Abstract
Myxococcus xanthus is a soil-dwelling, gram-negative bacterium that during nutrient deprivation is capable of undergoing morphogenesis from a vegetative rod to a spherical, stress-resistant spore inside a domed-shaped, multicellular fruiting body. To identify proteins required for building stress-resistant M. xanthus spores, we compared the proteome of liquid-grown vegetative cells with the proteome of mature fruiting body spores. Two proteins, protein S and protein S1, were differentially expressed in spores, as has been reported previously. In addition, we identified three previously uncharacterized proteins that are differentially expressed in spores and that exhibit no homology to known proteins. The genes encoding these three novel major spore proteins (mspA, mspB, and mspC) were inactivated by insertion mutagenesis, and the development of the resulting mutant strains was characterized. All three mutants were capable of aggregating, but for two of the strains the resulting fruiting bodies remained flattened mounds of cells. The most pronounced structural defect of spores produced by all three mutants was an altered cortex layer. We found that mspA and mspB mutant spores were more sensitive specifically to heat and sodium dodecyl sulfate than wild-type spores, while mspC mutant spores were more sensitive to all stress treatments examined. Hence, the products of mspA, mspB, and mspC play significant roles in morphogenesis of M. xanthus spores and in the ability of spores to survive environmental stress.
Many bacterial species have the capacity to differentiate into stress-resistant, dormant spores in order to survive in hostile environments. Spore production is common among gram-positive bacteria, such as Bacillus and Clostridium species, while sporulation is relatively rare in gram-negative bacteria. A notable exception is a group of gram-negative, spore-forming bacteria known as the myxobacteria. Like the end products of Bacillus and Clostridium spp. sporulation, the end products of myxobacterial sporulation are dormant cells that are surrounded by thick protective coats. However, a fundamental difference between these two groups of sporeformers is that gram-positive bacteria build stress-resistant endospores inside the sheltered environment of mother cells, while vegetative myxobacterial cells are physically converted into stress-resistant spores (the vegetative cell wall is altered without damaging its integrity).
In the myxobacteria, spore development has been characterized in the greatest detail in Myxococcus xanthus. When M. xanthus cells are deprived of nutrients and concentrated on solid media, rod-shaped cells cluster together in aggregation centers, these aggregates of cells build fruiting bodies, and the fruiting body cells differentiate into spherical spores (myxospores). Although the morphological changes that occur during M. xanthus sporulation have been well documented, relatively little is known about the corresponding molecular changes that allow cells inside fruiting bodies to differentiate into stress-resistant spores. Currently, only a few M. xanthus spore proteins have been identified, and most of these proteins are not required for sporulation or spore stress resistance (9, 11, 16, 17, 19, 24). The recently available M. xanthus genome sequence (GenBank accession no. CP000113) has made it possible to globally analyze gene expression by DNA microarray-based and proteomic-based approaches. Here we describe a comparison of the proteomes of M. xanthus vegetative cells and mature spores. We identified three previously uncharacterized proteins that are expressed at relatively high levels in mature spores compared to the levels in their vegetative cell counterparts. Inactivation of the genes encoding these three proteins by insertion mutagenesis does not alter cell aggregation into fruiting bodies, but it does affect the morphology of spores and their ability to resist environmental stress. This indicates that these novel major spore proteins are important for the M. xanthus sporulation process.
METHODS AND MATERIALS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. Strain DK1622 (13) is the wild-type, parental strain used to generate mutants. The mspA (MXAN_2269), mspB (MXAN_2432), and mspC (MXAN_6969) genes were inactivated in strain DK1622 by single-crossover recombination of kanamycin-resistant plasmids containing internal PCR-generated fragments of each gene, as described previously (4, 25). The PCR primers used to amplify internal fragments of the three genes are shown in Table 2. Plasmid pCR2.1-TOPO (Invitrogen) confers kanamycin resistance, and it is the vector into which the PCR fragments were cloned. Plasmids were propagated in Escherichia coli strain Top10F (Invitrogen). Strain Top10F was grown at 37°C in Luria broth (LB) containing 1% tryptone, 0.5% yeast extract, and 0.5% NaCl or on plates containing LB and 1.5% agar. LB and LB agar plates were supplemented with kanamycin (40 μg/ml) as needed. Plasmid insertions into the three M. xanthus genes were verified by PCR amplification, and the primers for these reactions are shown in Table 2. M. xanthus strains were grown at 32°C in CTTYE broth containing 1% Casitone, 0.5% yeast extract, 10 mM Tris-HCl (pH 8.0), 1 mM KH2PO4, and 8 mM MgSO4. Alternatively, cells were grown on CTTYE plates containing 1.5% agar. Fruiting body development occurred on plates containing TPM buffer (10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4, 8 mM MgSO4) and 1.5% agar incubated at 32°C, as previously described (4).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| DK1622 | Wild-type motility and development | 13 |
| AG681 | pAG681::mspA (plasmid insert in mspA) | This study |
| AG710 | pAG701::mspB (plasmid insert in mspB) | This study |
| AG810 | pAG810::mspC (plasmid insert in mspC) | This study |
| Plasmids | ||
| pCR2.1-TOPO | Kanr | Invitrogen |
| pAG681 | 294-bp fragment extending from bp 182 to 476 of the mspA gene | This study |
| pAG710 | 483-bp fragment extending from bp 251 to 734 of the mspB gene | This study |
| pAG810 | 262-bp fragment extending from bp 112 to 374 of the mspC gene | This study |
TABLE 2.
PCR primers
| Primer | Sequence | Use | Expected size (bp) |
|---|---|---|---|
| OAG 217 | 5′-GCT CGC AAG AGC CGT CGA TGC C-3′ | Insertion mutation in mspA gene | 294 |
| OAG 218 | 5′-GCC GGA GGT GAC TGT GGA TGT C-3′ | ||
| OAG 213 | 5′-TCG CCA CCA TCG CCA TGC TCG G-3′ | Insertion mutation in mspB gene | 483 |
| OAG 214 | 5′-TGT CCT GGT TGA TGG ACG AGT C-3′ | ||
| OAG 215 | 5′-AAG GCC GCC AAG GAC GTG ACA G-3′ | Insertion mutation in mspC gene | 262 |
| OAG 216 | 5′-CGA CCT CAT CGC CCT CCA ACT G-3′ | ||
| OAG 492 | 5′-GGA GAT TCG TGC GTT GGA TGC G-3′ | mspA mutation verification | 5,610 |
| OAG 493 | 5′-GAG CAT CTC TGT CGG CAA CGT G-3′ | ||
| OAG 488 | 5′-GTC TCC ACC AGC ATC TCC AGC A-3′ | mspB mutation verification | 5,899 |
| OAG 489 | 5′-CTC ACG TTG AGC AGC ATG GCG A-3′ | ||
| OAG 490 | 5′-ACG ACA CGA GCT TCG ACC GCA T-3′ | mspC mutation verification | 5,169 |
| OAG 491 | 5′-CCA CAG TCG TCG ATT CAC CGC T-3′ | ||
| OJD2U | 5′-CAT CGA TGC AGC ATC CAC CC-3′ | RT-PCR for mspA expression | 170 |
| OJD2D | 5′-CTC ATC CTT GAT GAA GAC C-3′ | ||
| OJD10U#2 | 5′-CAA CAA CCA CAG GAG AAC CTG-3′ | RT-PCR for mspB expression | 313 |
| OJD10D | 5′-GTC GAC GAA GAT CTT GTC GC-3′ | ||
| OJD9U | 5′-GTG ACA GGC ATC CAG ACG G-3′ | RT-PCR for mspC expression | 297 |
| OJD9D | 5′-CTG CTT CTT CTT GGA ACC C-3′ | ||
| OJD4U | 5′-CAA GGG AAC TGA GAG ACA GG-3′ | RT-PCR for 16S rRNA | 120 |
| OJD4D | 5′-CTC TAG AGA TCC ACT TGC G-3′ | ||
| OJD23U-1 | 5′-ATG GTG GAA CTG GAA GCC G-3′ | RT-PCR for MXAN_2431 gene | 500 |
| OJD23D-1 | 5′-GTC CTT TCG AAG TCT TGA CGG-3′ | ||
| OJD24U-2 | 5′-CAC CGT CAT CTG TGG TGG C-3′ | RT-PCR for MXAN_6970 gene | 500 |
| OJD24D-2 | 5′-CTT CTC GTT GAG CCA CAC CG-3′ |
Preparation of protein lysates and one-dimensional SDS-PAGE analysis.
