Abstract
The overall architecture of IncP-1 plasmids is very conserved in that the accessory genes are typically located in one or two specific regions: between oriV and trfA and between the tra and trb operons. Various hypotheses have been formulated to explain this, but none have been tested experimentally. We investigated whether this structural similarity is due to region-specific transposition alone or also is reliant on selection for plasmids with insertions limited to these two regions. We first examined the transposition of Tn21Km into IncP-1β plasmid pBP136 and found that most Tn21Km insertions (67%) were located around oriV. A similar experiment using the oriV region of IncP-1β plasmid pUO1 confirmed these results. We then tested the transferability, stability, and fitness cost of different pBP136 derivatives to determine if impairment of these key plasmid characters explained the conserved plasmid architecture. Most of the pBP136 derivatives with insertions in transfer genes were no longer transferable. The plasmids with insertions in the oriV-trfA and tra-trb regions were more stable than other plasmid variants, and one of these also showed a significantly lower fitness cost. In addition, our detailed sequence analysis of IncP-1 plasmids showed that Tn402/5053-like transposons are situated predominantly between the tra and trb operons and close to the putative resolution site for the ParA resolvase, a potential hot spot for those transposons. Our study presents the first empirical evidence that region-specific insertion of transposons in combination with selection for transferable and stable plasmids explains the structural similarity of IncP-1 plasmids.
Horizontal gene transfer is now well recognized as one of the key mechanisms of adaptive evolution of bacteria (20, 50, 53). Mobile genetic elements such as plasmids, transposons, and integrative and conjugative elements are known to promote horizontal gene transfer events and DNA rearrangements (18, 53, 57). Many of these elements carry so-called “accessory genes” that encode various functions such as resistance to antibiotics, degradation of xenobiotics, virulence, and symbiosis (18, 58). Conjugative plasmids are genetic elements that can efficiently transfer from cell to cell (50, 62). In addition to their so-called “backbone” genes, which encode the machineries for plasmid replication, regulation, stable inheritance, and transfer, they also serve as vectors for other mobile genetic elements such as transposons and integrons that carry the accessory genes (49). Comparative studies of conjugative plasmids have shown that the DNA sequence of plasmid backbones is conserved, which is in contrast to the broad genetic diversity of the accessory genes (2, 14, 35, 45, 59).
The incompatibility group P-1 (IncP-1) plasmids are among the best-studied plasmids of gram-negative bacteria and are of particular interest because of their broad host range, high transfer frequencies, and the wide variety of accessory genes they carry (2, 51). In contrast to their phenotypic diversity, their basic architecture is very conserved, in that the accessory fragments are usually located in one or both of two specific regions, i.e., between oriV and trfA and between the tra and trb operons (2, 51). To date, at least four subgroups of IncP-1 plasmids (α, β, γ, and δ) have been defined based on the phylogenetic relatedness of the trfA genes, which encode the plasmid replication initiation protein, and a fifth subgroup has been proposed (4). Strikingly, all these IncP-1 plasmids show the same typical architecture.
There are at least three hypotheses to explain the structural similarity of IncP-1 plasmids: (i) transposons specifically transpose into these regions of the IncP-1 backbone; (ii) transposition is completely random, but plasmids with insertions in the two specific regions are most stable or transferable, or least costly to their host, and therefore persist longer over evolutionary time than cognate plasmids with insertions in other sites (12, 41); or (iii) a combination of region-specific insertion and selection explains the common plasmid structure. Consistent with the first hypothesis, it has been proposed previously that 20-bp inverted repeats (IRs) with a consensus sequence of CATCGCCANNTCYGRCGATG found in the oriV-trfA and tra-trb regions of IncP-1β plasmids might be involved in the acquisition of transposons (24, 52). However, this hypothesis has not been tested experimentally, in part because there were no IncP-1 plasmids available that lacked insertions in both of the regions. Conversely, inconsistent with this first hypothesis is the observed transposon (Tn7) insertions in various backbone genes of IncP-1α plasmids, which had major effects on the plasmid phenotype (e.g., host range) (5, 36). However, these studies did not test the second and third hypotheses.
