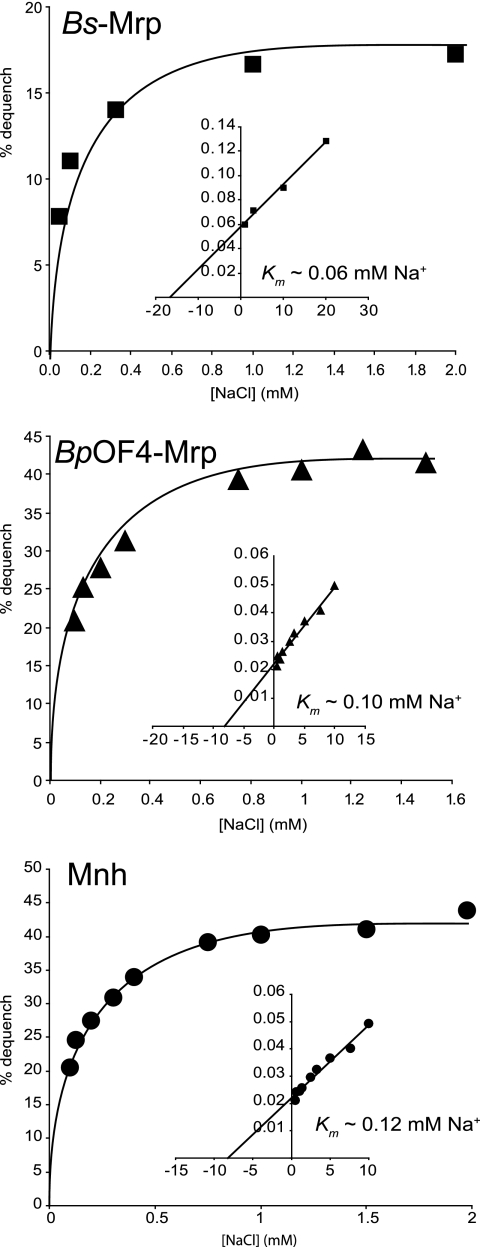
The Na+/H+ antiport of the three active Mrp antiporters was examined over a range of concentrations of added NaCl. Michaelis-Menten kinetics were observed (Fig. 2). The linear regression of a double-reciprocal plot (Fig. 2, inset) allowed calculation of apparent Km values at pH 8.5 (Fig. 2). Although the level of Na+-dependent MrpBs antiport was lower than that of the other two Mrp systems, apparent Km values were of a comparable order of magnitude for all three antiporters at pH 8.5, in the range of 0.06 to 0.12 mM for Na+ (Fig. 2). These low apparent Km values for Na+ (Fig. 2) are within the range of the lowest reported for a prokaryotic monovalent cation/H+ antiporter (9, 61). This property could facilitate acidification of cytoplasmic pH even at low concentrations of Na+ (20, 22).
FIG. 2.
Na+/H+ antiport activity of Mrp antiporters as a function of cation concentration at pH 8.5. Fluorescence-based assays of the Na+/H+ antiport activities of the Mrp antiporters in E. coli EP432 vesicles were conducted at pH 8.5 over a range of concentrations of added NaCl; the Michaelis-Menten plot is shown, and a reciprocal plot is shown as an inset in each panel. The assay protocol was identical to that described in Table 2, footnote a. The standard deviations of the values, which are derived from duplicate assays in at least three independent experiments, were less than 10% of the mean values.
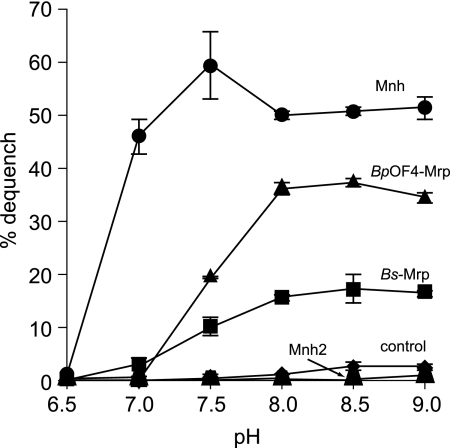
The activity profile of MrpBpOF4 as a function of pH indicated an optimum for Na+/H+ antiport between pH 8.0 and 9.0, with greatly reduced activity at pH 7.5 and no activity at pH 7.0 and below (Fig. 3). This profile is consistent with the inability of B. pseudofirmus OF4 to grow nonfermentatively at pH 7 (55) and its ability to maintain a cytoplasmic pH of around 8.2 while growing at pH 10.5 (55, 68). By contrast, Mnh exhibited Na+/H+ antiport activity over a broader range of pH values, from pH 7.0 through pH 9.0; S. aureus exhibits strong Na+ and alkali resistance that is consistent with this profile (63, 65). Significant MrpBs-dependent Na+/H+ antiport, albeit less active than the others, was observed at between pH 7.5 and 9.0, with retention of very modest activity relative to the control membranes at pH 7.0 but not 6.5 (Fig. 3). It is notable that a mrp null mutant of B. subtilis is profoundly sensitive to growth inhibition by Na+ at pH 7.0, probably accounting for the listing of mrp genes among essential genes in experiments conducted in LB medium which contains added Na+ (20, 30).