Two buffers were used to suspend M. xanthus vegetative cells and spores before lysis: TPM buffer (25) and DTT lysis buffer (0.3% sodium dodecyl sulfate [SDS], 200 mM dithiothreitol [DTT], 28 mM Tris-HCl, 22 mM Tris base, bacterial protease inhibitor cocktail [Sigma]) (2). Cells suspended in DTT lysis buffer were incubated in a 70°C water bath for 30 min before they were vortexed with glass beads. Cells were lysed by vortexing them with 0.1-mm-diameter glass beads (Biospec Products, Inc.) using a FastPrep FP120 device (Bio 101 Thermo Savant). Each sample was vortexed six times for 45 s at a setting of 6.5. Samples were incubated on ice for 5 min between vortex treatments. Following cell lysis, the samples were incubated at room temperature (25°C) for various times before they were boiled in 1× SDS sample buffer (125 mM Tris base, 20% glycerol, 2% SDS, 2% β-mercaptoethanol, 0.001% bromophenol blue), proteins were separated by 12% SDS-polyacrylamide gel electrophoresis (PAGE), and proteins were visualized by Coomassie blue staining. Spore coat proteins were released from 5-day-old spores by boiling the spores in 1% SDS, as previously described (16).
DIGE analysis.
Vegetative cell and spore proteomes were compared at the University of Georgia's Proteomics Resource Facility, Athens, GA, using minimal labeling differential in-gel electrophoresis (DIGE) analysis (26) and GE Healthcare products. Protein samples from cells lysed in DTT lysis buffer were concentrated, and ionic contaminants were removed by three changes of buffer containing 8 M urea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and 15 mM Tris (pH 8.3) using centrifugal filtration devices with a 10-kDa cutoff (Millipore). Covalent binding of the dyes does not significantly change the native molecular weights of proteins, nor does it change their charges (26). The Cy3 and Cy5 dyes used to label the protein lysates were limiting within the reactions so that approximately 1 to 2% of the total lysine residues were labeled. Fifty micrograms of each sample was labeled with 200 pmol of minimal Cy3 or Cy5 N-hydroxysuccinimide ester (GE Healthcare) at 4°C for 30 min. The labeling reaction was quenched with 10 nmol of lysine. Labeled proteins were mixed and subjected to denaturing isoelectric focusing on immobilized pH gradient gels (Immobiline DryStrips; length, 18 cm; pH range, pH 3 to 10) using an Ettan IPGphor isoelectric focusing system. Labeled protein mixtures were suspended in rehydration buffer (8 M urea, 2% CHAPS, 0.5% ampholytes, 0.28% DTT, 0.002% bromophenol blue), actively rehydrated at 30 V for 10 h, and focused for 33,000 V·h. Following separation in the first dimension, the Immobiline DryStrips were equilibrated in a solution containing 6 M urea, 2% SDS, 65 mM DTT, 30% glycerol, 50 mM Tris (pH 8.8), and 0.002% bromophenol blue for 15 min at room temperature. The IPG strips were then equilibrated with the buffer described above in which the DTT was replaced with 135 mM iodoacetamide for 15 min at room temperature. The IPG strips were transferred with a molecular weight standard to 8 to 15% gradient SDS-polyacrylamide gels (26 by 20 cm) and separated using the Ettan Dalt II large-format vertical system. Following separation in the second dimension, the gels were fixed in 30% ethanol and 7.5% acetic acid overnight at room temperature. Cy3- and Cy5-labeled proteins were visualized with a confocal laser scanner using a Typhoon 9400 with optimization of the photomultiplier voltage for each laser to achieve the broadest dynamic range, and differential expression was analyzed using the DeCyder 4.0 software. Cy3-labeled proteins were excited and falsely colored green, and Cy5-labeled proteins were excited and falsely colored red. Gels selected for picking were stained with Sypro Ruby (Molecular Probes) overnight, destained in 10% methanol and 6% acetic acid for 30 min at room temperature, imaged, and matched to the Cy images using the DeCyder software. The pick list was created based on the Sypro image using the DeCyder software. Processing of the gel plugs for mass spectrometry was performed at an Ettan spot handling workstation. This automated workstation was programmed to pick specific spots, reduce and alkylate cysteine residues, digest proteins in gel plugs with trypsin, extract peptides from gel plugs, and spot the peptides onto matrix-assisted laser desorption ionization (MALDI) target plates. Briefly, plugs were washed twice with 50 mM ammonium bicarbonate-50% methanol for 20 min at room temperature. Then the plugs were washed with 75% acetonitrile for 20 min at room temperature and dried at 40°C for 10 min. The plugs were incubated in 10 mM DTT-20 mM ammonium bicarbonate at 37°C for 1 h. The DTT solution was removed and immediately replaced with 100 mM iodoacetamide-20 mM ammonium bicarbonate, and the preparations were incubated at room temperature in the dark for 30 min. The plugs were washed as described above and then incubated with 200 ng of sequencing-grade trypsin (Promega) at 37°C for 2 h. The peptides were extracted twice with 50% acetonitrile-0.1% trifluoroacetic acid for 20 min at room temperature and dried at 40°C for 2 h. Approximately 25% of the resulting peptides were spotted with partially saturated α-cyano-4-hydroxycinnamic acid (Sigma). The extracted peptides were then subjected to peptide mass fingerprinting using a 4700 proteomics analyzer MALDI mass spectrometer (Applied Biosystems). Mass spectra were calibrated using two trypsin autolysis peaks (m/z 1045.45 and 2211.096). Mass lists were compared to an in silico tryptic digestion of an annotated M. xanthus genome database using a licensed copy of Mascot v. 1.9.05 (http://www.matrixscience.com/), considering fixed cysteine carbamidomethylation and partial methionine oxidation modifications, one missed tryptic cleavage, and 10 ppm mass accuracy. Identifications were cross-examined using mass accuracy, molecular weight, and pI.
RT-PCR analysis.
The relative level of mRNA for each transcribed gene was determined by performing a limiting-dilution reverse transcription (RT)-PCR analysis as previously described (3). Fruiting bodies of DK1622 were harvested from TPM buffer plates after starvation for various times. TPM buffer-washed pellets were suspended in 180 μl of RNase-free H2O (GeneMate) with 1% β-mercaptoethanol before SDS and sodium acetate (pH 5.2) were added to final concentrations of 0.9% and 90 mM, respectively. Samples were warmed to 65°C for 3 min before 200 μl of 65°C acid phenol (phenol-chloroform [5:1], pH 4.5; Ambion) was added and cells were incubated at 65°C for 5 min. The top aqueous phases were saved, and acid phenol extraction was repeated until the organic-aqueous interface was free of debris. A final extraction of the aqueous phases was performed with 150 μl of chloroform. Nucleic acids were recovered by adding sodium acetate (pH 5.2) to a final concentration of 300 mM and then adding 2.5 volumes of ethanol. Pellets were washed in 70% ethanol, dried under a vacuum without heat, and resuspended in RNase-free H2O. Samples were treated twice with RNase-free DNase I (Roche) at room temperature for 4 h before RNA was recovered using a QIAGEN RNeasy column. The RNA concentration was determined by determining the absorbance at 260 nm, and chromosomal contamination of preparations was determined by PCRs using primers for 16S rRNA and for the mspA, mspB, and mspC genes (Table 2). The RNA preparations were used to perform RT reactions using random hexamers and SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's protocol. RT-PCR primers specific for mspA, mspB, and mspC are shown in Table 2. Amersham PuReTaq Ready-to-Go PCR beads were used with the following thremocycler profile: 95°C for 2.5 min, followed by 35 cycles of 95°C for 30 s, 59°C for 30 s, and 72°C for 45 s. The PCR results were examined on 2% agarose gels.