Recently, the IncP-1β plasmid pBP136, isolated from the human pathogen Bordetella pertussis, was found not to carry any transposons or their footprints, yet it has two copies of the 20-bp IR in the oriV-trfA and tra-trb regions and one in klcB and between traM and kfrC (Fig. 1) (26). The objective of our study was to empirically test the three hypotheses by performing transposition experiments using a Tn21 derivative (designated Tn21Km) and the cryptic IncP-1β plasmid pBP136 and subsequently analyzing the behavior of plasmid variants with transposon insertions in various sites. Our empirical results and detailed sequence analysis of known IncP-1 plasmids support the third hypothesis, i.e., that region-specific insertion of transposons such as Tn21-like and Tn402/5053-like transposons, in combination with selection for transferable and stable plasmids, can account for the common architectural features of IncP-1 plasmids.
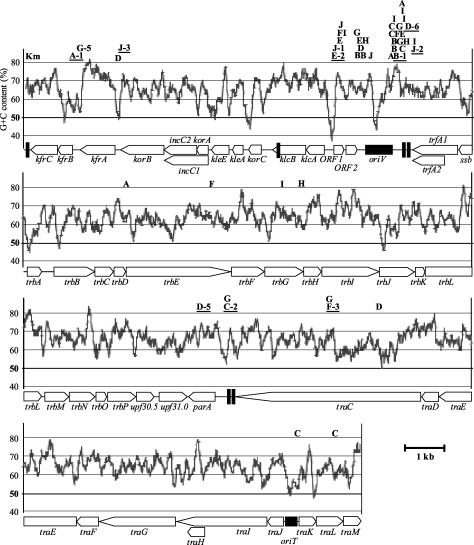
FIG. 1.
Physical map and G+C content of pBP136, showing the insertion sites of Tn21Km. The letters A to J indicate insertion sites in plasmids obtained from 10 independent mating experiments. Plasmids with insertions at the sites shown by underlined letters were used to investigate the plasmid stability and cost. Km indicates the location of the Kmr gene in pBP136Km. The black vertical bars represent the positions of the 20-bp IRs. The G+C content of pBP136 was calculated in an average calculation span of 100 bp. See reference 26 for detailed information on the genetic structure and function of the plasmid.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Escherichia coli strains used in this study were DH10B [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL nupG] (TaKaRa BIO), EC100 [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL nupG] (Epicentre Biotechnologies), S17-1 (recA1 endA1 thi hsdR, RP4-2 Tc::Mu::Tn7) (40), a Mu phage lysogen of DH1 (supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1) (designated DH1Mu), and a spontaneous rifampin-resistant (Rifr) mutant of MG1655 (designated K12R). The strains were cultured at 37°C. Plasmids used in this study are listed in Table 1. Plasmid pBP136 was provided to us in E. coli DH10B by K. Kamachi. Plasmid pBP136Km is a pBP136 derivative in which a kanamycin resistance (Kmr) gene derived from the cloning vector pUC4K (47) was inserted into the unique XbaI site of pBP136. Plasmid pMT1247 is a pACYC184 derivative carrying Tn21 (8, 56). This plasmid was digested with BamHI, and the pUC4K-derived Kmr gene was inserted into the BamHI site, generating pMT1247Km, which carried a Kmr Tn21 derivative (designated Tn21Km). Two regions of the IncP-1β plasmid pUO1 (positions 1 to 901 and 16,511 to 17,610; accession no. AB063332) (44) were amplified by PCR using appropriate primer sets, and the two PCR products were cloned together between EcoRI and HindIII sites of the cloning vector pARO190 (34). The resulting plasmid, pMS0220, carried the potentially ancestral oriV-trfA region of the plasmid, since the Tn21-like transposon TnHad2 between oriV and trfA (43) was removed. Luria-Bertani broth (LB) and LB agar (LBA) (3) were used throughout this study. The antibiotics added to the media were as follows: Km, 100 mg/liter; nalidixic acid (Nal), 20 mg/liter; Rif, 50 mg/liter; and streptomycin (Sm), 100 mg/liter.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristics | Reference |
|---|---|---|
| pARO190 | Apr, pMB1ori, pUC19 derivative with the RP4oriT sequence | 34 |
| pBP136 | IncP-1β, self-transmissible, no transposons or their footprints | 26 |