FIG. 3.
Na+/H+ antiport activity of Mrp antiporters as a function of pH. The assay protocol was as described in the legend to Fig. 2, with the pH of the buffers adjusted to the values indicated. The data shown are for assays with 2.5 mM NaCl. The error bars indicate standard deviations from duplicate assays from three independent experiments.
Mrp-dependent Na+/H+ antiport is electrogenic.
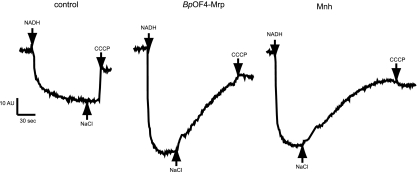
Secondary antiporters that catalyze net H+ accumulation in support of alkaline pH homeostasis are expected to be electrogenic (46), catalyzing an Na+/H+ exchange in which the inward H+ flux is greater than the outward Na+ flux during a single turnover. During such an electrogenic exchange, net positive charge is translocated inward. The antiport can be energized by the transmembrane electrical potential, i.e., the ΔΨ (negative inside in whole cells), thus partially dissipating ΔΨ during antiport. MrpBs antiport was previously inferred to be electrogenic from whole-cell experiments (19, 20), and its lower level of activity than the MrpBp and Mnh in the current study led us to test only MrpBpOF4 and Mnh antiport for electrogenicity in the in vitro assay. The fluorescent ΔΨ probe oxonol VI was used; this probe monitors the development of the ΔΨ, positive inside, that is established in the everted vesicle system upon addition of an electron donor to the respiratory chain. NADH was added to energize the vesicles under conditions in which the presence of nigericin prevents development of a transmembrane pH gradient. The resulting quench in oxonol fluorescence indicates the formation of the ΔΨ. After the ΔΨ reaches steady state, NaCl is added to evaluate whether a reversal of the fluorescence quench occurs in the Mrp-expressing vesicles. Such dequenching monitors Na+- and Mrp-dependent consumption of the ΔΨ, i.e., electrogenic Na+/H+ antiport, and was observed for membranes containing either MrpBpOF4 or Mnh (Fig. 4).
FIG. 4.
Electrogenicity of Mrp-dependent Na+/H+ antiport. Fluorescence-based assays of everted membrane vesicles from the control, MrpBpOF4, and Mnh transformants of E. coli EP432 were performed in reaction mixtures containing 200 μg vesicle protein/ml in a total of 2 ml. The reaction mixtures contained 50 mM BTP-sulfate, 5 mM MgSO4, 200 mM K2SO4, and 1 μM nigericin at pH 7.5, plus 1 μM oxonol VI. To initiate respiration, 1 mM Tris-NADH was added at the first arrow. Once the quenching achieved steady state, 10 mM NaCl was added at the second arrow. The final arrow indicates the addition of 10 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP). The traces shown are representative of at least three independent experiments.
MrpBpOF4 and Mnh increase ΔΨ generation by the E. coli EP432 host, as does plasmid-borne NhaA, an electrogenic E. coli antiporter.
In the fluorescence-based assays of electrogenicity, it was observed that both MrpBpOF4 and Mnh membranes consistently exhibit a larger initial NADH-dependent oxonol quench than the control membranes (see the magnitude of the middle and right traces shown in Fig. 4 compared to that of the control on the left during the interval between NADH addition and addition of NaCl). This indicates that membranes from these two Mrp-expressing transformants of E. coli EP432 cells produce a greater respiration-dependent ΔΨ than the control transformant. What might account for a greater capacity of the MrpBpOF4 and Mnh vesicles of E. coli EP432 to generate a ΔΨ than the control vesicles? The cells were all grown under the same conditions. The variable is the presence of electrogenic antiporters. These antiporters would have been very active under the conditions of growth, since the antiporters are expressed from multicopy plasmids, further alkalinization of the pH 7.5 medium occurs during growth (8), and there is sufficient contaminating Na+ to support MrpBpOF4- and Mnh-dependent antiport. We hypothesized that the following scenario leads to a higher capacity for ΔΨ generation under these conditions: (i) the high activity of ΔΨ-consuming antiporters depletes the ΔΨ; (ii) the ΔΨ depletion elicits a host response that leads to a compensatory increase in the capacity of the host to generate ΔΨ, i.e., by increasing levels of key respiratory chain components and/or enzymes that donate electrons to the respiratory chain; and (iii) the cell thus adapts to the high level of antiport activity without compromising other ΔΨ-dependent functions. Host responses of this type could be mediated by two-component signaling or other regulatory elements that respond directly to reduced ΔΨ or reduced total Δp. Alternatively, they could respond to a change in the redox poise of the quinone, NADH-NAD+, or flavin pools that would be secondary to an increase in the respiratory rate that is expected to occur as the ΔΨ is consumed. There are examples of regulatory elements that mediate responses to each of these signals in bacteria (5, 7, 10, 13, 39, 60).
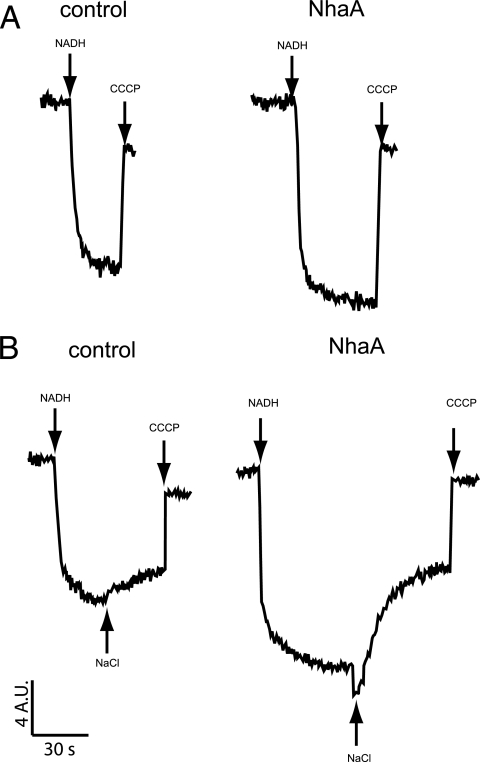
A major prediction of this hypothesis is that an effect similar to that of Mrp antiporters on the capacity of E. coli EP432 to generate ΔΨ should be observed if a native electrogenic Na+(Li+)/H+ antiporter is expressed from a plasmid in E. coli EP432 under the same growth conditions. NhaA was chosen because it is well established to be a secondary electrogenic antiporter that is a single gene product with a role in Na+ resistance at high pH (12, 59) and is structurally unrelated to Mrp-Mnh (6). The recombinant and control plasmids chosen for the NhaA experiments were those that have been used extensively in the characterization of wild-type and mutant NhaA activities in E. coli EP432 (26). The vector, pBR322, was a lower-copy-number plasmid than the pGEM-3Zf(+) used for the Mrp assays. Nonetheless, as shown in Fig. 5A, membranes containing NhaA generate a ΔΨ that is distinctly and consistently greater than that pf the control; the ΔΨ was an average of 20% greater in the NhaA-containing membranes than in control membranes in assays of three separate membrane preparations. Upon addition of Na+, the ΔΨ-consuming nature of NhaA-mediated electrogenic antiport was also evident (Fig. 5B).
FIG. 5.
NhaA-dependent effects on ΔΨ generation and Na+-dependent ΔΨ consumption. Fluorescence-based assays of everted membrane vesicles from control and NhaA transformants of E. coli EP432 were performed exactly as described in the legend to Fig. 4 except that the vector was pBR322 instead of pGEM3Z(f+) and only ΔΨ generation was assessed in the experiments shown in panel A, whereas both ΔΨ generation and Na+-dependent ΔΨ consumption were assessed in those shown in panel B. The traces shown are representative of at least three independent experiments. CCCP, carbonyl cyanide m-chlorophenylhydrazone.
Respiratory chain components are elevated in the MrpBpOF4 and Mnh transformants of E. coli EP432.
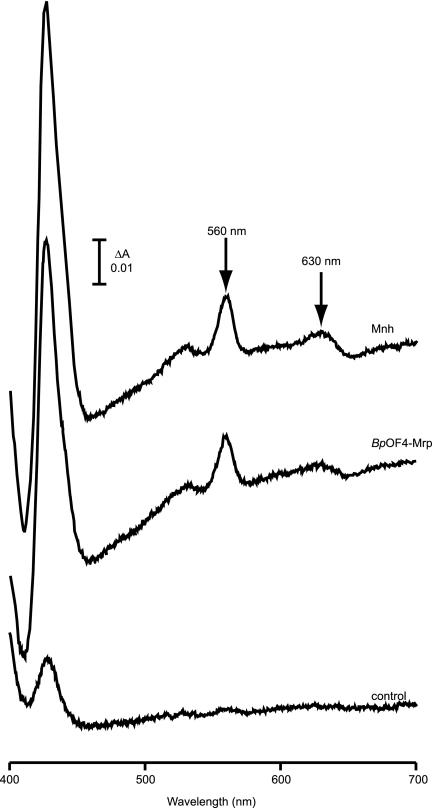
We next explored the hypothesized increase in activity and expression of respiratory chain components in MrpBpOF4 and Mnh vesicles versus control vesicles via enzymatic and spectral assays. NADH oxidation was first assayed by measurements of the decrease in A340 upon addition of NADH to oxygenated vesicles. This measured total respiratory chain activity from NADH oxidation through the terminal oxidases. NADH oxidation rates were significantly higher in both MrpBpOF4 and Mnh vesicles than in control vesicles (Table 3). NADH:ferricyanide reductase activity was then measured by addition of NADH or d-NADH, with ferricyanide reduction monitored via changes in A420; cyanide was present to inhibit the terminal oxidases. NADH-mediated activity assesses the total NADH dehydrogenase activity, i.e., activity of the proton-translocating Nuo complex plus the additional Ndh that does not pump protons (42), whereas d-NADH-mediated activity assesses only the proton-translocating Nuo complex I. No statistically significant difference in these two assay results was observed between the Mrp-containing membranes and control membranes in the average of four independent preparations (Table 3); however, it was noted that in each separate membrane preparation there was a 1.5-fold-higher activity of both NADH:ferricyanide and d-NADH:ferricyanide activity in the MrpBpOF4 and Mnh membranes relative to the control. A marked increase in succinate dehydrogenase activity was found in the MrpBpOF4 and Mnh membranes relative to the control using the DCPIP reduction assay. By contrast, no activity was detected for malate:quinone oxidoreductase (Table 3), whose activity had earlier been shown to be increased in an MrpBpOF4-dependent manner in the respiratory-deficient mutant E. coli ANN0222 (57). The cytochrome content of the membrane vesicle preparations was evaluated from dithionite reduced-minus-oxidized absorption spectra. Both MrpBpOF4 and Mnh membranes showed a threefold increase in the cytochrome b peak at 560 nm and a threefold increase in the cytochrome d peak at 630 nm relative to control membranes (Fig. 6 and Table 3). Since one of the elevated cytochromes, cytochrome d, is part of the terminal oxidase complement, it is likely that the overall increase in respiratory chain activity from NADH to oxygen (Table 3) involves increases in the levels of the terminal oxidases; the increased cytochrome b could correspond to cytochromes associated with both succinate dehydrogenase and the terminal oxidases, cytochromes bd and bo.
TABLE 3.
Effect of mrp-mnh-bearing plasmid on respiratory enzyme activities and cytochrome contents of the heterologous E. coli EP432 host cell
| Prepn | Activitya
|
Fold increasea
|
|||||
|---|---|---|---|---|---|---|---|
| NADH oxidase (nmol NADH/mg protein/min) | NADH:ferricyanide (nmol NADH/mg protein/min) | d-NADH:ferricyanide (nmol d-NADH/mg protein/min) | Succinate dehydrogenase (nmol succinate/mg protein/min) | Malate:quinone oxidoreductase (nmol l-malate/mg protein/min) | Cytochrome b content | Cytochrome d content | |
| Control | 226 ± 81 | 1,097 ± 323 | 629 ± 65 | 113 ± 113 | 0.2 ± 0.1 | 1.0 | 1.0 |
| MrpBpOF4 | 500 ± 81*** | 1,258 ± 81 | 677 ± 81 | 1,387 ± 806*** | 0.2 ± 0.3 | 3.1 ± 1.5 | 3.0 ± 1.7 |
| Mnh | 484 ± 97*** | 1,177 ± 129 | 661 ± 210 | 1,210 ± 968** | 0.2 ± 0.1 | 3.5 ± 1.2** | 3.3 ± 1.0** |
**, P > 0.05 using a two-tailed Student t test; ***, P > 0.005 using a two-tailed Student t test. Values are means and standard deviations.
FIG. 6.
Sodium dithionite reduced-minus-air-oxidized spectra of everted membrane vesicles of control or Mrp-Mnh-expressing E. coli EP432. Everted membrane vesicles (2 mg/ml) were suspended in 50 mM Tricine (pH 8.0) containing 0.1% dodecyl maltoside. Dithionite was added, and spectra were recorded after 1 min of incubation at room temperature. The traces shown are representative of at least three independent experiments. The peak at 560 nm corresponds to cytochrome b, and the peak at 630 nm corresponds to cytochrome d.
To specifically assess whether an increase in transcription of respiratory chain components is part of the host cell response to the presence of an active Mrp system, the succinate dehydrogenase locus (sdh) was chosen because of the large overall increase in its enzymatic activity (Table 3). RT-PCR measurements were conducted on sdh in control cells and in cells expressing either MrpBpOF4 or Mnh, as described in Materials and Methods. The results of three independent experiments showed that the average level of sdh mRNA is threefold higher in cells expressing MrpBpOF4 and Mnh than in control cells, when normalized to levels of 16S rRNA rrsA (data not shown). The assays demonstrate that transformants of E. coli EP432 expressing MrpBpOF4 or Mnh from a multicopy plasmid express and incorporate higher levels of respiratory chain components into their membranes than a control transformant during growth on media that support high Mrp activity. The electrogenic properties shown for Mrp antiporters in this study lead to the expectation that these antiporters will have a significant ΔΨ-consuming effect when active and present in multicopy. It is a plausible hypothesis, therefore, that it is this antiporter-mediated ΔΨ depletion that elicits a host response that counters the ΔΨ depletion by enhancing the host capacity for ΔΨ generation.
Is it also plausible that analogous ΔΨ depletion and compensation are part of the normal physiologic adaptation to an increase in pH and/or Na+ in which a major chromosomally encoded antiporter is activated? Several observations are consistent with this scenario in E. coli. First, E. coli with a wild-type complement of antiporters exhibits an alkali-sensitive phenotype in Na+-containing media upon deletion of the ArcAB two-component system (69). ArcAB responds to the redox poise of the quinone pool and derepresses respiratory components, including sdh and cyd, when the pool is relatively oxidized (10, 39); this could occur as ΔΨ depletion by activated NhaA accelerates respiration, leading to a more oxidized quinone pool. Second, in E. coli that is wild type for two-component systems, alkaline pH is lethal for E. coli expressing a mutant nhaA that has very high Na+/H+ antiport activity across a broad pH spectrum, i.e., has lost a characteristic feature whereby its own activity is down-modulated as a function of pH (51). Padan and colleagues have suggested that ΔΨ depletion that exceeds the cell's capacity for compensation is the basis for the alkali-lethal phenotype. The reduced ΔpH at high pH lowers the total Δp and thus unregulated ΔΨ depletion would be especially detrimental (46, 51). In S. aureus cells, too, there is an observation that best fits a scenario in which Mnh activity leads to a host response to the threat of ΔΨ depletion, i.e., a host response that increases the ΔΨ. Bayer et al. (3) observed that the ΔΨ generated by wild-type S. aureus in an Na+-replete medium is larger than that of a mutant with a disrupted mnh locus. They suggested that Mnh itself functions as a ΔΨ-generating NADH dehydrogenase rather than as a secondary, ΔΨ-consuming antiporter, thus directly accounting for the greater ΔΨ. However, Mnh and Mrp have been found to be devoid of NADH dehydrogenase activity themselves (14, 17, 21, 57). This is expected, since they lack an apparent NADH binding site or flavin motifs that are usually found in all types of NADH dehydrogenases. We cannot rule out the possibility that in the natural host, Mnh recruits a cytoplasmic module containing the requisite redox centers and NADH binding site to transform into a primary ΔΨ-generating element as modeled by Bayer et al. (3). However, our data here show that a bona fide secondary, electrogenic antiporter, NhaA, increases the ΔΨ-generating capacity in a manner similar to that of Mnh and Mrp. Moreover, succinate dehydrogenase activity increases most substantially, not NADH dehydrogenase activity. In an earlier study, MrpBpOF4 was shown to increase ΔΨ generation in an E. coli strain lacking both NADH dehydrogenases; Mrp did not restore NADH dehydrogenase activity but led to a large increase in malate:quinone oxidoreductase (57). We thus consider it likely that in both natural and heterologous settings, activated (and/or overexpressed) electrogenic antiporters elicit responses to ΔΨ depletion that lead to gene expression changes. The net result is maintenance of, or even an increase in, ΔΨ under conditions in which the antiporters are most active. It should be possible to test this hypothesis of a “systems” response to ΔΨ depletion in S. aureus and alkaliphilic Bacillus pseudofirmus OF4 and, if it is supported, to identify the signaling path(s) involved.
Acknowledgments
This research was supported by grant GM28454 from the National Institute of General Medical Sciences (to T.A.K.) and grants from the 21st Century Center of Excellence program of the Ministry of Education, Culture, Sports, Science and Technology of Japan and from the Inoue Enryo Memorial Foundation for Promoting Science (to M.I.).
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Ambudkar, S. V., G. W. Zlotnick, and B. P. Rosen. 1984. Calcium efflux from Escherichia coli. Evidence for two systems. J. Biol. Chem. 259:6142-6146. [PubMed] [Google Scholar]
- 2.Armstrong, J. M. 1964. The molar extinction coefficient of 2,6-dichlorophenol indophenol. Biochim. Biophys. Acta 86:194-197. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, A. S., P. McNamara, M. R. Yeaman, N. Lucindo, T. Jones, A. L. Cheung, H. G. Sahl, and R. A. Proctor. 2006. Transposon disruption of the complex I NADH oxidoreductase gene (snoD) in Staphylococcus aureus is associated with reduced susceptibility to the microbicidal activity of thrombin-induced platelet microbicidal protein 1. J. Bacteriol. 188:211-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco-Rivero, A., F. Leganes, E. Fernandez-Valiente, P. Calle, and F. Fernandez-Pinas. 2005. mrpA, a gene with roles in resistance to Na+ and adaptation to alkaline pH in the cyanobacterium Anabaena sp. PCC7120. Microbiology 151:1671-1682. [DOI] [PubMed] [Google Scholar]
- 5.Brekasis, D., and M. S. Paget. 2003. A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). EMBO J. 22:4856-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busch, W., and M. H. Saier, Jr. 2002. The transporter classification (TC) system, 2002. Crit. Rev. Biochem. Mol. Biol. 37:287-337. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, J. C., M. S. Johnson, and B. L. Taylor. 2006. Differentiation between electron transport sensing and proton motive force sensing by the Aer and Tsr receptors for aerotaxis. Mol. Microbiol. 62:823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrell, M. J., and S. E. Finkel. 2003. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J. Bacteriol. 185:7044-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galili, L., A. Rothman, L. Kozachkov, A. Rimon, and E. Padan. 2002. Transmembrane domain IV is involved in ion transport activity and pH regulation of the NhaA-Na+/H+ antiporter of Escherichia coli. Biochemistry 41:609-617. [DOI] [PubMed] [Google Scholar]
- 10.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the Arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 11.Gerchman, Y., A. Rimon, M. Venturi, and E. Padan. 2001. Oligomerization of NhaA, the Na+/H+ antiporter of Escherichia coli in the membrane and its functional and structural consequences. Biochemistry 40:3403-3412. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg, E. B., T. Arbel, J. Chen, R. Karpel, G. A. Mackie, S. Schuldiner, and E. Padan. 1987. Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 84:2615-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gyan, S., Y. Shiohira, I. Sato, M. Takeuchi, and T. Sato. 2006. Regulatory loop between redox sensing of the NADH/NAD+ ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J. Bacteriol. 188:7062-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamamoto, T., M. Hashimoto, M. Hino, M. Kitada, Y. Seto, T. Kudo, and K. Horikoshi. 1994. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol. Microbiol. 14:939-946. [DOI] [PubMed] [Google Scholar]
- 15.Hatefi, Y., and D. L. Stiggall. 1978. Preparation and properties of succinate: ubiquinone oxidoreductase (complex II). Methods Enzymol. 53:21-27. [DOI] [PubMed] [Google Scholar]
- 16.Hicks, D. B., R. J. Plass, and P. G. Quirk. 1991. Evidence for multiple terminal oxidases, including cytochrome d, in facultatively alkaliphilic Bacillus firmus OF4. J. Bacteriol. 173:5010-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiramatsu, T., K. Kodama, T. Kuroda, T. Mizushima, and T. Tsuchiya. 1998. A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J. Bacteriol. 180:6642-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunte, C., M. Screpanti, M. Venturi, A. Rimon, E. Padan, and H. Michel. 2005. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 534:1197-1202. [DOI] [PubMed] [Google Scholar]
- 19.Ito, M., A. A. Guffanti, B. Oudega, and T. A. Krulwich. 1999. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J. Bacteriol. 181:2394-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, M., A. A. Guffanti, W. Wang, and T. A. Krulwich. 2000. Effects of nonpolar mutations in each of the seven Bacillus subtilis mrp genes suggest complex interactions among the gene products in support of Na+ and alkali but not cholate resistance. J. Bacteriol. 182:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, M., A. A. Guffanti, and T. A. Krulwich. 2001. Mrp-dependent Na+/H+ antiporters of Bacillus exhibit characteristics that are unanticipated for completely secondary active transporters. FEBS Lett. 496:117-120. [DOI] [PubMed] [Google Scholar]
- 22.Ito, M., H. Xu, A. A. Guffanti, Y. Wei, L. Zvi, D. E. Clapham, and T. A. Krulwich. 2004. The voltage-gated Na+ channel NavBP has a role in motility, chemotaxis, and pH homeostasis of an alkaliphilic Bacillus. Proc. Natl. Acad. Sci. USA 101:10566-10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivey, D. M., A. A. Guffanti, J. Zemsky, E. Pinner, R. Karpel, E. Padan, S. Schuldiner, and T. A. Krulwich. 1993. Cloning and characterization of a putative Ca2+/H+ antiporter gene from Escherichia coli upon functional complementation of Na+/H+ antiporter-deficient strains by the overexpressed gene. J. Biol. Chem. 268:11296-11303. [PubMed] [Google Scholar]
- 24.Ji, Y., B. Zhang, S. F. Van, Horn, P. Warren, G. Woodnutt, M. K. Burnham, and M. Rosenberg. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266-2269. [DOI] [PubMed] [Google Scholar]
- 25.Kao, M. C., E. Nakamaru-Ogiso, A. Matsuno-Yagi, and T. Yagi. 2005. Characterization of the membrane domain subunit NuoK (ND4L) of the NADH-quinone oxidoreductase from Escherichia coli. Biochemistry. 44:9545-9554. [DOI] [PubMed] [Google Scholar]
- 26.Karpel, R., Y. Olami, D. Taglicht, S. Schuldiner, and E. Padan. 1988. Sequencing of the gene ant which affects the Na+/H+ antiporter activity in Escherichia coli. J. Biol. Chem. 263:10408-10414. [PubMed] [Google Scholar]
- 27.Kashyap, D. R., L. M. Botero, C. Lehr, D. J. Hassett, and T. R. McDermott. 2006. A Na+:H+ antiporter and a molybdate transporter are essential for arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kita, K., K. Konishi, and Y. Anraku. 1986. Purification and properties of two terminal oxidase complexes of Escherichia coli aerobic respiratory chain. Methods Enzymol. 126:94-113. [DOI] [PubMed] [Google Scholar]
- 29.Kitada, M., S. Kosono, and T. Kudo. 2000. The Na+/H+ antiporter of alkaliphilic Bacillus sp. Extremophiles 4:253-258. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosono, S., S. Morotomi, M. Kitada, and T. Kudo. 1999. Analyses of a Bacillus subtilis homologue of the Na+/H+ antiporter gene which is important for pH homeostasis of alkaliphilic Bacillus sp. C-125. Biochim. Biophys. Acta 1409:171-175. [DOI] [PubMed] [Google Scholar]
- 32.Kosono, S., Y. Ohashi, F. Kawamura, M. Kitada, and T. Kudo. 2000. Function of a principal Na+/H+ antiporter, ShaA, is required for initiation of sporulation in Bacillus subtilis. J. Bacteriol. 182:898-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosono, S., K. Haga, R. Tomizawa, Y. Kajiyama, K. Hatano, S. Takeda, Y. Wakai, M. Hino, and T. Kudo. 2005. Characterization of a multigene-encoded sodium/hydrogen antiporter (Sha) from Pseudomonas aeruginosa: its involvement in pathogenesis. J. Bacteriol. 187:5242-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosono, S., Y. Kajiyama, S. Kawasaki, T. Yoshinaka, K. Haga, and T. Kudo. 2006. Functional involvement of membrane-embedded and conserved acidic residues in the ShaA subunit of the multigene-encoded Na+/H+ antiporter in Bacillus subtilis. Biochim. Biophys. Acta 1758:627-635. [DOI] [PubMed] [Google Scholar]
- 35.Li, Q., L. Li, T. Rejtar, D. J. Lessner, B. L. Karger, and J. G. Ferry. 2006. Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J. Bacteriol. 188:702-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) Method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 37.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 38.Macnab, R. M., and A. M. Castle. 1987. A variable stoichiometry model for pH homeostasis in bacteria. Biophys. J. 52:637-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malpica, R., B. Franco, C. Rodriguez, O. Kwon, and D. Georgellis. 2004. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. USA 101:13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathiesen, C., and C. Hägerhäll. 2002. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim. Biophys. Acta 1556:121-132. [DOI] [PubMed] [Google Scholar]
- 41.Mathiesen, C., and C. Hägerhäll. 2003. The ‘antiporter module’ of respiratory chain complex I includes the MrpC/NuoK subunit—a revision of the modular evolution scheme. FEBS Lett. 5459:7-13. [DOI] [PubMed] [Google Scholar]
- 42.Matsushita, K., T. Ohnishi, and H. R. Kaback. 1987. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry 26:7732-7737. [DOI] [PubMed] [Google Scholar]
- 43.Nozaki, K., K. Inaba, T. Kuroda, M. Tsuda, and T. Tsuchiya. 1996. Cloning and sequencing of the gene for Na+/H+ antiporter of Vibrio parahaemolyticus. Biochem. Biophys. Res. Commun. 222:774-779. [DOI] [PubMed] [Google Scholar]
- 44.Padan, E., M. Venturi, Y. Gerchman, and N. Dover. 2001. Na+/H+ antiporters. Biochim. Biophys. Acta 1505:144-157. [DOI] [PubMed] [Google Scholar]
- 45.Padan, E., T. Tzubery, K. Herz, L. Kozachkov, A. Rimon, and L. Galili. 2004. NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+antiporter. Biochim. Biophys. Acta 1658:2-13. [DOI] [PubMed] [Google Scholar]
- 46.Padan, E., E. Bibi, M. Ito, and T. A. Krulwich. 2005. Alkaline pH homeostasis in bacteria: new insights. Biochim. Biophys. Acta 1717:67-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinner, E., Y. Kotler, E. Padan, and S. Schuldiner. 1993. Physiological role of nhaB, a specific Na+/H+ antiporter in Escherichia coli. J. Biol. Chem. 268:1729-1734. [PubMed] [Google Scholar]
- 48.Putnoky, P., A. Kereszt, T. Nakamura, G. Endre, E. Grosskopf, P. Kiss, and A. Kondorosi. 1998. The pha gene cluster of Rhizobium meliloti involved in pH adaptation and symbiosis encodes a novel type of K+ efflux system. Mol. Microbiol. 28:1091-1101. [DOI] [PubMed] [Google Scholar]
- 49.Quirk, P. G., A. A. Guffanti, R. J. Plass, S. Clejan, and T. A. Krulwich. 1991. Protonophore-resistance and cytochrome expression in mutant strains of the facultative alkaliphile Bacillus firmus OF4. Biochim. Biophys. Acta 1058:131-140. [DOI] [PubMed] [Google Scholar]
- 50.Radchenko, M. V., K. Tanaka, R. Waditee, S. Oshimi, Y. Matsuzaki, M. Fukuhara, H. Kobayashi, T. Takabe, and T. Nakamura. 2006. Potassium/proton antiport system of Escherichia coli. J. Biol. Chem. 281:19822-19829. [DOI] [PubMed] [Google Scholar]
- 51.Rimon, A., Y. Gerchman, Z. Kariv, and E. Padan. 1998. A point mutation (G338S) and its suppressor mutations affect both the pH response of the NhaA-Na+/H+ antiporter as well as the growth phenotype of Escherichia coli. J. Biol. Chem. 273:26470-26476. [DOI] [PubMed] [Google Scholar]
- 52.Rosen, B. P. 1986. Ion extrusion systems in E. coli. Methods Enzymol. 125:328-386. [DOI] [PubMed] [Google Scholar]
- 53.Safferling, M., H. Griffith, J. Jin, J. Sharp, M. De Jesus, C. Ng, T. A. Krulwich, and D. N. Wang. 2003. TetL tetracycline efflux protein from Bacillus subtilis is a dimer in the membrane and in detergent solution. Biochemistry 42:13969-13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saier, M. H., Jr. 2000. Families of proteins forming transmembrane channels. J. Membr. Biol. 175:165-180. [DOI] [PubMed] [Google Scholar]
- 55.Sturr, M. G., A. A. Guffanti, and T. A. Krulwich. 1994. Growth and bioenergetics of alkaliphilic Bacillus firmus OF4 in continuous culture at high pH. J. Bacteriol. 176:3111-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swartz, T. H., S. Ikewada, O. Ishikawa, M. Ito, and T. A. Krulwich. 2005. The Mrp system: a giant among monovalent cation/proton antiporters. Extremophiles 9:345-354. [DOI] [PubMed] [Google Scholar]
- 57.Swartz, T. H., M. Ito, D. B. Hicks, M. Nuqui, A. A. Guffanti, and T. A. Krulwich. 2005. The Mrp Na+/H+ antiporter increases the activity of the malate:quinone oxidoreductase of an Escherichia coli respiratory mutant. J. Bacteriol. 187:388-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swartz, T. H. 2006. Mrp systems of gram-positive bacteria: properties of the monovalent cation/proton antiport. Ph.D. dissertation. Mount Sinai School of Medicine of New York University, New York, NY.
- 59.Taglicht, D., E. Padan, and S. Schuldiner. 1993. Proton-sodium stoichiometry of NhaA, an electrogenic antiporter from Escherichia coli. J. Biol. Chem. 268:5382-5387. [PubMed] [Google Scholar]
- 60.Taylor, B. L., I. B. Zhulin, and M. S. Johnson. 1999. Aerotaxis and other energy-sensing behavior in bacteria. Annu. Rev. Microbiol. 53:103-128. [DOI] [PubMed] [Google Scholar]
- 61.Tzubery, T., A. Rimon, and E. Padan. 2004. Mutation E252C increases drastically the Km value for Na+ and causes an alkaline shift of the pH dependence of NhaA Na+/H+ antiporter of Escherichia coli. J. Biol. Chem. 279:3265-3272. [DOI] [PubMed] [Google Scholar]
- 62.Verkhovskaya, M. L., B. Barquera, and M. Wikstrom. 2001. Deletion of one of two Escherichia coli genes encoding putative Na+/H+ exchangers (ycgO) perturbs cytoplasmic alkali cation balance at low osmolarity. Microbiology 147:3005-3013. [DOI] [PubMed] [Google Scholar]
- 63.Vijaranakul, U., M. J. Nadakavukaren, D. O. Bayles, B. J. Wilkinson, and R. K. Jayaswal. 1997. Characterization of an NaCl-sensitive Staphylococcus aureus mutant and rescue of the NaCl-sensitive phenotype by glycine betaine but not by other compatible solutes. Appl. Environ. Microbiol. 63:1889-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waggoner, A. S. 1979. Dye indicators of membrane potential. Annu. Rev. Biophys. Bioeng. 8:47-68. [DOI] [PubMed] [Google Scholar]
- 65.Wilkinson, B. J. 1997. Biology, p. 1-38. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, London, United Kingdom.
- 66.Yagi, T. 1986. Purification and characterization of NADH dehydrogenase complex from Paracoccus denitrificans. Arch. Biochem. Biophys. 250:302-311. [DOI] [PubMed] [Google Scholar]
- 67.Yang, L., J. Jiang, W. Wei, B. Zhang, L. Wang, and S. Yang. 2006. The pha2 gene cluster involved in Na+ resistance and adaption to alkaline pH in Sinorhizobium fredii RT19 encodes a monovalent cation/proton antiporter. FEMS Microbiol. Lett. 262:172-177. [DOI] [PubMed] [Google Scholar]
- 68.Yumoto, I. 2002. Bioenergetics of alkaliphilic Bacillus spp. J. Biosci. Bioeng. 93:342-353. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, L., X. H. Lei, B. R. Bochner, and B. L. Wanner. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185:4956-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]