SEM analysis of fruiting bodies.
Scanning electron microscopy (SEM) was performed as previously described (27), with the following modifications. A paper hole punch was used to cut small disks of Millipore 0.22-μm-diameter filters, and the disks were autoclaved. The disks were then dipped into molten TPM agar and placed on TPM agar plates, aliquots of concentrated M. xanthus strains were spotted on top of the disks, and the plates were incubated at 32°C for 5 days to allow mature fruiting bodies to form on the disks. After 5 days of development, the disks were gently removed from the agar plates and fixed by floating them on drops of 50% gluteraldehyde for 2 h. Fixation was performed in 24-well tissue culture plates with Parafilm around the seam of each plate to reduce evaporation of liquids. Disks were then removed and floated on drops of H2O for 5 min. Disks were frozen in liquid nitrogen before lyophilization overnight using a Virtis lyophilizer (Virtis, Gardner, NY). Samples were fixed onto aluminum SEM mounts, sputter coated with gold (Techniques Hummer II), and analyzed using a Hitachi S570 SEM. Images were captured with the PCI quartz imaging program.
TEM analysis of spores.
Five-day-old fruiting bodies were harvested from TPM agar plates and suspended in TPM buffer. Pelleted cells were resuspended in 2% paraformaldehyde-2% glutaraldehyde in 0.1 M cacodylate buffer containing 0.2 M sucrose for 12 h at 4°C. The cells were rinsed three times with 0.1 M cacodylate buffer containing 0.2 M sucrose before they were postfixed in 2% OsO4 in 0.1 M cacodylate buffer for 2 h at room temperature. The cells were rinsed three times with H2O and stained with 1% tannic acid for 1 h at room temperature. This procedure helped ensure that the arrangement of spores within the fruiting bodies remained intact. Samples were then dehydrated with a series of 10-min steps using 30, 50, 70, and 95% ethanol before the final three 10-min dehydration steps using 100% ethanol. Samples were then infiltrated in a stepwise fashion as follows: 100% acetone twice for 10 min each time, acetone-Spurrs resin (1:1) for 1 h at room temperature, 100% Spurrs resin for 2 h at room temperature, and then fresh 100% Spurrs resin overnight at room temperature. Postsectioning staining was performed with 4% uranyl acetate for 10 min and with lead citrate for 5 min. Transmission electron microscopy (TEM) was performed with a JOEL 1200 EX (JOEL, Tokyo, Japan).
Stress resistance assays.
Wild-type and mutant cells that developed on TPM agar plates incubated for 5 days were placed in 1-ml aliquots of TPM buffer. Before and after all stress treatments, cells were subjected to three 10-s bursts with a model 100 Sonic Dismembrator (Fisher) using an intensity setting of 1.5. One minute of incubation at room temperature followed each 10-s burst. This gentle disruption did not aberrantly reduce the viabilities of mutant spores compared to the viability of DK1622 spores. Assays for resistance to heat (50 and 55°C) and sonication were performed as previously described (23), except that cells were harvested from TPM agar plates and were sonicated (setting, 1.5) before and after heat treatments in 50 and 55°C water baths for 2 h. To test for resistance to sonication, cells were sonicated with three 10-s bursts at an intensity setting of 4.0. To examine SDS sensitivity, SDS (Fisher) was added to suspensions of harvested spores to a final concentration of 1%. The cells were incubated for 2 h at room temperature with continuous agitation using a rotating belly dancer. To examine lysozyme sensitivity, lysozyme (Sigma) was added to spore cell suspensions to a final concentration of 250 μg/ml. Cells were then incubated at room temperature with continuous agitation for 12 h. To test for sensitivity to UV irradiation, aliquots of spore cells were first placed into tissue culture wells containing 1 ml of TPM buffer. The tissue culture plates were rotated on a belly dancer rotator, which was positioned 56 cm from a UV 97505 series lamp (Cole Palmer). Cells were exposed to UV light at a wavelength of 254 nm and an intensity of 31 μW/cm2 for 1 min. During UV exposure, the cells in the tissue culture plates were agitated continuously. Following all of the stress treatments described above, aliquots of cells were diluted into CTTSA (1% Casitone, 10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4, 8 mM MgSO4, 0.7% agar) and poured onto CTTYE medium plates with or without kanamycin sulfate (40 μg/ml). Colonies were counted after 5 days of growth, and the number of colonies represented the number of viable spores.
RESULTS
Stability of M. xanthus proteome after cell lysis.
Based on the intrinsically high levels of proteases in myxobacteria (20), in any study of the M. xanthus proteome workers must first consider the stability of protein lysates after cells are disrupted. Assuming that metabolically active vegetative cells are more prodigious protease producers than spores, we analyzed the stability of protein lysates obtained from cells grown in CTTYE broth to the mid-log phase. Before vegetative cells were lysed, they were suspended in one of two buffers: TPM buffer or DTT lysis buffer. In addition, vegetative cells resuspended in DTT lysis buffer were heated at 70°C for 30 min to further inactivate any proteases. Proteins from cells lysed in TPM buffer were relatively stable for 48 h at room temperature, with the exception of a few bands (Fig. 1A). Heat and the presence of DTT inactivated most of the vegetative cell proteases, as shown by the uniform appearance of proteins in each lane (Fig. 1B). Because DTT lysis buffer was superior in terms of both the quantity and quality of the protein extracts, this buffer was used for subsequent DIGE experiments in which we compared vegetative and spore cell extracts.
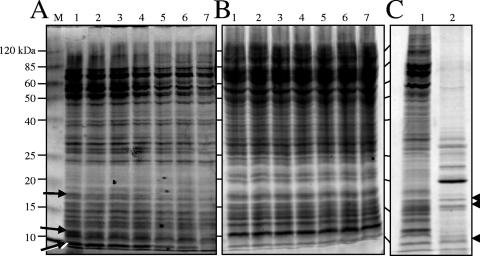
FIG. 1.
Lysis of M. xanthus cells with glass beads. Vegetative cells grown in CTTYE broth to the mid-log phase were resuspended in either TPM buffer (A) or DTT lysis buffer (B), vortexed with 0.1-mm-diameter glass beads, separated by 12% SDS-PAGE, and stained with Coomassie blue. Before protein mixtures were separated by electrophoresis, they were allowed to sit at room temperature to examine the potential of endogenous proteases to hydrolyze proteins. Lane M contained the molecular weight standard, and protein molecular masses are indicated on the left. Lane 1, zero time at room temperature; lane 2, 2 h; lane 3, 4 h; lane 4, 6 h; lane 5, 16 h; lane 6, 24 h; lane 7, 48 h. The arrows in panel A indicate protein species that were degraded over time in the TPM buffer lysate. (C) Vegetative cells (100 μg) (lane 1) and 5-day-old myxospores (10 μg) (lane 2) were lysed in DTT lysis buffer by vortexing with glass beads, separated by 12% SDS-PAGE, and stained with Coomassie blue. The arrowheads indicate protein species appearing at similar locations and in similar amounts in the two lysates.