| pBP136Km | Kmr, pBP136 derivative with a Kmr determinant at the unique XbaI site between traM and kfrC | 26 |
| pBPTn/A-1 | Kmr, pBP136 derivative carrying Tn21Km in kfrB (position 1209-1210 of pBP136, 26 bp downstream from the 5′ end of kfrB) | This study |
| pBPTn/B-1 | Kmr, pBP136 derivative carrying Tn21Km between oriV and trfA (position 9085-9086 of pBP136) | This study |
| pBPTn/C-2 | Kmr, pBP136 derivative carrying Tn21Km between parA and traC (position 26974-26975 of pBP136) | This study |
| pBPTn/D-5 | Kmr, pBP136 derivative carrying Tn21Km in parA (position 26320-26321 of pBP136, near center of parA) | This study |
| pBPTn/D-6 | Kmr, pBP136 derivative carrying Tn21Km between oriV and trfA (position 9369-9370 of pBP136) | This study |
| pBPTn/E-2 | Kmr, pBP136 derivative carrying Tn21Km between klcA and ORF1 (position 7595-7596 of pBP136, 156 bp upstream of the 5′ end of klcA) | This study |
| pBPTn/F-3 | Kmr, pBP136 derivative carrying Tn21Km in traC (position 29473-29474 of pBP136, near center of traC) | This study |
| pBPTn/G-5 | Kmr, pBP136 derivative carrying Tn21Km in kfrA (position 1410-1411 of pBP136, 7 bp upstream from the 3′ end of kfrA) | This study |
| pBPTn/J-1 | Kmr, pBP136 derivative carrying Tn21Km in ORF1 (position 7628-7629 of pBP136, 33 bp downstream from the 5′ end of ORF1) | This study |
| pBPTn/J-2 | Kmr, pBP136 derivative carrying Tn21Km between oriV and trfA (position 9524-9525 of pBP136) | This study |
| pBPTn/J-3 | Kmr, pBP136 derivative carrying Tn21Km between kfrA and korB (position 2367-2368 of pBP136, 95 bp upstream from the 5′ end of kfrA) | This study |
| pMS0220 | Apr, pARO190 derivative that carry the 2-kb oriV-trfA fragment of pUO1 (see text and Fig. 2) | This study |
| pMT1247 | Cmr, p15Aori, pACYC184 derivative carrying Tn21 | 56 |
| pMT1247Km | Cmr, Kmr, pMT1247 derivative in which a Kmr determinant is inserted at the BamHI site of Tn21 (designated Tn21Km) | This study |
| pUC4K | Apr, Kmr, cloning vector | 47 |
| pUO1 | IncP-1β, haloacetate degradation, mercury resistance | 44 |
DNA methodology.
Standard methods were used for extraction of the plasmid DNA, DNA digestion with restriction endonucleases, ligation, gel electrophoresis, and transformation of E. coli cells (3). PCR amplification was carried out with ExTaq DNA polymerase (TAKARA BIO) or AccuPrime Pfx DNA polymerase (Invitrogen). Purification of the PCR-amplified DNA fragments was done with GFX PCR DNA and the Gel Band purification kit (GE Healthcare) according to the manufacturer's protocol.
Transposition assays.
Transposition of Tn21Km into pBP136 was detected by the “mating-out” experiments described previously (55). For this purpose, we introduced pMT1247Km into the DH10B derivative harboring pBP136. Ten transformants were selected and cultured to the stationary phase in LB containing Km [LB(Km)] and then mixed with K12R on membrane filters. After 6 hours of incubation at 37°C, the cells were harvested, and appropriate dilutions were plated on LBA(Km, Rif) to select for Kmr K12R transconjugants. Plasmids from three to six Kmr transconjugants in each of the 10 mating experiments were analyzed to identify the insertion sites of Tn21Km in pBP136, using the primer Tn21-F19170 (5′-TTTGGATTGGATAGCGTAAC-3′), which annealed to the tnpA-distal end of Tn21 (accession no. AF071413) in the outward direction. Transposition of Tn21Km into pMS0220 was also detected by mating-out experiments. Plasmids pMT1247Km and pMS0220 were introduced into E. coli S17-1 (40), and the resulting strain was mated with DH1Mu. The steps after the mating were done as described above except for the use of Nal instead of Rif.
Plasmid stability experiment.
Stability of pBP136 derivatives carrying Tn21Km (pBPTn plasmids) was investigated as follows. The EC100 derivative harboring a pBPTn plasmid was grown to the stationary phase in 5 ml LB(Km) (defined as generation 0 cells). The cells were collected by centrifugation, rinsed in saline, and resuspended in 5 ml of saline; 4.88 μl of this cell suspension was transferred into 5 ml LB without Km, the cells were cultured for 16 h, and this procedure was repeated daily (ca. 10 generations per day). At regular time points the cells were plated on LBA, and colonies thus formed were picked onto LBA(Km) to determine the fraction that showed the Kmr phenotype.
Estimates of plasmid maintenance costs.
Pairwise competition experiments were performed between EC100 and EC100(pBPTn). Both strains were precultured for 12 h in LB(Sm) and LB(Km, Sm), respectively. After being rinsed in saline, the cells were inoculated into LB(Sm) at a 1:1 ratio (calculated from optical density at 600 nm) and were grown for 24 h at 30°C and 200 rpm. Appropriate dilutions of the initial and final culture media were plated on LBA(Sm) and LBA(Km, Sm) using Autoplate 4000 (Spiral Biotech), and the number of colonies was counted with QCount (Spiral Biotech). The relative fitness (W) was determined as described previously (7, 10) using the following formula: Wij = log2[Ni(1)/Ni(0)]/log2[Nj(1)/Nj(0)], where Ni(0) and Nj(0) are the initial densities of EC100(pBPTn) and EC100, respectively, and Nj(1) and Nj(1) are their corresponding final densities.
Statistical analyses.
The standard Student t test with Bonferroni correction was used to determine the significance of any differences between means of the plasmid costs. Differences between means of plasmid stability were assessed with a Dunnett multiple-comparison test (15). The χ-square test was used for testing differences between observed and expected frequencies of transposon insertions (25). Since the χ-square test requires an expected frequency of at least five in each category and there were 48 plasmids with transposon insertions, the 41-kb sequence of pBP136 (accession no. AB237782) was divided into eight regions, representing eight categories (expected frequency = 6 per region). In each analysis, P values of <0.05 were considered significant.
DNA sequence analysis.
The nucleotide sequencing was performed with an ABI PRISM model 3730 sequencer (Applied Biosystems). The computer analysis of the DNA sequences was performed with the software programs GENETYX 13 (SDC Inc.) and BLAST (National Center for Biotechnology Information).
RESULTS
Transposition of Tn21Km to pBP136.
To test the null hypothesis that insertion of Tn21 transposons in IncP-1 plasmids is random, we conducted transposition experiments using the transposon-free IncP-1β plasmid pBP136 (26) as the target for Tn21Km, a Kmr derivative of Tn21 (13). A total of 48 plasmids were analyzed to identify the insertion sites of Tn21Km. Thirty-two of the 48 plasmids (67%) had the Tn21Km insertion near the oriV sequence, while Tn21Km was found in the control and maintenance regions (kfrA, kfrB, between kfrA and kfrB, and between kfrA and korB) of four plasmids, in the transfer genes (trbD, trbE, trbG, traC, oriT, and traL) of nine plasmids, and between the tra and trb operons (parA, between parA and traC) of three plasmids (Fig. 1). A χ-square test (25) confirmed that there were significantly more insertions in the oriV region than under the assumption of random insertions and that there were significantly fewer insertions in the two transfer regions and in the control and maintenance regions (P < 0.001). Therefore, our results rejected the null hypothesis that transposition was random throughout the plasmid. However, since not all insertions were found around oriV or between the tra and trb operons, the hypothesis that the current IncP-1 plasmid structures can be completely explained by region-specific transposition is also rejected.
At least two hypotheses have previously been formulated to explain the specific architecture of IncP-1β plasmids. First, it has been hypothesized that the 20-bp IRs may attract transposons (24, 52). However, although pBP136 has two 20-bp IRs in both the oriV-trfA and the tra-trb intergenic regions and one in klcB and between traM and kfrC, the Tn21Km insertions were clustered only in the oriV-trfA region (Fig. 1). It has also been shown that Tn3-like transposons such as Tn3 and Tn21 tend to transpose into AT-rich sequences (38). Therefore, we plotted the G+C content of pBP136 (the average calculation span was 100 bp) and indicated the insertion sites on the plot (Fig. 1). Although the pBP136 sequence has several AT-rich regions (G+C content of <50%), the Tn21Km insertions were not clustered in these regions but in rather high-G+C (>65%) regions. The region with the highest density of insertions (downstream of oriV) had >70% G+C content (Fig. 1). These results suggest that (i) the 20-bp IRs are not directly involved in acquisition of transposons and (ii) region-specific location of transposons is not correlated with the G+C content of pBP136.
Experimental bias.
During the transposition and subsequent mating experiments that were performed, only plasmid variants that could still replicate and transfer with or without the help of coresident plasmids would have been retrieved in the transconjugants. Therefore, our method may not have detected every possible plasmid derivative, but there may have been a bias for those plasmids that still efficiently replicated and transferred. Due to the short incubation times (24 h or less) and the presence of antibiotics in the selection media, we assume that there was no bias for plasmid variants with higher stability or lower fitness cost (see below). We tested the transferability of pBP136 derivatives with insertions in trbD, trbE, trbG, traC, and traL and near oriT. The plasmids with an insertion in traC or near oriT were able to transfer into recipient cells at a frequency similar to that of pBP136Km (ca. 10−2 per recipient), but those with an insertion in trbD, trbE, trbG, or traL were not transferable (<10−8 per recipient). These results indicate that our experimental system can detect transfer-deficient pBP136 derivatives even if Tn21Km was inserted into the regions encoding plasmid transfer, probably due to mobilization by coresident plasmid variants. Thus, only nonreplicating plasmids and plasmids that are no longer transferable even in the presence of transfer-proficient plasmids would have been lost from our detection system. Therefore, we conclude that the high concentration of the Tn21Km insertions found in the oriV region (67%) cannot be entirely due to experimental bias but must at least in part be due to region specificity of the transposition itself.
Can selection of specific plasmid variants explain the common IncP-1 plasmid architecture?
There are three major factors that contribute to the long-term persistence of self-transmissible plasmids: (i) a low segregational loss rate, (ii) a small plasmid cost, and (iii) a horizontal plasmid transfer rate that is high enough to compensate for plasmid loss and cost (12, 41). We examined whether impairment of these key characters in some plasmid variants can explain the conserved plasmid architecture. We already showed that most of the pBP136 derivatives with insertions in tra and trb genes were no longer transferable in subsequent conjugation assays (see “Experimental bias” above). Such transfer-deficient plasmids would thus likely be lost from bacterial communities and not be found in the extant IncP-1 plasmid pool.
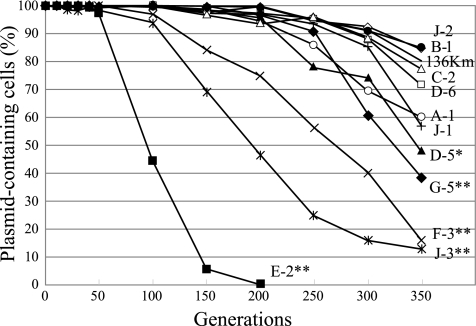
To test the effect of the location of transposon insertions on the stability of plasmids, we compared the stability in E. coli EC100 of 11 pBPTn plasmids during 350 generations of growth. These plasmids carried the transposon in kfrA (possible plasmid maintenance; pBPTn/G-5), kfrB (possible plasmid maintenance; pBPTn/A-1), parA (plasmid-multimer resolution; pBPTn/D-5), traC (DNA primase; pBPTn/F-3), ORF1 (pBPTn/J-1), between klcA and ORF1 (pBPTn/E-2), between kfrA and korB (pBPTn/J-3), between parA and traC (pBPTn/C-2), and between oriV and trfA (pBPTn/B-1, pBPTn/D-6, and pBPTn/J-2) (Table 1 and Fig. 1). Of the 11 plasmids tested, those with an insertion between oriV and trfA (pBPTn/B-1, pBPTn/D-6, and pBPTn/J-2) or between the tra and trb operons (pBPTn/C-2) were significantly more stable than at least four of the seven plasmids with insertions outside of these regions (Fig. 2). Three of these seven plasmids, pBPTn/E-2, pBPTn/F-3, and pBPTn/J-3, which had an insertion between klcA and ORF1, in traC, and between kfrA and korB, respectively, were very unstable. These results indicate that pBP136 derivatives with transposons inserted in the two typical regions for IncP-1 plasmids, i.e., oriV-trfA and tra-trb, are much more likely to persist longer in bacterial communities than plasmid variants with insertions in other regions. This may explain why these other IncP-1 plasmid variants have not been detected so far among natural bacterial populations.
FIG. 2.
Stability of pBP136 derivatives with Tn21Km insertions. The detailed insertion sites of Tn21Km in pBP136 are shown in Table 1 and Fig. 1. The percentages of plasmid-containing cells are averages from five independent stability assays. Standard deviations are not shown in this figure for simplicity. Asterisks indicate statistically significant differences (P < 0.05) between the means of stabilities as assessed by Dunnett's multiple-comparison test (15): *, significantly different from the four most stable plasmids, pBP136Km, pBPTn/B-1, pBPTn/C-2, and pBPTn/J-2; **, significantly different from the five most stable plasmids, pBP136Km, pBPTn/B-1, pBPTn/C-2, pBPTn/J-2, and pBPTn/D-6.
Since the fitness cost that a plasmid confers to its host negatively affects long-term plasmid persistence in the absence of selection for plasmid-encoded traits (6, 7, 10, 29), pBP136 derivatives with the lowest cost would be more likely to persist and thus represent the IncP-1β plasmids we know today. To test whether insertion sites of Tn21Km in pBP136 affected the plasmid cost, we estimated the relative fitness (W) of plasmid-containing versus plasmid-free isogenic E. coli EC100 hosts in pairwise competition experiments during 10 generations and attributed the relative decrease in host fitness to the plasmid cost (1 − W). Since all pBPTn plasmids used were stably maintained in EC100 within this period (>99.6% on average [Fig. 2]), instability did not confound the plasmid cost data. For 9 of the 12 plasmids tested, the relative fitness of the plasmid-containing host was 3 to 9% lower than that of plasmid-free cells, indicating a plasmid cost (Table 2). Of the three plasmids with insertions between oriV and trfA, pBPTn/D-6 was the only one that conferred no cost to its host (it even conferred a potential benefit), whereas the other two variants decreased the host fitness by more than 3%. The cost of plasmid pBPTn/D-6 was also significantly lower than that of plasmids carrying inserts outside the two typical regions (except for the traC insertion). Interestingly, one other pBP136 derivative that did not significantly lower the host fitness (W = 0.99) carried an insertion in the tra-trb region (Table 2). These results indicate that some but not all of the Tn21Km insertions in the two typical insertion regions of IncP-1 plasmids resulted in a lower plasmid cost than insertions in other regions. Selection for low plasmid cost can thus only partially explain the conserved architecture of currently known IncP-1 plasmids.
TABLE 2.
Relative fitnesses for pBP136 derivatives carrying Tn21Km
| Plasmid | Insertion site of Tn21Km or Kmr determinant | Mean relative fitness of plasmid-carrying to plasmid-free cells (SE, no. of replicates)a |
|---|---|---|
| pBPTn/A-1 | kfrB | 0.95235 (0.01365, 10)** |
| pBPTn/B-1 | Between oriV and trfA | 0.96315 (0.01316, 10)* |
| pBPTn/C-2 | Between parA and traC | 0.99008 (0.01469, 10) |
| pBPTn/D-5 | parA | 0.90980 (0.01560, 10)*** |
| pBPTn/D-6 | Between oriV and trfA | 1.03237 (0.01194, 20) |
| pBPTn/E-2 | Between klcA and ORF1 | 0.93581 (0.01134, 20)*** |
| pBPTn/F-3 | traC | 1.00948 (0.01675, 10) |
| pBPTn/G-5 | kfrA | 0.95019 (0.01588, 10)*** |
| pBPTn/J-1 | ORF1 | 0.95113 (0.01540, 10)*** |
| pBPTn/J-2 | Between oriV and trfA | 0.96912 (0.01327, 10)* |
| pBPTn/J-3 | Between kfrA and korB | 0.93896 (0.01563, 10)*** |
| pBP136Km | Between traM and kfrC | 0.96757 (0.00918, 10)* |
The relative fitness was measured by pairwise competition experiments using EC100(pBPTn) and EC100. Asterisks indicate P values from Student's t test with Bonferroni correction for pairwise comparisons of the relative fitness between pBPTn/D-6 and one of the others: *, P < 0.05; **, P < 0.01; ***, P < 0.001. No asterisk, not significantly different from the value for pBPTn/D-6.
Transposition of Tn21Km to the oriV-trfA region of pUO1.
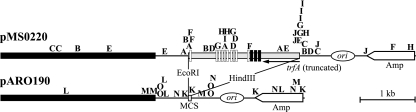
To investigate whether the finding that Tn21Km insertions are concentrated around oriV is limited to plasmid pBP136 or is more generally true for IncP-1β plasmids, a similar transposition experiment was performed using pMS0220. This vector is a pUC19-based plasmid that carries the 2-kb potential oriV-trfA region from the IncP-1β plasmid pUO1 (44), which shows 92% nucleotide identity to the corresponding sequence of pBP136. E. coli S17-1(pMS0220)(pMT1247Km) was mated with DH1Mu, and Kmr Nalr transconjugants were selected. Forty pMS0220::Tn21Km plasmids from 10 independent experiments were sequenced to identify the insertion sites of the transposon. In 29 plasmids (73%), Tn21Km was inserted in or near (±100 bp) the 2-kb fragment. Sixteen of these were close to iteron sequences, and 13 were near a truncated trfA gene (Fig. 3). As was the case with pBP136, the 20-bp IRs of pUO1 did not seem to be directly involved in acquisition of Tn21Km (Fig. 3). In the control experiment using the cloning vector pARO190 as the Tn21Km target, the transposon was inserted randomly into the pUC19-derived portion of the plasmid (Fig. 3). These results together with those for pBP136 suggest that the oriV-trfA sequence of IncP-1 plasmids attracts insertions of Tn21Km.
FIG. 3.
Transposition of Tn21Km into plasmids with and without the oriV-trfA region of pUO1. The meaning of the letters is the same as described in the legend of Fig. 1. The shaded bar indicates the potential oriV-trfA region of pUO1 from which the Tn21-like transposon TnHad2 (43), located in this region, was removed. The horizontal black bar represents the RP4-derived fragment containing its oriT sequence in the pUC19-based vector pARO190 (34). The vertical white and black boxes show the iteron sequences and 20-bp IRs, respectively. Plasmid pUO1 has three copies of the 20-bp IRs, whereas pBP136 has two in the corresponding sequence. The replication origin of pARO190 is shown by the oval. Abbreviations: MCS, multicloning site; Amp, gene for resistance to ampicillin.
DISCUSSION
One of the most striking features of the more than 18 complete nucleotide sequences of IncP-1 plasmids deposited in databases today is the location of transposons and/or their remnants in one or both of two specific regions, namely, the regions between oriV and trfA (oriV-trfA) and between the tra and trb operons (tra-trb). Our study provides the first empirical evidence that region-specific insertion of transposons in combination with selection for transferable and stable plasmids explains this conserved architecture. We first showed that after transposition of Tn21Km into plasmids pBP136 and pMS0220, followed by plasmid transfer, a very large proportion of plasmids carried the transposon in the oriV region. We then demonstrated that several of the pBP136 plasmid derivatives with Tn21Km insertions outside the two typical regions were either not self-transferable, less stable, or more costly to the host than the plasmids with insertions limited to these two regions. Although our studies employed only IncP-1β plasmids, we believe most of the results can be generalized to all subgroups of IncP-1 plasmids. All IncP-1 plasmids have the same conserved architecture, and they differ only in terms of the accessory genes they carry, sequence divergence and presence or absence of particular backbone genes, and presence of the 20-bp IRs.
In the past, three hypotheses have been formulated that may explain the conserved IncP-1 plasmid architecture. One possible explanation for the region-specific insertions into IncP-1β plasmids is the involvement of the 20-bp IRs present in these two regions in the acquisition of transposons (24, 52). A proposed role of the IRs is to provide cleavage of DNA somewhere within the 20-bp sequence or at their flanking restriction sites, which may facilitate insertion of other DNA fragments (52). At present, we have no idea whether such cleavage took place in pBP136 before transposition. However, our experiments indicated that none of the six 20-bp IRs were directly involved in the acquisition of transposons. This is consistent with the fact that only the β subgroup of IncP-1 plasmids has the 20-bp IRs, while the plasmid architecture is also conserved within the other IncP-1 subgroups. A second hypothesis was the preferential insertion of class II transposons in AT-rich regions. Although class II transposons are known to transpose into AT-rich sequences (38), our data showed that there was no relationship between the insertion sites of Tn21Km and the G+C content of the pBP136 sequence (Fig. 1). Conformational change of plasmids is a third possible factor that may allow transposons to insert in specific regions. For example, another type of transposon, Tn7, inserted near the oriT region of the IncF plasmid pOX-G during conjugation (60). However, conformational change is probably not the case in our study, since Tn21Km transposed near the oriV sequence regardless of its activity (Fig. 3). Given that a different factor(s) must attract Tn21-like transposons into this region, we propose that the unique secondary structure of the oriV-trfA region, resulting from repeated iteron sequences, may be very important for the target specificity of Tn21-like transposons (Fig. 3). This idea is consistent with previous reports that that DNA secondary structure plays an important role in target site selection for transposons (11, 19, 60).
To allow transposon insertion in a limited number of specific regions might be beneficial for promiscuous and frequently transferable plasmids to persist in bacterial communities, because these plasmids may often be targeted by various transposons. The currently known oriV-rep (replication protein gene) regions may thus have evolved as “attractants” for transposons. Although the oriV-trfA region is vital for vegetative replication, it is also the most flexible region of the IncP-1 plasmids in that (i) it has a much longer noncoding sequence (ca.1.5 kb) than other open reading frame-intervening sequences and (ii) it is independent from operons encoding plasmid maintenance and transfer and their tight regulatory circuits. This “attractant” hypothesis is supported by the fact that in many promiscuous plasmids of the IncP-1, IncQ, IncW, and IncN groups (but not those of the IncP-7 and IncP-9 groups, which have a less wide host range and lower transfer frequencies), the oriV sequence is found downstream of the rep gene (17, 35, 39, 45). This enables these plasmids to normally express the Rep proteins even if transposons are inserted near oriV. In the case of the IncP-1 group, these ideas are consistent with the complicated structures of accessory fragments in these regions, which show many past DNA rearrangements caused by various transposons and are in stark contrast with the strongly conserved backbones.
Since several Tn21Km insertions in pBP136 were also found outside the two typical regions and since others may have escaped our detection system, we postulate that the absence of similar plasmid constructs in the currently known IncP-1 plasmid pool is due to impairment of key plasmid characteristics, such as stability, fitness cost, and self-transferability. First, immobilization of a plasmid by insertion of transposons into an essential transfer gene, as shown for several pBP136 variants, gives a direct competitive disadvantage in bacterial communities, since conjugative transfer is thought to contribute to plasmid persistence (41). Second, plasmids that are unstable in the absence of selection would have a disadvantage to persist in bacterial communities. As we have shown, pBP136 derivatives with Tn21Km insertions in the central control region (pBPTn/E-2 and pBPTn/J-3) were much less stable than those with insertions in other positions (Fig. 2), even though they did not disrupt any genes. The low stability of these two plasmids might be due to the disturbance of the tight regulation of the plasmids (28, 49, 61). In fact, in pBPTn/J-3, