Abstract
Escherichia coli strains that cause disease outside the intestine are known as extraintestinal pathogenic E. coli (ExPEC) and include human uropathogenic E. coli (UPEC) and avian pathogenic E. coli (APEC). Regardless of host of origin, ExPEC strains share many traits. It has been suggested that these commonalities may enable APEC to cause disease in humans. Here, we begin to test the hypothesis that certain APEC strains possess potential to cause human urinary tract infection through virulence genotyping of 1,000 APEC and UPEC strains, generation of the first complete genomic sequence of an APEC (APEC O1:K1:H7) strain, and comparison of this genome to all available human ExPEC genomic sequences. The genomes of APEC O1 and three human UPEC strains were found to be remarkably similar, with only 4.5% of APEC O1's genome not found in other sequenced ExPEC genomes. Also, use of multilocus sequence typing showed that some of the sequenced human ExPEC strains were more like APEC O1 than other human ExPEC strains. This work provides evidence that at least some human and avian ExPEC strains are highly similar to one another, and it supports the possibility that a food-borne link between some APEC and UPEC strains exists. Future studies are necessary to assess the ability of APEC to overcome the hurdles necessary for such a food-borne transmission, and epidemiological studies are required to confirm that such a phenomenon actually occurs.
Escherichia coli is among the world's most well-studied organisms and is often found at the forefront of advancing technology. Not surprisingly, E. coli is on the leading edge of an ongoing shift in the field of genomics (3, 6, 65). Now that at least one representative organism per species has been sequenced for most pathogens of interest, the focus in genomics has reoriented towards obtaining multiple sequences within a species. With more genomic sequences available for E. coli than for any other species, it leads this trend (3). Thus far, all of the pathogenic E. coli strains sequenced have originated from human hosts (6, 8, 10, 24, 47, 68). This bias has left a gap in our knowledge, as various E. coli strains cause significant and widespread disease in animals, including in those raised for human consumption (2, 13, 41). Consequently, while the genomic analysis of E. coli strains from animals can be justified solely on the basis of E. coli's detrimental impact on animal agriculture, a broader justification would also include the potential link between animal-source E. coli and human disease.
Links between human and animal disease caused by E. coli are well established in some instances but remain speculative in others. For instance, recent reports of outbreaks of human urinary tract infections (UTIs) have stimulated interest in the potential that E. coli from animals has to cause human UTIs via the food supply (28, 41, 49). Since UTIs are among the world's most common bacterial infections (20), cause significant morbidity, and cost the health care system of the United States over a billion dollars annually (20, 55), this putative link deserves attention. Although it is generally agreed that the immediate source of uropathogenic E. coli (UPEC) causing human UTIs is an individual's own colonic flora (27), it is not completely understood how these virulent clones come to inhabit the colon. One hypothesis is that retail poultry harboring avian pathogenic E. coli (APEC) represents a food-borne source of E. coli clones capable of causing human UTIs (50, 52).
APEC cause colibacillosis in production birds, a widespread disease which is responsible for multimillion dollar annual losses for the world's poultry industries (2). APEC strains are a subset of extraintestinal pathogenic E. coli (ExPEC), a pathotypic category which also includes UPEC (55) and E. coli causing neonatal meningitis and septicemia (27, 34). As ExPEC, APEC and UPEC both cause extraintestinal disease, share virulence-associated traits, and have overlapping O serogroups and phylogenetic types (50). Additionally, there is a well-documented history of transfer of E. coli strains and their plasmids from poultry to humans (37, 38, 44, 67), and a recent report has shown that APEC plasmids can contribute to the urovirulence of E. coli for mammalian hosts (62). These observations have led to further speculation that APEC might be a source of UPEC, which gains access to the human colon following ingestion of contaminated poultry (50). Here, we begin to test the hypothesis that some APEC strains are a source of human UPEC by generating the first APEC genome sequence and comparing it to all of the human UPEC genomes currently available. This newly sequenced ExPEC strain, APEC O1 (an O1:K1:H7 strain), was chosen for study because its serotype, phylogenetic group, and virulence genotype were representative of 1,000 well-characterized APEC and human UPEC isolates.
MATERIALS AND METHODS
Choice of strain for sequencing. (i) Bacterial strains.
The choice of APEC O1 for sequencing was based on its possession of traits characteristic of strains causing extraintestinal disease in poultry and humans. APEC isolates were obtained from lesions of chickens and turkeys clinically diagnosed with colibacillosis from various locations within the United States. The UPEC strains were isolated from cases of human UTIs; they were kindly provided by Paul Carson (Meritcare Hospital, Fargo, ND) or were from the VA Medical Center, Minneapolis, MN. A subset of both groups of isolates has been described previously (50, 51). For embryo lethality studies, three additional APEC isolates were used: APEC V1 (O nontypeable) and APEC V2 and V7 (both O2). For 1-day-old chick studies, three strains previously characterized as being of high, intermediate, and low lethality towards birds were used (53). All isolates were serogrouped through the E. coli Reference Center (Pennsylvania State University, University Park, PA) and were screened for traits associated with APEC and UPEC virulence, using the techniques described below. Organisms were stored at −80°C in brain heart infusion broth (Difco Laboratories, Detroit, MI) with 10% (vol/vol) glycerol until use (57).
(ii) Virulence gene and phylogenetic typing.
Test and control organisms were examined for the presence of a variety of genes known for their association with APEC or UPEC virulence by using multiplex PCR procedures which have been previously described (31, 32, 50, 51).
Genomic analysis. (i) Genomic sequencing of APEC O1.
Two random shotgun libraries of APEC O1 were created: a 2- to 3-kb small-insert library and a 35- to 40-kb fosmid library. Shotgun sequencing was performed on the small-insert library by MWG Biotech (Hedersberg, Germany), using ABI 3700 and ABI 3730xl capillary sequencers. The fosmid library was also end sequenced. Finishing was accomplished using the pooled primer technique described by Tettelin et al. and used elsewhere (31, 32, 66). The final gaps were closed using standard PCR. Sequencing reads were assembled using SeqMan software from DNASTAR (Madison, WI). Open reading frames (ORFs) within the plasmid sequence were identified using GeneQuest from DNASTAR (Madison, WI), followed by manual inspection. The initiation codon giving the longest coding region was used, including GUG and UUG. ORFs corresponding to proteins larger than 66 amino acids in size were identified. Translated ORFs were then compared to known protein sequences using by BLAST (NCBI, September 2006). Those with greater than 90% amino acid sequence identity over greater than 90% of the sequence were considered matches. The DNA sequence was also analyzed to identify noncoding features. The G+C content of individual ORFs was analyzed using EMBOSS (58). Insertion sequences and repetitive elements were identified using IS FINDER (http://www-is.biotoul.fr/). tRNAs were identified using tRNA-Scan (39).
(ii) Comparative genomics.
Each ORF of APEC O1 was compared to the predicted proteins of E. coli strains K-12 MG1655, CFT073, 536, and UTI89 by using the BLAST tool (1). The APEC O1 chromosome was aligned to the CFT073, 536, and UTI89 chromosomes by using MAVID (7), and these alignments were visualized by using GenomeViz (22). The GenomeViz output was incorporated into a circular genome map created by using GenVision from DNASTAR.
(iii) MLST.
Multilocus sequence typing (MLST) was used to determine the genetic relatedness of all the fully sequenced E. coli genomes that are available to the public (http://www.mlst.net). Additionally, Fred Blattner and Guy Plunkett III of the University of Wisconsin, Madison, kindly provided access to the pertinent sequences of neonatal meningitis-associated E. coli strain RS218 for use in this study (http://www.genome.wisc.edu). Seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) were extracted from the sequence database for all these genomic sequences, concatenated to produce a contiguous sequence of 9,015 bp, and aligned by the neighbor-joining method with 1,000 bootstrap iterations using ClustalW. A dendrogram of the groupings based on this alignment was created using MegAlign from DNASTAR. Sequences were also assigned sequence types based upon the scheme described at the E. coli MLST database (http://web.mpiib-berlin.mpg.de/mlst/dbs/Ecoli), which assigns allele designations to unique variants based on ∼500-bp internal regions of each gene.
Virulence assays. (i) Embryo lethality assay.
APEC O1 and several other well-characterized APEC strains were assessed for lethality in chicken embryos by inoculation of 500 CFU of overnight washed bacterial cultures into the allantoic cavities of 12-day-old, embryonated, specific-pathogen-free eggs (42). Phosphate-buffered saline-inoculated and uninoculated embryos were used as controls. Embryo deaths were recorded for 4 days.
(ii) Day-old chick lethality.
The ability of APEC O1 to cause avian colibacillosis was confirmed in chicks, as described previously (35, 46, 53). Day-old chicks, vaccinated only against Marek's disease, were obtained from a commercial source and were divided into groups of 10 chickens each in Horsfall units at the Livestock and Infectious Disease Facility, Iowa State University. Chicks were provided food and water ad libitum. Chicks were inoculated intratracheally with 0.1 ml of a bacterial suspension containing ∼107 CFU/ml of APEC O1 or with 0.5 ml of phosphate-buffered saline subcutaneously in the back of the neck. Chicks were monitored for 7 days. Deaths were recorded, and the survivors were euthanatized and examined for macroscopic lesions. Organisms were classified as being of low, intermediate, or high virulence based upon mortalities and gross lesions related to colibacillosis. Three APEC strains representing each of the pathogenicity groups, as kindly supplied by Sandra Cloud (University of Delaware), were used as controls (53).
Biostatistics.
A cluster analysis of the genotyping results for the isolates tested was performed using the average linkage method based upon Jaccard's dissimilarity coefficient calculated from the presence of virulence genes (SAS Institute, Inc., 2004). In order to better discern patterns among the isolates, results of the cluster analysis along with the isolates' virulence genotypes were used to construct a single figure as described previously (50). Differences in embryo lethality between the strains were evaluated for statistical significance using a z-test for the equality of two binomial proportions (63). Differences in day-old-chick lethality were examined using Fisher's exact test (63).
Nucleotide sequence accession number.
The complete sequence and annotation of APEC O1 has been deposited in GenBank under accession number NC_008563.
RESULTS AND DISCUSSION
Choice of strain.
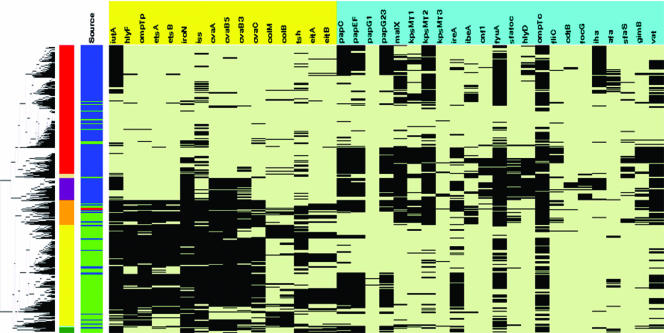
Extensive virulence genotyping was performed on large populations of APEC and UPEC strains in order to identify an APEC strain for sequencing. This was done in an effort to identify an APEC strain that was classified in a serogroup, phylogenetic group, and virulence genotype characteristic of both APEC and UPEC. Serogrouping, phylogenetic typing, and virulence genotyping for nearly 40 APEC- and ExPEC-associated genes were performed on 500 APEC and 500 UPEC isolates (Fig. 1). The genes examined included those encoding plasmid-associated virulence factors, such as the increased serum survival gene (iss) (19, 25, 43, 48), the aerobactin and salmochelin siderophore system genes (16, 31, 32), the autotransporter gene tsh (17, 31, 32), and certain colicin-associated genes (31, 32). Chromosomal virulence-associated genes were also sought, such as the vacuolating autotransporter gene (vat) (45), the pap (pyelonephritis-associated pilus) operon (36), the yersiniabactin siderophore system genes (55), and the ireA (iron-responsive element) gene (26, 36, 54). A cluster analysis of these genotyping results separated the APEC and UPEC isolates into two primary clusters and several subclusters (Fig. 1). One of the two major clusters contained primarily APEC strains, characterized by their possession of plasmid-associated virulence genes. The second major cluster contained primarily UPEC strains, characterized by virulence genes which have been localized to chromosome-encoded pathogenicity islands (PAIs). The remaining isolates fell into mixed multiple clusters, containing both APEC and UPEC strains and possessing both plasmid- and chromosome-encoded virulence traits. APEC O1 was selected from one of these mixed clusters (Fig. 1). Within this mixed cluster were 85 isolates, of which 67% were found to be of the B2 phylotype. The most commonly occurring serogroups within this cluster were O1, O2, and O18.
FIG. 1.
Cluster analysis of 500 APEC and 500 UPEC strains for 39 ExPEC-associated traits. The leftmost portion of the figure is the dendrogram created from the average linkage cluster analysis on the presence of virulence factors. Just to the right of the dendrogram is column 1, which shows cluster membership based upon the dendrogram: red, cluster 1; cream, cluster 2; purple, cluster 3; orange, cluster 4; yellow, cluster 5; light blue, cluster 6; and green, cluster 7. Column 2 identifies an isolate as a UPEC strain (blue) or an APEC strain (green). Also, APEC O1 is identified in this column with a red bar. Columns 3 to 41 show the virulence genotype of each isolate tested. Each column in this group shows the results for a single gene. Black, gene is present; light green, gene is absent. Labels across the top of columns 3 to 41 show the APEC plasmid-linked genes (yellow) and the ExPEC chromosome-associated virulence genes (sky blue). ompTp, plasmid-encoded ompT; cvaB5, 5′ end of the cvaB gene; cvaB3, 3′ end of the cvaB gene; colM, cma; colB, cbi; papG1, papG allele 1; papG23, papG alleles 2 and 3. This method of analysis was first described by Rodriguez-Siek et al. (50) and has since been used in similar form elsewhere (8).
APEC O1 was selected because it contained traits typical of both APEC and UPEC (50). APEC O1 is an O1:K1:H7 strain that was originally isolated from the lung of a chicken clinically diagnosed with colisepticemia. The O1 serogroup is one of the more commonly occurring serogroups among both APEC and UPEC strains (2, 29, 50, 51). O1 strains are also known to cause other important conditions in animals (5, 18, 21) and humans (4, 60), and a recent report of interhousehold spread of a UPEC O1:K1:H7 strain involving family members and their dog suggests that such strains may have zoonotic potential (30). Despite the widespread contributions of O1 strains to human and animal disease, no genomic sequence of an E. coli O1 strain had been described prior to this report.
APEC O1 is also classified in the B2 phylogenetic group, which is closely associated with extraintestinal virulence in E. coli (12, 50). Prior to sequencing, the ability of APEC O1 to cause disease in birds was confirmed by intratracheal inoculation of 1-day-old broiler chicks, according to the scheme used by Rosenberger et al. (53). Using this model, APEC O1 was categorized as being highly virulent for chickens (Table 1).
TABLE 1.
Lethality for 1-day-old chicks
| Organisma | Mean lesion scoreb | P valuec |
|---|---|---|
| APEC O1 | 2.69 | |
| Controls | ||
| High | 2.75 | 0.54 |
| Medium | 0.89 | <0.0001 |
| Low | 0.17 | <0.0001 |
APEC O1 and the control organisms (one each for high, intermediate, and low pathogenicity) were classified by their virulence towards chicks (53).
Average of lesion scores (ranked from 0 to 3 based upon occurrence of airsacculitis, pericarditis, and perihepatitis) for 12 birds for each organism tested.
P value generated using Fisher's exact test (63). There was no statistically significant difference between the lesion scores following inoculation with APEC O1 and the high-pathogenicity group control. However, the lesion scores obtained with APEC O1 did differ significantly from those in birds inoculated with the medium- and low-pathogenicity control organisms. Collectively, these results demonstrate that APEC O1 is classified as a “high-pathogenicity” organism.
Overview of the APEC O1 genome.
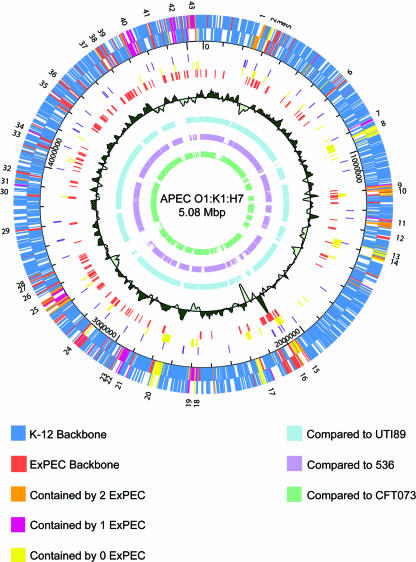
The genome of APEC O1 was assembled and finished at approximately eightfold coverage. It contained a 5,082,025-bp chromosome (Fig. 2) and, unlike other sequenced ExPEC strains, contained four plasmids totaling 565,600 bp in size (Table 2), giving it a total genome size of 5,647,625 bp. The presence of these plasmids was not surprising, considering that plasmids are a defining trait of the APEC pathotype (32, 33, 51). Such plasmids have also been identified among UPEC strains, although to a lesser extent (64). Two of APEC O1's plasmids have been previously described: a 174,231-bp IncFIB, colicin-encoding virulence plasmid (32) and a 241,348-bp IncHI2 resistance plasmid encoding resistance to eight antimicrobial agents (33). The remaining two plasmids, pAPEC-O1-Cryp1 and pAPEC-O1-Cryp2, are 101 and 49 kb in size, respectively. These plasmids share limited identity with Yersinia-type plasmids, encode mostly hypothetical proteins, and do not confer any apparent phenotype.
FIG. 2.
Map of the APEC O1 chromosome and comparison of APEC O1's genome to those of other ExPEC strains. The outer ring shows genomic islands, as numbers (1 to 43) corresponding to those in Table 5; the 2nd and 3rd rings are coding regions in forward and reverse orientation (blue, backbone ORFs also present in K-12; dark orange, ORFs absent from K-12 but present in all other sequenced ExPEC strains; light orange, ORFs absent from K-12 but present in APEC O1 and two other ExPEC strains; lavender, ORFs absent from K-12 but present in APEC O1 and one other ExPEC strain; yellow, ORFs present only in APEC O1); the 4th ring depicts the scale in base pairs; the 5th ring depicts tRNA genes in dark purple; the 6th ring depicts ORFs unique to APEC O1 in yellow; the 7th ring depicts ORFs common to all sequenced ExPEC strains in orange; the 8th ring shows sliding G+C content compared to the overall average of 50.6%; and the 9th, 10th, and 11th rings show genome alignments of APEC O1 with UTI89 (light blue), 536 (light purple), and CFT073 (light green), respectively.
TABLE 2.
Overview of APEC O1 genome
| Structure | Size (bp) | G+C content (%) | No. (%) of ORFsa
|
|||
|---|---|---|---|---|---|---|
| Total | Backbone | Common ExPEC | APEC O1 specific | |||
| Chromosome | 5,082,025 | 50.55 | 4,467 | 3,497 (78.3) | 3,890 (87.1) | 202 (4.5) |
| Plasmids | ||||||
| pAPEC-O1-ColBM | 174,241 | 49.65 | ||||
| pAPEC-O1-R | 241,387 | 46.43 | ||||
| pAPEC-O1-Cryp1 | 105,834 | 46.55 | ||||
| pAPEC-O1-Cryp2 | 46,870 | 44.62 | ||||
| Total genomeb | 5,650,357 | 5,042 | 3,497 (69.4) | 3,901 (77.4) | 721 (14.3) | |
ORFs are defined as backbone, common ExPEC, or APEC O1 specific based on their comparison with E. coli strains K-12 MG1655, CFT073, UTI89, and 536.
The total genome includes 93 tRNA genes and 22 rRNA genes.
The G+C content of the APEC O1 chromosome was found to be 50.5%, which is comparable to that of other E. coli strains (3). However, the plasmids of APEC O1 had a variable G+C content, ranging from 44.6% to 49.6% (Table 2). Within the chromosome, regional differences in G+C content could be discerned, with the differences occurring primarily within the genomic islands, which are defined here as regions of APEC O1 greater than 4 kb that are not present in E. coli K-12 MG1655. Codon adaptation indices reflected such differences in G+C content, with differential codon usage also occurring within the genomic islands.
APEC O1 is highly similar to human ExPEC.
At the time of this publication, the genomic sequences of three human ExPEC strains were available for comparative genomics: UPEC strain CFT073 (O6:K2:H1), isolated from a patient with pyelonephritis (68); UPEC strain 536 (O6:K15:H31), isolated from a patient with a complicated UTI (8); and UPEC UTI89 (O18:K1:H7), isolated from a patient with uncomplicated cystitis (10). Comparison of APEC O1 to these E. coli sequences was quite revealing (Table 3). Overall, APEC O1 shared the greatest nucleotide and protein similarities with UTI89, followed by 536 and CFT073. APEC O1 has a chromosome which is 149 kb smaller than that of CFT073, 143 kb larger than that of 536, and 16 kb larger than that of UTI89 (Table 3). Within the APEC O1 chromosome, 4,467 ORFs were identified, 78.3% of which were a part of a backbone common among all sequenced E. coli strains. This comparison also revealed the presence of a common ExPEC backbone (K-12-like sequences plus common ExPEC sequences), accounting for 87.1% of the ORFs of APEC O1. Of these ORFs, 9% were common to all sequenced ExPEC strains but absent from E. coli K-12 MG1655. Such a backbone of genes might be essential in the ability of ExPEC to cause extraintestinal disease or to survive in the extraintestinal environment, as genes within this ExPEC backbone include loci thought to play a role in ExPEC virulence and/or fitness. Some of the regions identified within this ExPEC backbone include the yersiniabactin and pap operons, ireA, vat, the lipopolysaccharide core synthesis region, the sit iron/manganese transport system, and the auf fimbrial operon (9, 36, 54, 56, 59) (Table 4). In addition to the characterized islands within the common ExPEC backbone, 16 other islands were identified and were simply termed “ExPEC islands,” encoding mostly hypothetical proteins (Table 5). Future studies will be needed to ascertain their functions and roles in virulence and persistence, if any.
TABLE 3.
Comparison of the fully sequenced ExPEC strains
| Strain | Chromosome size (bp) | Plasmid size(s) (kb) | Content of APEC O1 ORFs (%) |
|---|---|---|---|
| APEC O1 | 5,082,025 | 240, 174,106, 47 | 100 |
| UPEC UTI89 | 5,065,741 | 114 | 93 |
| UPEC CFT073 | 5,231,428 | 90 | |
| UPEC 536 | 4,938,875 | 87 | |
| ExPECa | 3,942,842 | NAb | 86 |
| E. coli K-12 MG1655 | 4,639,221 | 78 |
All completely sequenced ExPEC strains.
NA, not applicable.
TABLE 4.
Virulence-associated genes of APEC O1
| Gene(s) | Description | Presence in:
|
|||
|---|---|---|---|---|---|
| APEC O1 | CFT073 | UTI89 | 536 | ||
| papI to -G | pap fimbrial operon | + | + | + | + |
| ireA | Iron-regulated element | + | + | − | − |
| fyuA | Yersiniabactin siderophore gene | + | + | + | + |
| iutA | Aerobactin siderophore gene | + | + | − | − |
| iroN | Salmochelin siderophore gene | + | + | + | + |
| sitA to -D | Iron/manganese transport genes | + | + | + | + |
| iss | Serum survival gene | + | − | − | − |
| vat | Vacuolating autotransporter gene | + | + | + | + |
| tsh | Autotransporter/adhesin gene | + | − | − | − |
| cdtB | Cytolethal distending toxin gene | + | − | − | − |
| ibeA | Invasion gene | + | − | + | − |
| tia | Invasion gene | + | − | − | − |
TABLE 5.
Genomic islands of APEC O1
| Island no. | Start (bp) | Stop (bp) | Description | Size (bp) | Presence in strain:
|
|||
|---|---|---|---|---|---|---|---|---|
| K-12 MG1655 | CFT073 | UTI89 | 536 | |||||
| 1 | 271626 | 243408 | PAI IIAPEC O1 near tRNA-Asp | 28,218 | − | − | + | + |
| 2 | 314347 | 295179 | PAI IIIAPEC O1 near tRNA-Thr; contains vat | 19,168 | − | + | + | + |
| 3 | 328228 | 339123 | ExPEC island | 10,895 | − | + | + | + |
| 4 | 347107 | 353093 | ExPEC island | 5,986 | − | + | + | + |
| 5 | 370099 | 375196 | ExPEC island | 5,097 | − | + | + | − |
| 6 | 682227 | 691198 | ExPEC island | 8,971 | − | + | + | + |
| 7 | 888045 | 897875 | ExPEC island | 9,830 | − | − | + | − |
| 8 | 919803 | 958265 | Prophage | 38,462 | − | − | − | − |
| 9 | 1182688 | 1221584 | Prophage | 38,896 | − | + | + | − |
| 10 | 1224376 | 1229282 | ExPEC island containing sitABCD operon | 4,906 | − | + | + | + |
| 11 | 1336568 | 1377916 | Prophage | 41,348 | Partial | Partial | + | − |
| 12 | 1414877 | 1421687 | ExPEC island | 6,810 | − | + | + | + |
| 13 | 1470904 | 1515743 | Prophage | 44,839 | Partial | − | − | − |
| 14 | 1515743 | 1522998 | ExPEC island containing cdt locus | 7,255 | − | − | − | − |
| 15 | 2054786 | 2080866 | Prophage | 26,080 | − | Partial | Partial | Partial |
| 16 | 2080867 | 2168373 | PAI IVAPEC O1 near tRNA-Asn; contains yersiniabactin operon | 87,506 | − | + | + | + |
| 17 | 2281456 | 2255598 | Prophage | 25,858 | − | − | − | − |
| 18 | 2554458 | 2560406 | ExPEC island | 5,948 | − | + | + | + |
| 19 | 2608989 | 2570656 | Prophage | 38,333 | − | − | Partial | − |
| 20 | 2777929 | 2738458 | Prophage | 39,471 | − | − | − | − |
| 21 | 2879008 | 2888332 | ExPEC island near tRNA-Arg | 9,324 | − | − | + | − |
| 22 | 2947835 | 2914107 | Prophage | 33,728 | − | − | + | − |
| 23 | 2950455 | 2957275 | ExPEC island | 6,820 | − | − | + | − |
| 24 | 3154123 | 3116205 | ExPEC island near tRNA-Met | 37,918 | − | Partial | + | Partial |
| 25 | 3387657 | 3304326 | PAI IAPEC O1 near tRNA-Phe; contains ireA, tia, pap operon | 83,331 | − | Partial | Partial | Partial |
| 26 | 3404606 | 3413814 | ExPEC island | 9,208 | − | + | + | + |
| 27 | 3443933 | 3450091 | ExPEC island | 6,158 | − | + | + | + |
| 28 | 3461178 | 3471122 | ExPEC island | 9,944 | − | + | + | + |
| 29 | 3688691 | 3696219 | ExPEC island | 7,528 | − | + | + | + |
| 30 | 3857622 | 3848199 | ExPEC island containing auf fimbrial operon | 9,423 | − | + | + | + |
| 31 | 3893727 | 3897975 | ExPEC GimB island | 4,248 | − | − | − | + |
| 32 | 3944861 | 3953813 | ExPEC island containing chu locus | 8,952 | − | + | + | + |
| 33 | 4089354 | 4098487 | Lipopolysaccharide core synthesis region | 9,133 | − | + | + | + |
| 34 | 4122439 | 4135802 | ExPEC island | 13,363 | − | + | + | + |
| 35 | 4311724 | 4334676 | ExPEC island | 22,952 | − | + | + | + |
| 36 | 4377338 | 4385895 | ExPEC island | 8,557 | − | + | + | + |
| 37 | 4548317 | 4553013 | ExPEC island | 4,696 | − | + | + | + |
| 38 | 4591752 | 4607443 | ExPEC island | 15,691 | − | + | + | + |
| 39 | 4636683 | 4642553 | ExPEC island | 5,870 | − | + | + | + |
| 40 | 4769233 | 4711722 | Ethanolamine utilization island near tRNA-Phe | 57,511 | − | + | − | − |
| 41 | 4833450 | 4842949 | ExPEC island | 9,499 | − | + | + | + |
| 42 | 4955192 | 4931724 | ExPEC island containing ibeA | 23,468 | − | − | + | − |
| 43 | 5047045 | 5002685 | Prophage | 44,360 | − | − | + | − |
The APEC O1 chromosome contained a total of 43 genomic islands greater than 4 kb in size that were not present in the K-12 genome. Of these, a total of 21 islands were greater than 10 kb in size and 17 were greater than 20 kb (Table 5). Of the 43 total islands, only 4 (9%) were APEC specific compared to other sequenced ExPEC strains, while 25 islands (58%) were possessed by all ExPEC strains. All of the APEC-specific islands either were composed of prophage-like elements or were found adjacent to prophage-like elements. These phage-related, APEC O1-specific regions will require further study in order to determine if they might be used as markers to distinguish APEC from human ExPEC. Failure to identify a marker specific to avian ExPEC which is not present in human strains would lend credence to the hypothesis that some APEC strains possess zoonotic potential. Additionally, these APEC O1-specific regions might harbor unknown virulence factors that contribute to this strain's ability to cause extraintestinal disease. Consequently, future studies to ascertain the prevalence and distribution of these APEC O1-specific regions among ExPEC strains and determine their functions may prove helpful in testing the hypothesis that APEC strains are linked to human disease.
Among the 43 chromosomal islands and four plasmids of APEC O1, five putative PAIs were identified by their possession of multiple virulence genes, differential G+C content, and size (Table 5) (23). Four of these were chromosomal and contained genes of the yersiniabactin operon, vat, ireA, the pap operon, and an invasion determinant, tia. A fifth PAI occurred within the virulence plasmid of APEC O1, pAPEC-O1-ColBM, and contained iss, the sitABCD operon, the aerobactin operon, the salmochelin operon, and hlyF (32). The occurrence of these traits on a plasmid-encoded PAI is one of few differences between APEC O1 and the human ExPEC examined, although many of the traits of this plasmid-located APEC PAI, such as the aerobactin, sit, and salmochelin operons, have been found in chromosomal PAIs of human ExPEC (8, 10, 14, 15, 68).
There have been previous reports of virulence-associated genes and/or operons occurring at specific locations in the APEC genome. Comparison of these regions with the genomic islands of APEC O1 and other sequenced ExPEC strains helps to further define these groups. For instance, Lymberopoulos et al. (40) recently described the Stg fimbrial operon occurring in an APEC O78 strain. This operon contributed to the ability of that strain to colonize the respiratory tract and was present in 45% of the APEC strains examined. Also, this operon was inserted between the glmS and pstS genes in APEC χ7122. However, this operon was absent in APEC O1 and the other sequenced ExPEC strains, and no other insertions were found within this region. Similarly, Chouikha et al. (11) described a virulence-associated genomic island at the selC locus in an APEC O2 strain. In APEC O1 and the three other sequenced ExPEC strains, though, a different island is inserted at this location, whereas the island described by Chouikha et al. is absent (genomic island 34 in Table 5).
Parreira and Gyles described a novel PAI near the thrW locus of an APEC O2 strain, which contained the vat gene (45). Interestingly, vat is present in APEC O1 and all of the other sequenced ExPEC strains at the same locus. However, in all of these sequenced strains, vat occurs within different genomic islands (genomic island 2 in Table 5). Further investigation of this and the other genomic islands will likely reveal much about their roles in the evolution of ExPEC virulence.
Based upon virulence genotyping, APEC O1 had one of the highest counts of virulence genes of all the ExPEC strains we examined (Fig. 1; Table 6). Despite its complement of virulence genes, it was significantly less virulent in an embryo assay of virulence than other APEC isolates (61), which contained fewer ExPEC-associated virulence genes (Table 6). Therefore, it would seem that the number of PAIs and/or virulence genes in an ExPEC strain is, in itself, insufficient to predict virulence, at least in this model. Instead, these results may be related to some minimal ExPEC backbone that is required for virulence. Additionally, virulence gene regulation is likely to play an important role in ExPEC's ability to cause disease, and studies to determine if correlations between gene expression and virulence can be discerned are under way. Certainly, such results make it clear that the genomic basis of ExPEC virulence remains incompletely understood.
TABLE 6.
Embryo lethality at 4 days post infection
As knowledge of the E. coli genome has expanded, the E. coli pan-genome (total nonredundant proteins occurring among all E. coli genomes) has increased, although the number of new ORFs contributed by each newly sequenced strain is rapidly declining. Also, the number of ORFs thought to be strain specific has decreased. For instance, CFT073 was estimated to have nearly 20% strain-specific ORFs in its chromosome at the time of publication of its sequence (68), but with the completion of subsequent ExPEC genomes, it has become apparent that many of these ORFs are shared (8, 10). Only 4.5% of APEC O1's chromosome is absent from other sequenced E. coli genomes. Thus, comparative analysis of the APEC O1 and other ExPEC genomes suggests that as more ExPEC sequences become available, fewer differences between individual ExPEC strains will be found. As a result, it seems likely that a common ExPEC backbone, containing sequences not found in other pathotypes, will become increasingly better defined.
APEC O1 and other sequenced ExPEC are phylogenetically related.
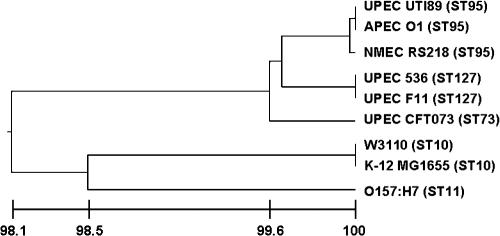
Protein BLAST searches of the ORFs of APEC O1 indicated that it is very closely related to other ExPEC strains. MLST was used to determine the potential phylogenetic relationships among all sequenced E. coli strains (Fig. 3). APEC O1 was found to be very closely related to E. coli UTI89, a strain isolated from a case of human cystitis (10); 536, a strain isolated from a case of human UTI (8); and F11, also originating from a human cystitis case (GenBank draft sequence; accession number AAJU00000000). E. coli RS218, an O18:K1:H7 strain implicated in human neonatal meningitis, was also very closely related to APEC O1 (69). Not surprisingly, E. coli strains K-12 MG1655, K-12 W3110 (a second K-12 strain recently sequenced), and O157 EDL933 were more distantly related to APEC O1 than the aforementioned genomes (6, 47). CFT073, a human pyelonephritis isolate (68), appeared to be divergent from the other sequenced ExPEC strains based on the MLST results. This divergence is also illustrated by genome alignment of APEC O1 with other sequenced ExPEC. That is, of the sequenced ExPEC strains, CFT073 shared the least homology with APEC O1 (Fig. 2). Overall, these results demonstrate that the sequenced APEC and UPEC genomes are more closely related to each other than to other E. coli subtypes in terms of sequence identity and phylogeny. Such results suggest that there is no convincing genetic support for host- or syndrome-specific pathotypes (e.g., APEC and UPEC) within the broader ExPEC group. These results also further emphasize the importance of pathogenomics in better understanding highly heterogeneous groups such as ExPEC.
FIG. 3.
Results of MLST of fully sequenced E. coli genomes. The bottom line indicates percent similarity between strains. Sequence types (STs) are indicated to the right of the strain's name.
Conclusions.
Overall, this study provides compelling evidence that the genomes of APEC O1 and several sequenced human ExPEC strains are very similar, thereby providing suggestive support for the hypothesis that at least some APEC strains are a source of UPEC causing human UTI. However, these findings must be interpreted within their context. That is, APEC O1 was selected because it belonged to a serogroup and phylogenetic group implicated in both human and avian disease, and it belonged to a virulence genotyping cluster (Fig. 1) containing a mixture of APEC and UPEC strains that were typified by possession of several common ExPEC virulence factors. This work makes no claim that all, or even most, human extraintestinal infections are derived from poultry-source E. coli. Rather, this work provides evidence that at least some human and avian ExPEC strains are highly similar to one another, and it supports the possibility that a food-borne link exists between some APEC and UPEC strains. However, validating this hypothesis requires an assessment of the ability of APEC to survive and persist on retail poultry, to traverse the human intestinal tract, and to ascend and colonize the urinary tract. Furthermore, epidemiological studies will be required to confirm that such a phenomenon actually occurs.
While APEC O1 was highly similar to other sequenced human ExPEC strains, it was not identical to them. As observed with all E. coli strains sequenced thus far, APEC O1 contained a percentage of “unique” genomic regions not occurring in the other sequenced E. coli genomes. Before definitive conclusions can be drawn about the validity of the hypothesis that some APEC strains are involved in zoonotic disease, the distribution and function of these APEC O1-specific regions should be addressed. It is possible that these regions limit the disease-causing ability of APEC O1, and perhaps other APEC strains, to avian hosts. However, the sequencing data available at this point imply that all E. coli strains possess at least some unique regions, regardless of their source. A case in point is the four UPEC genomes which have been sequenced. While the chromosomes of these strains were quite heterogeneous, they were isolated from patients with similar types of disease. Thus, a better understanding of ExPEC genomics and a more lucid definition of the ExPEC “subpathotypes” are necessary in the future.
In conclusion, this study provides the first genomic sequence of an APEC isolate and of any animal source E. coli isolate. Based on comparative genomic analyses, it appears that APEC O1 shares extensive similarities with human ExPEC, and study of its sequence has resulted in a refined understanding of the E. coli pan-genome and the ExPEC backbone. These extensive comparisons of APEC and UPEC leave open the possibility that some APEC strains represent a potential food-borne source of human UPEC. Regardless, this study provides an APEC genomic sequence on which further testing of this hypothesis can be effectively based. Thus, the sequence of APEC O1 is another stepping stone towards controlling ExPEC-caused diseases in avian and human hosts.
Acknowledgments
This work was supported by the Roy J. Carver Charitable Trust Fund (L.K.N. and T.J.J.) and the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.R.J.).
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, H. J., J. P. Vaillancourt, and W. B. Gross. 2003. Diseases of poultry, p. 631-652. Iowa State University Press, Ames, IA.
- 3.Binnewies, T. T., Y. Motro, P. F. Hallin, O. Lund, D. Dunn, T. La, D. J. Hampson, M. Bellgard, T. M. Wassenaar, and D. W. Ussery. 2006. Ten years of bacterial genome sequencing: comparative-genomics-based discoveries. Funct. Integr. Genomics 6:165-185. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, M., J. E. Blanco, M. P. Alonso, and J. Blanco. 1994. Virulence factors and O groups of Escherichia coli strains isolated from cultures of blood specimens from urosepsis and non-urosepsis patients. Microbiologia 10:249-256. [PubMed] [Google Scholar]
- 5.Blanco, M., J. Blanco, J. E. Blanco, and J. Ramos. 1993. Enterotoxigenic, verotoxigenic, and necrotoxigenic Escherichia coli isolated from cattle in Spain. Am. J. Vet. Res. 54:1446-1451. [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Bray, N., and L. Pachter. 2004. MAVID: constrained ancestral alignment of multiple sequences. Genome Res. 14:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brzuszkiewicz, E., H. Bruggemann, H. Liesegang, M. Emmerth, T. Olschlager, G. Nagy, K. Albermann, C. Wagner, C. Buchrieser, L. Emody, G. Gottschalk, J. Hacker, and U. Dobrindt. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. USA 103:1287912884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckles, E. L., F. K. Bahrani-Mougeot, A. Molina, C. V. Lockatell, D. E. Johnson, C. B. Drachenberg, V. Burland, F. R. Blattner, and M. S. Donnenberg. 2004. Identification and characterization of a novel uropathogenic Escherichia coli-associated fimbrial gene cluster. Infect. Immun. 72:3890-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, S. L., C. S. Hung, J. Xu, C. S. Reigstad, V. Magrini, A. Sabo, D. Blasair, T. Bieri, R. R. Meyer, P. Ozersky, J. R. Armstrong, R. S. Fulton, J. P. Latreille, J. Spieth, T. M. Hooten, E. R. Mardis, S. J. Hultgren, and J. I. Gordon. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA 103:5977-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chouikha, I., P. Germon, A. Bree, P. Gilot, M. Moulin-Schouleur, and C. Schouler. 2006. A selC-associated genomic island of the extraintestinal avian pathogenic Escherichia coli strain BEN2908 is involved in carbohydrate uptake and virulence. J. Bacteriol. 188:977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:55-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DebRoy, C., and C. W. Maddox. 2001. Identification of virulence attributes of gastrointestinal Escherichia coli isolates of veterinary significance. Anim. Health Res. Rev. 2:129-140. [PubMed] [Google Scholar]
- 14.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobrindt, U., G. Blum-Oehler, T. Hartsch, G. Gottschalk, E. Z. Ron, R. Funfstuck, and J. Hacker. 2001. S-Fimbria-encoding determinant sfa(I) is located on pathogenicity island III(536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 69:4248-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dozois, C. M., F. Daigle, and R. Curtiss III. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dozois, C. M., M. Dho-Moulin, A. Bree, J. M. Fairbrother, C. Desautels, and R. Curtiss III. 2000. Relationship between the tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the tsh genetic region. Infect. Immun. 68:4145-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fecteau, G., J. M. Fairbrother, R. Higgins, D. C. VanMetre, J. Pare, B. P. Smith, C. A. Holmberg, and S. Jang. 2001. Virulence factors in Escherichia coli isolated from the blood of bacteremic neonatal calves. Vet. Microbiol. 78:241-249. [DOI] [PubMed] [Google Scholar]
- 19.Foley, S. L., S. M. Horne, C. W. Giddings, T. R. Gustad, E. D. Handegard, M. Robinson, and L. K. Nolan. 2003. Monoclonal antibodies to avian Escherichia coli Iss. Avian Dis. 47:79-86. [DOI] [PubMed] [Google Scholar]
- 20.Foxman, B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl. 1A):5S-13S. [DOI] [PubMed] [Google Scholar]
- 21.Garabal, J. I., E. A. Gonzalez, F. Vazquez, J. Blanco, and J. E. Blanco. 1996. Serogroups of Escherichia coli isolated from piglets in Spain. Vet. Microbiol. 48:113-123. [DOI] [PubMed] [Google Scholar]
- 22.Ghai, R., T. Hain, and T. Chakraborty. 2004. GenomeViz: visualizing microbial genomes. BMC Bioinformatics 5:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi, K., N. Morooka, Y. Yamamoto, K. Fujita, K. Isono, S. Choi, E. Ohtsubo, T. Baba, B. L. Wanner, H. Mori, and T. Horiuchi. 21 February 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2:2006.0007. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horne, S. M., S. J. Pfaff-McDonough, C. W. Giddings, and L. K. Nolan. 2000. Cloning and sequencing of the iss gene from a virulent avian Escherichia coli. Avian Dis. 44:179-184. [PubMed] [Google Scholar]
- 26.Johnson, J. R., and J. J. Brown. 1996. A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal(alpha 1-4)Gal-binding PapG adhesins of Escherichia coli. J. Infect. Dis. 173:920-926. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, J. R., P. Delavari, T. T. O'Bryan, K. E. Smith, and S. Tatini. 2005. Contamination of retail foods, particularly turkey, from community markets (Minnesota, 1999-2000) with antimicrobial-resistant and extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 2:38-49. [DOI] [PubMed] [Google Scholar]
- 29.Johnson, J. R., K. L. Owens, C. R. Clabots, S. J. Weissman, and S. B. Cannon. 2006. Phylogenetic relationships among clonal groups of extraintestinal pathogenic Escherichia coli as assessed by multi-locus sequence analysis. Microbes Infect. 8:1702-1713. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, J. R., and C. R. Clabots. 2006. Sharing of virulent Escherichia coli clones among household members of a woman with acute cystitis. Clin. Infect. Dis. 43:e101-108. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, T. J., K. E. Siek, S. J. Johnson, and L. K. Nolan. 2006. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188:745-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, T. J., S. J. Johnson, and L. K. Nolan. 2006. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J. Bacteriol. 188:5975-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, T. J., Y. M. Wannemuehler, J. A. Scaccianoce, S. J. Johnson, and L. K. Nolan. 2006. Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob. Agents Chemother. 50:3929-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 35.Kariyawasam, S., B. N. Wilkie, and C. L. Gyles. 2004. Construction, characterization, and evaluation of the vaccine potential of three genetically defined mutants of avian pathogenic Escherichia coli. Avian Dis. 48:287-299. [DOI] [PubMed] [Google Scholar]
- 36.Kariyawasam, S., T. J. Johnson, and L. K. Nolan. 2006. The pap operon of avian pathogenic Escherichia coli strain O1:K1 is located on a novel pathogenicity island. Infect. Immun. 74:744-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy, S. B., G. B. Fitzgerald, and A. B. Macone. 1976. Spread of antibiotic-resistant plasmids from chicken to chicken and from chicken to man. Nature 260:40-42. [DOI] [PubMed] [Google Scholar]
- 38.Linton, A. H., K. Howe, P. M. Bennett, M. H. Richmond, and E. J. Whiteside. 1977. The colonization of the human gut by antibiotic resistant Escherichia coli from chickens. J. Appl. Bacteriol. 43:465-469. [DOI] [PubMed] [Google Scholar]
- 39.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lymberopoulos, M. H., S. Houle, F. Daigle, S. Leveille, A. Bree, M. Moulin-Schouleur, J. R. Johnson, and C. M. Dozois. 2006. Characterization of Stg fimbriae from an avian pathogenic Escherichia coli O78:K80 strain and assessment of their contribution to colonization of the chicken respiratory tract. J. Bacteriol. 188:6449-6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mainil, J. 1999. Shiga/verocytotoxins and Shiga/verotoxigenic Escherichia coli in animals. Vet. Res. 30:235-257. [PubMed] [Google Scholar]
- 42.Nolan, L. K., R. E. Wooley, J. Brown, K. R. Spears, H. W. Dickerson, and M. Dekich. 1992. Comparison of a complement resistance test, a chicken embryo lethality test, and the chicken lethality test for determining virulence of avian Escherichia coli. Avian Dis. 36:395-397. [PubMed] [Google Scholar]
- 43.Nolan, L. K., S. M. Horne, C. W. Giddings, S. L. Foley, T. J. Johnson, A. M. Lynne, and J. Skyberg. 2003. Resistance to serum complement, Iss, and virulence of avian Escherichia coli. Vet. Res. Commun. 27:101-110. [DOI] [PubMed] [Google Scholar]
- 44.Ojeniyi, A. A. 1989. Direct transmission of Escherichia coli from poultry to humans. Epidemiol. Infect. 103:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parreira, V. R., and C. L. Gyles. 2003. A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect. Immun. 71:5087-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peighambari, S. M., and C. L. Gyles. 1998. Construction and characterization of avian Escherichia coli cya crp mutants. Avian Dis. 42:698-710. [PubMed] [Google Scholar]
- 47.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, J. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 48.Pfaff-McDonough, S. J., S. M. Horne, C. W. Giddings, J. O. Ebert, C. Doetkott, M. H. Smith, and L. K. Nolan. 2000. Complement resistance-related traits among Escherichia coli isolates from apparently healthy birds and birds with colibacillosis. Avian Dis. 44:23-33. [PubMed] [Google Scholar]
- 49.Riley, M., and A. R. Manges. 2005. Epidemiologic versus genetic relatedness to define an outbreak-associated uropathogenic Escherichia coli group. Clin. Infect. Dis. 41:567-568. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, M. K. Fakhr, and L. K. Nolan. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151:2097-2110. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, and L. K. Nolan. 2005. Characterizing the APEC pathotype. Vet. Res. 36:241-256. [DOI] [PubMed] [Google Scholar]
- 52.Ron, E. Z. 2006. Host specificity of septicemic Escherichia coli: human and avian pathogens. Curr. Opin. Microbiol. 9:28-32. [DOI] [PubMed] [Google Scholar]
- 53.Rosenberger, J. K., P. A. Fries, and S. S. Cloud. 1985. In vitro and in vivo characterization of avian Escherichia coli. III. Immunization. Avian Dis. 29:1108-1117. [PubMed] [Google Scholar]
- 54.Russo, T. A., U. B. Carlino, and J. R. Johnson. 2001. Identification of a new iron-regulated virulence gene, ireA, in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 69:6209-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo, T. A., and J. R. Johnson. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5:449-456. [DOI] [PubMed] [Google Scholar]
- 56.Sabri, M., S. Leveille, and C. M. Dozois. 2006. A SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology 152:745-758. [DOI] [PubMed] [Google Scholar]
- 57.Sanderson, K. E., and D. R. Zeigler. 1991. Storing, shipping, and maintaining records on bacterial strains. Methods Enzymol. 204:248-264. [DOI] [PubMed] [Google Scholar]
- 58.Sarachu, M., and M. Colet. 2005. wEMBOSS: a web interface for EMBOSS. Bioinformatics 21:540-541. [DOI] [PubMed] [Google Scholar]
- 59.Schubert, S., A. Rakin, and J. Heesemann. 2004. The Yersinia high-pathogenicity island (HPI): evolutionary and functional aspects. Int. J. Med. Microbiol. 294:83-94. [DOI] [PubMed] [Google Scholar]
- 60.Siitonen, A., A. Takala, Y. A. Ratiner, A. Pere, and P. H. Makela. 1993. Invasive Escherichia coli infections in children: bacterial characteristics in different age groups and clinical entities. Pediatr. Infect. Dis. J. 12:606-612. [DOI] [PubMed] [Google Scholar]
- 61.Skyberg, J., S. M. Horne, C. W. Giddings, R. E. Wooley, P. S. Gibbs, and L. K. Nolan. 2003. Characterizing avian Escherichia coli isolates with multiplex polymerase chain reaction. Avian Dis. 47:1441-1447. [DOI] [PubMed] [Google Scholar]
- 62.Skyberg, J. A., T. J. Johnson, J. R. Johnson, C. Clabots, C. M. Logue, and L. K. Nolan. 2006. Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect. Immun. 74:6287-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Snedecor, G. W., and W. G. Cochran. 1980. Statistical methods, 7th ed. Iowa State University Press, Ames, IA.
- 64.Sorsa, L. J., S. Dufke, J. Heesemann, and S. Schubert. 2003. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of uropathogenic Escherichia coli: evidence for horizontal transfer of a chromosomal virulence factor. Infect. Immun. 71:3285-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tatum, E. L., and D. Bonner. 1944. Indole and serine in the biosynthesis and breakdown of tryptophane. Proc. Natl. Acad. Sci. USA 30:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tettelin, H., D. Radune, S. Kasif, H. Khouri, and S. L. Salzberg. 1999. Optimized multiplex PCR: efficiently closing a whole-genome shotgun sequencing project. Genomics 62:500-507. [DOI] [PubMed] [Google Scholar]
- 67.Van Den Bogaard, A. E., N. London, C. Driessen, and E. E. Stobberingh. 2001. Antibiotic resistance of faecal Escherichia coli in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemother. 47:763-771. [DOI] [PubMed] [Google Scholar]
- 68.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie, Y., V. Kolisnychenko, M. Paul-Satyaseela, S. Elliot, G. Parthasarathy, Y. Yao, G. Plunkett III, F. R. Blattner, and K. S. Kim. 2006. Identification and characterization of Escherichia coli RS218-derived islands in the pathogenesis of E. coli meningitis. J. Infect. Dis. 194:358-364. [DOI] [PubMed] [Google Scholar]