Figure 1C shows that it was possible to prepare high-quality, soluble proteins from myxospores and vegetative cells that were to be labeled with cyanine dyes for DIGE analysis. Furthermore, Fig. 1C shows the paucity of extractable myxospore proteins compared to the extractable proteins of the vegetative cells. This was expected because of the difficulty in breaking open stress-resistant spores. Three proteins bands at identical positions were obtained for the two lysates (Fig. 1C). To obtain roughly equivalent amounts of these three proteins, myxospore protein lysates and vegetative cell protein lysates were loaded at a ratio of 10:1 (vol/vol). These equivalent proteins appeared as yellow spots in the DIGE analysis discussed below (Fig. 2B).
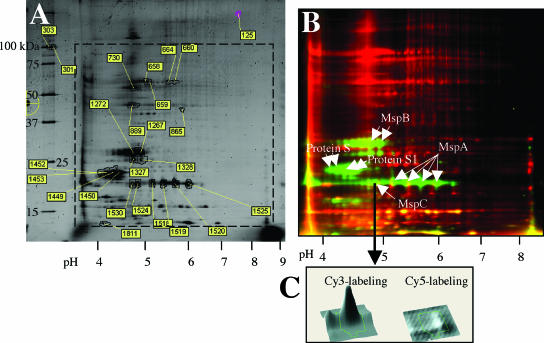
FIG. 2.
DIGE comparison of proteomes of vegetative cells and 5-day-old myxospores. Protein samples were prepared, fluorescently labeled, mixed, and separated in two dimensions, as described in Materials and Methods. (A) Sypro Ruby staining of the two-dimensional gel. Proteins picked for identification are numbered, and the numbers correspond to the protein spots in Table 3. (B) Dual-channel fluorescence of Cy3-labeled myxospore proteins (green) and Cy5-labeled vegetative cell proteins (red). The positions of known myxospore proteins S and S1 are indicated, as are the positions of novel proteins MspA, MspB, and MspC. (C) DeCyder software analysis of one protein spot (spot 1524; MspC) indicated that the level of expression of this protein was 44-fold higher in the myxospore lysate than in the vegetative cell lysate.
Identification of proteins differentially expressed in M. xanthus spores.
DIGE analysis eliminates the inherent variability of running two separate two-dimensional gels side by side. This technique relies on the ability to differentially label two protein mixtures with two fluorflors, run the protein mixtures together in the same two-dimensional gel, and then excite the gel with two wavelengths of light to visualize and analyze the two sets of proteins. Figure 2A shows a typical DIGE comparison of vegetative and spore lysates that were stained with Sypro Ruby to visualize the two sets of proteins in vegetative and spore lysates. The protein spots picked for analysis by MALDI mass spectrometry were numbered, and the identities of some spots are shown in Table 3. Figure 2B shows a fluorescent image of Cy3-labeled spore proteins (green spots) overlaid with a fluorescent image of Cy5-labeled vegetative proteins (red spots). Proteins whose expression was similar in the two cell types produced yellow spots. The intensity of protein labeling was determined using the DeCyder software, which could directly compare levels of fluorescence of single protein species and present a three-dimensional display of Cy3 labeling or Cy5 labeling for each protein. A three-dimensional display of one protein, later identified as MspC, is shown in Fig. 2C. The level of the MspC protein was 44-fold higher in myxospores (Cy3 labeled) than in vegetative cells (Cy5 labeled). More than 5,000 M. xanthus protein spots were visualized with fluorescent labeling and enumerated with the DeCyder software. The ratios of the Cy5 label to the Cy3 label for several proteins that were identified are shown in Table 3. Several proteins were excised from the gel, and their identities were determined by peptide mass fingerprinting (Fig. 2A and Table 3). Proteins S and S1 are two previously characterized spore-specific proteins. Protein S (encoded by the tps gene) is the major protein found in the M. xanthus spore coat, while protein S1 (encoded by the ops gene) seems to be located primarily in the core (24). Proteins S and S1 exhibit 90% amino acid sequence homology, are immunologically cross-reactive, and can both self-assemble on the spore surface in a Ca2+-dependent fashion (24). The fact that we identified proteins S and S1 in spores provides a proof of principle for DIGE analysis for identification of relatively abundant proteins in spores. In addition to proteins S and S1, three other prominent Cy3-labeled proteins were identified: MspA, MspB, and MspC. One explanation for the finding that each protein occurred at multiple positions (“spot trains”) on the x axis is that there can be multiple forms of a protein with different isoelectric points. For example, four different spots of the same molecular weight species were all identified as MspA. A number of posttranslational events, including deamidation, phosphorylation, oxidation, and exogenous chemical modification, are known to affect a protein's pI and resulting migration in an IPG strip. However, because the three Msp proteins are novel, there are no previously published data indicating that such in vivo modifications occur in these proteins. Searches of the GenBank database revealed no proteins with homology to MspA, MspB, or MspC. However, based on the primary sequences the computer program CELLO (28) predicted that MspB is an extracellular protein and that both MspA and MspC are located in the periplasmic space.
TABLE 3.
Proteins identified by peptide mass fingerprinting
| Spota | Fold differenceb | Accession no. | Protein | No. of peptides matching databasec | Mascot scored | % Sequence coveragee |
|---|---|---|---|---|---|---|
| 658 | 4.04 | MXAN_4467 | Chaperonin GroEL | 18 | 152 | 38 |
| 659 | 4.11 | MXAN_4467 | Chaperonin GroEL | 18 | 187 | 36 |
| 660 | 3.26 | MXAN_7028 | ATP synthase α chain | 9 | 111 | 20 |
| 664 | 3.06 | MXAN_7028 | ATP synthase α chain | 8 | 100 | 18 |
| 730 | 2.28 | MXAN_6923 | ATP synthase β chain | 11 | 123 | 31 |
| 889 | 4.56 | MXAN_3068 | Translation elongation factor TU | 16 | 184 | 51 |
| 1267 | −240 | MXAN_2432 | Major spore protein B | 7 | 84 | NDf |
| 1272 | −167 | MXAN_2432 | Major spore protein B | 7 | 80 | ND |
| 1449 | −383 | MXAN_5430 | Developmental-specific protein S | 4 | 52 | 29 |
| MXAN_5432 | S homolog (S1) | 6 | 89 | 48 | ||
| 1450 | −352 | MXAN_5430 | Developmental-specific protein S | 4 | 49 | 29 |
| MXAN_5432 | S homolog (S1) | 6 | 84 | 48 | ||
| 1452 | −178 | MXAN_5430 | Development-specific protein S | 4 | 62 | ND |
| 1453 | −366 | MXAN_5430 | Development-specific protein S | 3 | 61 | ND |
| MXAN_5432 | S homolog (S1) | 5 | 31 | ND | ||
| 1518 | −22 | MXAN_2269 | Major spore protein A | 5 | 66 | ND |
| 1519 | −69 | MXAN_2269 | Major spore protein A | 5 | 63 | ND |
| 1520 | −655 | MXAN_2269 | Major spore protein A | 5 | 69 | ND |
| 1524 | −44 | MXAN_6969 | Major spore protein C | 6 | 85 | 32 |
| 1525 | −208 | MXAN_2269 | Major spore protein A | 6 | 80 | ND |
The numbers indicate Sypro Ruby-stained spots labeled in Fig. 2A.
Positive numbers are ratios for vegetative cell/spore protein fold differences, and negative numbers are ratios for spore/vegetative cell protein fold differences.
Number of peptides matching the database entry. The general consensus is that at least four or five peptides matching the predicted mass fingerprint are needed for a match to be considered a good match (“hit”).
The Mascot score represents the probability that the database entry “hit” is actually a random “hit.” The Mascot score was log transformed so that a high score represented a low probability of being random.
Percentage of the protein sequence of the “hit” that was covered by all the matching peptides. The general consensus is that a minimum of 20% sequence coverage for peptide mass fingerprinting is needed before a “hit” is considered a real “hit.”
ND, not determined.
Analysis of gene expression.
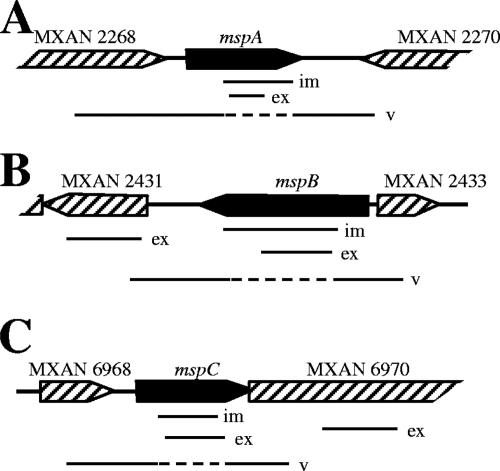
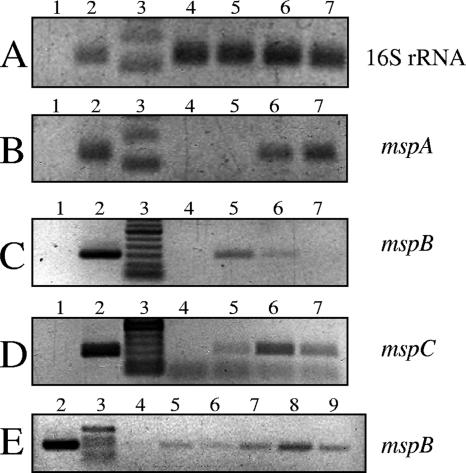
DIGE analysis showed that the MspA, MspB, and MspC proteins are relatively abundant in M. xanthus spores. Therefore, it is likely that the expression of the genes coding for these proteins increases when fruiting body development is induced. To examine the expression of these genes in strain DK1622 during fruiting body development, an RT-PCR analysis was performed. Figure 3 shows schematic diagrams of the three msp genes with surrounding open reading frames (ORFs) and the predicted PCR fragments used to generate insertion mutations or to determine levels of gene expression. We extracted mRNA from wild-type cells that were grown vegetatively in CTTYE broth and from wild-type cells that were allowed to develop on TPM agar for 24, 48, or 72 h. The relative levels of expression of mspA, mspB, and mspC are shown in Fig. 4. Using 16S rRNA as the template, similar levels of expression were detected in vegetatively growing cells (Fig. 4A, lane 4) and developing cells (Fig. 4A, lanes 5 to 7). For mspA, mspB, and mspC (Fig. 4B, C, and D, respectively) gene expression was not detected in vegetative cells (lanes 4) but was detected in sporulating cells (lanes 5 to 7). The levels of mspA and mspC gene expression were highest after 48 to 72 h of development on TPM agar. However, the level of expression of the mspB gene was highest after 24 h of starvation and then decreased with further development (Fig. 4C, compare lane 5 with lanes 6 and 7). To determine if mspB expression occurred prior to 24 h after starvation, the levels of mRNA for this gene were determined after 0, 6, 12, 18, 24, and 30 h of starvation (Fig. 4E, lanes 4 to 9). mspB gene expression occurred as early as 6 h after starvation, peaked at 24 h, and then decreased.
FIG. 3.
Genomic organization of msp genes: schematic diagrams for mspA (A), mspB (B), and mspC (C) (solid bars). Surrounding ORFs (striped bars) in the M. xanthus chromosome are also included. The relative positions are indicated for PCR products used to generate insertional mutations (im), to measure gene expression by RT-PCR (ex), and to verify insertion of the Kanr plasmid into the site of the gene (v). The dashed lines for the verification PCR products indicate potential sites into which the Kanr plasmid has integrated.
FIG. 4.
mRNA expression from msp genes during sporulation. RT-PCR was used to determine the relative levels of expression of 16S rRNA (A), mspA (B), mspB (C and E), and mspC (D) genes during myxospore development. Lanes 1 contained negative control PCR mixtures in which different primer sets were used to amplify products from RNA preparations that had been treated with DNase but not reverse transcribed. Lanes 2 contained positive control PCR mixtures in which chromosomal DNA was used as the PCR template. Lanes 3 contained DNA ladders. One microliter of each DNase-treated, reverse-transcribed reaction mixture was used as a template for PCRs (lanes 4 to 9). (A to D) Lane 4, zero time (just before aliquots of vegetative cells were spotted onto TPM starvation plates); lane 5, starvation for 24 h; lane 6, starvation for 48 h; lane 7, starvation for 72 h. (E) Lane 4, zero time; lane 5, starvation for 6 h; lane 6, starvation for 12 h; lane 7, starvation for 18 h; lane 8, starvation for 24 h; lane 9, starvation for 30 h.
Inactivation of sporulation genes and analysis of the effects on aggregation and myxospore formation.
To characterize the functions of MspA, MspB, and MspC in myxospore physiology, the corresponding genes were inactivated using single-crossover plasmid insertions, as previously described (4). PCR analysis was used to verify that the plasmid insertions occurred at the correct sites in the M. xanthus chromosome (Fig. 3). To confirm that the insertions in mspA, mspB, and mspC did not have polar effects on the transcription of downstream genes, we performed an RT-PCR analysis of downstream gene expression. The gene immediately downstream of mspA is in the opposite orientation, and so it was not tested for a possible polar effect. The MXAN_2431and MXAN_6970 genes are downstream of the mspB and mspC genes, respectively. The RT-PCR analysis showed that expression of these downstream genes was not affected by insertions in the corresponding upstream genes (data not shown).
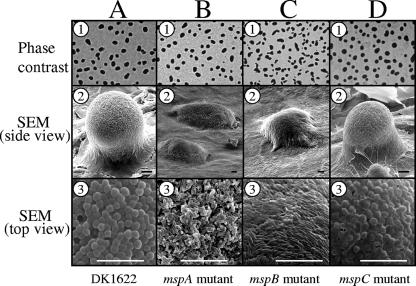
After verifying that the insertion mutations did not have polar effects, we assayed the strains to determine whether there were aggregation defects on TPM starvation agar. Phase-contrast microscopy (Fig. 5, row 1) revealed that after 5 days of development, wild-type strain DK1622 and the three mutant strains all produced phase-dark fruiting bodies. Furthermore, the timing of fruiting body formation was similar for all four strains (data not shown). However, high-resolution images of 5-day-old fruiting bodies obtained by SEM revealed differences between DK1622 and two of the msp mutants (Fig. 5, rows 2 and 3). The fruiting bodies of the mspA and mspB insertion mutants were relatively stunted (Fig. 5B2 and 5C2). In contrast to the mspA and mspB mutant fruiting bodies, the mspC mutant resembled those of wild-type strain DK1622 (Fig. 5D2). In addition to the overall appearance of the fruiting bodies, there were also differences in the morphologies of cells on the surfaces of the fruiting bodies (Fig. 5, row 3). DK1622 and mspC mutant fruiting bodies had spherical spores on their surfaces (Fig. 5A3 and D3, respectively). However, the cells on the surfaces of mspA mutant fruiting bodies appeared to be shortened rods whose widths resembled the widths of typical vegetative cells (Fig. 5B3). Surface-exposed cells on the mspB mutant fruiting bodies were indistinguishable from typical vegetative rod-shaped cells (Fig. 5C3). It is important to point out that despite the outer appearance of mspA and mspB mutant fruiting bodies, these clusters of cells did in fact contain spherical spores, as determined by phase-contrast microscopy of dispersed spores (data not shown) and TEM analysis of intact fruiting bodies (Fig. 6). Furthermore, mspA and mspB mutant fruiting body spores exhibited full or partial resistance to a variety of environmental stresses (Table 4).
FIG. 5.
Comparison of fruiting body formation on TPM agar. Five-day-old fruiting bodies of wild-type strain DK1622 (column A) and the mspA (column B), mspB (column C), and mspC (column D) mutants are shown. Fruiting bodies were viewed by either phase-contrast microscopy (row 1) or SEM (row 2, side view; row 3, top surface of fruiting bodies). Bars = 10 μm.
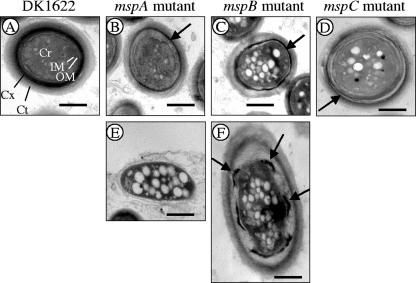
FIG. 6.
TEM comparison of ultrastructures of spores from 5-day-old fruiting bodies. Spores from fruiting bodies that had been fixed with glutaraldehyde and paraformaldehyde but had not been disrupted by sonication were examined. (A) DK1622 spore. Cr, core; IM, inner membrane; OM, outer membrane; Cx, cortex; Ct, coat. (B to D) Typical spores from mspA (B), mspB (C), and mspC (D) mutant fruiting bodies. These spores tended to have thin cortex layers (arrows). (E) Cell from the periphery of an mspA mutant fruiting body. In some spores of the mspB mutant a thin, intact cortex was observed (panel D, arrow), but in other spores the cortex appeared to be a series of disconnected patches (panel F, arrows). Bars = 500 nm.
TABLE 4.
Resistance of wild-type and msp mutant spores to heat, SDS, lysozyme, and sonication
| Strain | % Viable spores followinga:
|
|||||
|---|---|---|---|---|---|---|
| Heat treatment
|
SDS treatment
|
Lysozyme treatment (12 h) | Sonication (setting, 4.0) | |||
| 50°C, 2 h | 55°C, 2 h | 1% SDS, 1 h | 1% SDS, 2 h | |||
| DK1622 | 100 ± 37.8 | 100 ± 16.9 | 100 ± 27.1 | 100 ± 16.6 | 100 ± 19 | 100 ± 7.7 |
| AG681 | 50.1 ± 10 | 9.2 ± 1.9 | 2.2 ± 0.4 | 1.7 ± 0.8 | 136 ± 40 | 101.6 ± 12.3 |
| AG701 | 115.3 ± 5 | 41.4 ± 6.2 | 32.9 ± 12.7 | 15.3 ± 0.6 | 147 ± 45 | 100 ± 4.7 |
| AG810 | 4.6 ± 9 | 0.03 ± 0.02 | 5.6 ± 0.01 | 5.7 ± 0.01 | 14.6 ± 9.2 | 1.7 ± 0.3 |
The spore resistance assays were performed at least three times for each strain. The means ± standard deviations for the assays are expressed as percentages of the results for DK1622 (wild type), which were defined as 100%. The numbers of wild-type spores that survived the stress treatments ranged from 4.9 × 105 to 5.3 × 107 spores.
We previously reported that an M. xanthus strain having an insertion in a gene for cortex biosynthesis (cbgA mutant) generated aberrant fruiting bodies containing spores that lacked the type of electron-dense cortex layers typically seen in wild-type spores (25). Therefore, we examined the msp mutant spores described here for possible alterations in the cellular ultrastructure. Figure 6 shows the results of a TEM analysis of spores of wild-type strain DK1622 (Fig. 6A) and of the mspA, mspB, and mspC mutants (Fig. 6B, C, and D, respectively). Subcellular regions are indicated in Fig. 6A. Wild-type spores had characteristically thick, electron-dense cortex regions and outer coats with a lighter appearance (Fig. 6A). However, the spores of each of the three mutants examined had relatively thin cortexes compared to the cortexes of DK1622 spores. mspA mutant spores are shown in Fig. 6B and E. Fixation and imbedding of unsonicated fruiting bodies for TEM analysis allowed the multicellular architecture of the fruiting body to be viewed intact. Figure 6B shows a spore in the interior of a fruiting body, while Fig. 6E shows a cell on the perimeter of the same fruiting body. Spores in the interior of an mspA mutant fruiting body were spherical, but they had cortex layers that were much thinner than the cortex layers of wild-type spores (Fig. 6B). Cells on the exterior of mspA mutant fruiting bodies appeared to lack the ability to fully differentiate from vegetative cells into spores (Fig. 6E). This is consistent with the SEM data showing that the surfaces of mspA mutant fruiting bodies contained short rods that had not undergone complete cellular morphogenesis into spherical spores (Fig. 5B3). The interior of mspB mutant fruiting bodies also contained spores with relatively thin cortex layers (Fig. 6C). In addition, many of these interior mutant spores appeared to have cortex layers segmented into patches (Fig. 6F). The potential significance of these patches is addressed in the Discussion. TEM analysis of cells obtained from the mspC mutant fruiting bodies revealed spherical cells that had cortex layers that were separated from one another (Fig. 6D). This cortex phenotype was seen regardless of whether mspC mutant spores were on the surface or in the interior of fruiting bodies. Because this separation was not seen with wild-type spores prepared under identical conditions, we believe that it was not an artifact of TEM analysis. mspC mutant spores also had thinner coat layers than DK1622 spores. Collectively, these TEM and SEM results show that while the mspA, mspB, and mspC mutants are capable of forming aggregation centers, two of the mutant strains are defective for fruiting body morphogenesis and all three mutants have defects in spore morphogenesis.
Stress resistance properties of myxospores.
M. xanthus spores are more resistant to environmental stresses than their vegetative cell counterparts (22, 23). Previously, we reported that an M. xanthus strain in which a cortex biosynthesis gene (cbgA) was inactivated was still capable of forming fruiting bodies, but the resulting spores had decreased resistance to heat and SDS (25). The three sporulation mutants described here were subjected to the same stress analyses, and the results are shown in Table 4. Before and after each stress treatment, sonication at a low-intensity setting was used to disperse cells from the fruiting bodies. The viabilities of mutant spores were not affected by the relatively gentle sonication treatments.
When subjected to an elevated temperature (50°C) for 2 h, mspB mutant spores were relatively unaffected compared to spores of the wild-type DK1622 strain. However, after exposure to 50°C for 2 h, the numbers of viable mspA and mspC spores were 50 and 5% of the number of wild-type spores, respectively. After 2 h of exposure to 55°C, spores produced by all three msp mutants exhibited more pronounced losses of viability. At this elevated temperature, the numbers of viable spores of the mspA, mspB, and mspC mutants were 9, 41, and 0.03% of the number of wild-type spores, respectively.
Spores produced by all three mutants showed significant SDS sensitivity. We previously described a possible correlation between an intact spore cortex and resistance to heat and SDS treatment (25). The cortex formation defects (Fig. 6) and increased heat and detergent sensitivities of the msp mutants described here are consistent with this idea. In addition, we found that the mspC mutant spores exhibited increased sensitivity to sonication at a higher energy setting of the sonicator (Sonic Dismembrator [Fisher] at an intensity setting of 4.0 instead of 1.5). The mspC mutant also exhibited increased sensitivity to lysozyme. All three mutant strains showed wild-type levels of resistance to UV radiation (data not shown). In general, spores lacking MspC appeared to be the most compromised with regard to spore stress tolerance.
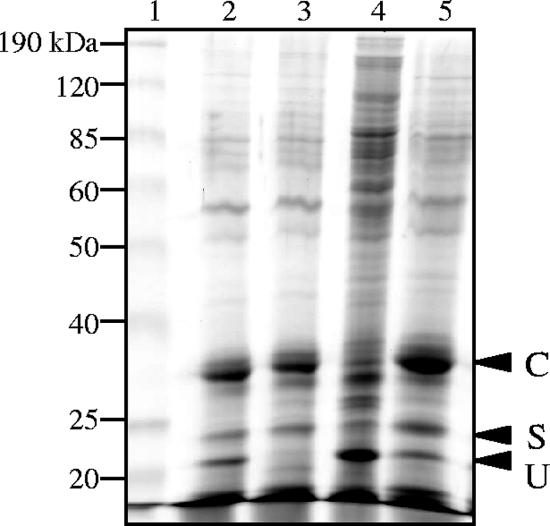
Analysis of spore surface proteins.
It is possible that the increased SDS sensitivity of the three msp mutants and the increased lysozyme sensitivity of the mspC mutant were due to altered spore coats since the coats are believed to act as molecular sieves (21). To test this hypothesis, the surface protein compositions of the three mutant spores were compared with that of DK1622 spores. By boiling spores without mechanically breaking them open, proteins C, S, and U could be released from wild-type spores, and they appeared as three prominent bands (Fig. 7, lane 2). Not all of these bands were present or prominent, however, in the msp mutant spore coat analyses. Spores lacking MspA also lacked protein U (Fig. 7,lane 3). mspB mutant spores had reduced levels of protein C compared to the levels in DK1622, and they appeared to release many more high-molecular-weight proteins (Fig. 7, lane 4). These larger proteins could mean that the mspB mutant spores were lysed during the SDS extraction and that many of the cells in the mspB mutant fruiting body retained a vegetative cell-like state (Fig. 5C3). The pattern of spore surface proteins released from the mspC mutant spores is indistinguishable from the pattern of spore surface proteins released from the DK1622 spores (Fig. 7, lane 5). This is in spite of the observation that mspC mutant spores have thinner coats than their wild-type counterparts (Fig. 6).
FIG. 7.
Coomassie blue-stained 12% polyacrylamide-SDS gel showing spore coat proteins extracted by boiling with 1% SDS. Lane 1, molecular weight standard; lane 2, DK1622; lane 3, mspA mutant; lane 4, mspB mutant; lane 5, mspC mutant. The positions of proteins C, S, and U are indicated, and their identities are based on similar positions observed in a previous study (16).
DISCUSSION
We report here the discovery of three novel M. xanthus spore proteins (major spore proteins) and the effects that inactivation of the corresponding genes have on fruiting body architecture and on spore morphology and stress resistance. These proteins were discovered using DIGE technology and the recently available M. xanthus annotated genome. Despite the production of large amounts of proteases, the M. xanthus protein lysates of vegetative cells were relatively stable when they were prepared in DTT lysis buffer (Fig. 1B). All three of the M. xanthus Msp proteins were present at relatively high levels in the spores (Fig. 2). Spores in this study were prepared from fruiting bodies that developed on TPM starvation agar for 5 days. However, it should be possible to use DIGE technology to examine the protein profiles of glycerol-induced spores. In agreement with the appearance of Msp proteins in spores, we observed that expression of the mspA, mspB, and mspC genes occurred when fruiting body development was induced on TPM agar (Fig. 4).
In an early study of myxospore protein production, Inouye et al. (10) used radioactive pulse-labeling of developing fruiting bodies. These authors showed that a variety of proteins are differentially expressed during fruiting body and spore morphogenesis; these proteins included 10 soluble proteins (designated proteins M toV) and five membrane-bound proteins (designated proteins 1 to 5). Genes for most of these spore proteins, however, have never been identified or characterized by mutational analysis. Two of the soluble proteins (proteins S and U) and a protein later identified by McCleary et al. (protein C) (16) have been examined, and protein S is the best characterized of the three. Production of protein S begins 5 to 11 h after initiation of starvation, reaches a peak at 24 h, and continues for as long as 48 h (10, 17). M. xanthus produces a paralog of protein S, called protein S1, that is produced at 52 to 168 h during development, as determined by Western blot analysis (24). lacZ fusions to the tps (protein S) and ops (protein S1) promoters also confirmed these temporal patterns for the two protein S paralogs (5). Like protein S, protein C is a major spore surface protein of M. xanthus. Western blot analysis showed that protein C production begins as early as 6 h after starvation initiation (16). Although protein U is also a spore surface protein, its synthesis begins after spore formation begins (40 to 45 h) (10). Protein U may appear on spore surfaces since this protein, unlike protein S, has a signal peptide sequence for secretion (9). Deletion analysis of the tps and ops genes revealed that despite a delay in morphogenesis, there is no defect in aggregation, fruiting body morphogenesis, or spore resistance to heat or sonication (8, 14, 21). Mutational analyses of the genes for protein C and protein U have not been reported. The lack of a significant phenotype for protein S- and S1-deficient spores is in contrast to the results obtained for all three msp mutants described here. Whereas all three mutant strains aggregate normally, the mspA and mspB mutants both show defects in fruiting body morphogenesis (Fig. 5). All three msp mutants show reduced resistance to heat and SDS detergent. The mspC mutant spores also show reduced resistance to sonication and lysozyme treatment (Table 4).
In the annotated M. xanthus genome, the three Msp proteins are encoded by hypothetical ORFs. These proteins could have enzymatic or regulatory functions, but because high concentrations of them are present in M. xanthus spores, it seems likely that they have structural roles. Gram-negative bacteria, such as M. xanthus, have five major subcellular localization sites. These sites are the cytoplasm, the inner membrane, the periplasm, the outer membrane, and the extracellular region. Various computer programs have been developed to help predict the subcellular locations of proteins. Using the CELLO program (28), MspB was predicted to be an extracellular protein, while MspA and MspC were both predicted to be periplasmic proteins. CELLO predictions are based on bacterial proteins whose sequences are known and whose subcellular localizations were determined empirically. Despite the strength of the CELLO program to predict subcellular locations, experimental studies are needed to determine the actual locations of the three Msp proteins in M. xanthus spores.
The mspA and mspB mutants both produced fruiting bodies with rod-shaped cells on their surfaces (Fig. 5, row 3), while inside the mounds the cells were spherical (Fig. 6). This may have been due to a developmental delay in spore formation, like the delay that has been observed for M. xanthus mutants in which protein S has been inactivated (8). Perhaps if mspA and mspB fruiting bodies had been allowed to develop for more than 5 days they would have had spherical cells on their surfaces. The mspA and mspB mutant fruiting bodies also did not form mounds that were the height of DK1622 fruiting bodies. This might have been due to reduced adhesion of spores inside each mound that allowed spores to be packed higher. Inouye et al. (12) have speculated that the function of protein S on spore surfaces is to act as an adhesive to allow connectivity between adjacent spores. Lack of cell-to-cell cohesiveness seems especially possible for mspB mutant spores since they are more easily dispersed from fruiting bodies by hand vortexing (data not shown). Figure 7 shows that mspA mutant spores lack protein U and mspB mutant spores have reduced levels of protein C. The absence of these cell surface proteins might affect cell adhesion in the fruiting bodies. mspC mutant fruiting bodies, however, appear to have the same spore coat proteins as DK1622 spores, and correspondingly, mspC mutant spores form fruiting bodies whose appearance is identical to that the wild-type strain fruiting bodies (Fig. 5). However, mspC mutant spores have the thinnest coats and the greatest environmental sensitivities of the three msp mutant strains.
Future studies of M. xanthus spore Msp proteins will involve generating polyclonal antibodies to the three proteins. Such antibodies should allow temporal studies of Msp production, like those that have been performed for proteins S and C using Western blot analysis (16, 24). These antibodies should also help us determine if the Msp proteins are expressed in peripheral rods, as proteins S and C are (18), and they should allow localization studies using subcellular spore fractions or immunoelectron microscopy.
DIGE technology allowed us to compare the proteomes of vegetative cells and spores, but another approach is needed to compare the polysaccharide compositions of these two cell types. After 8 h of glycerol induction of sporulation, up to 75% of the spore coat's dry weight is accounted for by carbohydrates containing glucose and galactosamine (15). Even more dramatic is the observation that the total level of cellular polysaccharides increased 200% during glycerol induction (1). Pulse-chase labeling has revealed dramatic alterations in polysaccharide metabolism as the M. xanthus spore coat develops (6, 7). The observation that the coat thickness of the mspC mutant is reduced (Fig. 6D) but amounts of coat proteins are not diminished (Fig. 7, lane 5) suggests that the carbohydrate content in the coat may be reduced. Thus, the carbohydrate composition of myxospores is probably important for the structural and stress resistance properties of the spores, and this hypothesis awaits further analysis.
Acknowledgments
This research was made possible by National Science Foundation award MCB 031674 to J. L. Dahl and A. G. Garza.
We thank the Monsanto Company and TIGR for providing access to the M. xanthus genome sequence prior to submission to GenBank (accession number CP000113). We thank Christine Davitt for her assistance with the electron microscopy studies. Kriti Arora helped in editing the manuscript, and John Wyrick assisted in the bioinformatics analysis.
This research is dedicated in loving memory of Robert J. Kadner, a mentor and friend.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Bacon, K., D. Clutter, R. H. Kottel, M. Orlowski, and D. White. 1975. Carbohydrate accumulation during myxospore formation in Myxococcus xanthus. J. Bacteriol. 124:1635-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betts, J. C., P. Dodson, S. Quan, A. P. Lewis, P. J. Thomas, K. Duncan, and R. A. McAdam. 2000. Comparison of the proteome of Mycobacterium tuberculosis strain H37Rv with clinical isolate CDC 1551. Microbiology 146:3205-3216. [DOI] [PubMed] [Google Scholar]
- 3.Butcher, P. D., J. A. Mangan, and I. M. Monahan. 1998. Intracellular gene expression, p. 285-306. In T. Parish and N. G. Stoker (ed.), Mycobacterial protocols, 1st ed. Humana Press, Totowa, NJ.
- 4.Caberoy, N. B., R. D. Welch, J. S. Jakobsen, S. C. Slater, and A. G. Garza. 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J. Bacteriol. 185:6083-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Downard, J. S., D. Kupfer, and D. R. Zusman. 1984. Gene expression during development of Myxococcus xanthus. Analysis of the genes for protein S. J. Mol. Biol. 175:469-492. [DOI] [PubMed] [Google Scholar]
- 6.Filer, D., S. H. Kindler, and E. Rosenberg. 1977. Myxospore coat synthesis in Myxococcus xanthus: enzymes associated with uridine 5′ diphosphate-N-acetylgalactosamine formation during myxospore development. J. Bacteriol. 131:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filer, D., D. White, S. H. Kindler, and E. Rosenberg. 1977. Myxospore coat synthesis in Myxococcus xanthus: in vivo incorporation of acetate and glycine. J. Bacteriol. 131:751-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuichi, T., T. Komano, M. Inouye, and S. Inouye. 1985. Functional complementation between two homologous genes, ops and tps, during differentiation of Myxococcus xanthus. Mol. Gen. Genet. 199:434-439. [DOI] [PubMed] [Google Scholar]
- 9.Gollop, R., M. Inouye, and S. Inouye. 1991. Protein U, a late-developmental spore coat protein of Myxococcus xanthus, is a secretory protein. J. Bacteriol. 173:3597-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inouye, M., S. Inouye, and D. R. Zusman. 1979. Gene expression during development of Myxococcus xanthus; pattern of protein synthesis. Dev. Biol. 68:579-591. [DOI] [PubMed] [Google Scholar]
- 11.Inouye, M., S. Inouye, and D. R. Zusman. 1979. Biosynthesis and self-assembly of protein S, a development specific protein of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inouye, S., T. Franceschini, and M. Inouye. 1983. Structural similarities between the development-specific protein S from a Gram-negative bacterium, Myxococcus xanthus, and calmodulin. Proc. Natl. Acad. Sci. USA 80:6829-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komano, T., T. Furuichi, M. Teintze, M. Inouye, and S. Inouye. 1984. Effects of deletion of the gene for the development-specific protein S on differentiation of Myxococcus xanthus. J. Bacteriol. 158:1195-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kottel, R. H., K. Bacon, D. Clutter, and D. White. 1975. Coats of Myxococcus xanthus: characterization and synthesis during myxospore differentiation. J. Bacteriol. 124:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCleary, W. R., B. Esmon, and D. R. Zusman. 1991. Myxococcus xanthus protein C is a major spore surface protein. J. Bacteriol. 173:2141-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson, D. R., and D. R. Zusman. 1983. Transport and localization of protein S, a spore coat protein, during fruiting body formation by Myxococcus xanthus. J. Bacteriol. 150:547-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connor, K. A., and D. R. Zusman. 1991. Analysis of Myxococcus xanthus cell types by two-dimensional polyacrylamide gel electrophoresis. J. Bacteriol. 173:3334-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otani, M., K. Satoshi, X. Chunying, C. Umezawa, K. Sano, and S. Inouye. 1998. Protein W, a spore-specific protein in Myxococcus xanthus, formation of a large electron-dense particle in a spore. Mol. Microbiol. 30:57-66. [DOI] [PubMed] [Google Scholar]
- 20.Rosenbluh, A., and E. Rosenberg. 1993. Developmental lysis and autocides, p. 213-233. In M. Dworkin and D. Kaiser (ed.), Mycobacteria II. American Society for Microbiology, Washington, DC.
- 21.Scherrer, R., and P. Gerhardt. 1964. Molecular sieving by cell membranes of Bacillus megaturium. Nature 204:649-650. [DOI] [PubMed] [Google Scholar]
- 22.Sudo, S., and M. Dworkin. 1973. Comparative biology of prokaryotic resting cells. Adv. Microb. Physiol. 9:153-224. [DOI] [PubMed] [Google Scholar]
- 23.Sudo, S. Z., and M. Dworkin. 1969. Resistance of vegetative cells and microcysts of Myxococcus xanthus. J. Bacteriol. 98:883-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teintze, M., M. Inouye, and S. Inouye. 1985. Two homologous genes coding for spore-specific proteins are expressed at different times during development of Myxococcus xanthus. J. Bacteriol. 163:121-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tengra, F. K., J. L. Dahl, D. Dutton, L. Coyne, and A. G. Garza. Identification and characterization of a cortex biosynthesis gene involved in Myxococcus xanthus spore stress resistance. J. Bacteriol., in press.
- 26.Tonge, R., J. Shaw, B. Middleton, R. Rowlinson, S. Rayner, J. Young, F. Pognan, E. Hawkins, I. Currie, and M. Davison. 2001. Validation and development of fluorescence two-dimensional gel electrophoresis proteomics technology. Proteomics 1:377-396. [DOI] [PubMed] [Google Scholar]
- 27.Vasquez, G. M., F. Qualls, and D. White. 1985. Morphogenesis of Stigmatella aurantiaca fruiting bodies. J. Bacteriol. 163:515-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, C. S., C. J. Lin, and J. K. Hwang. 2004. Predicting subcellular localization of proteins for gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13:1402-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]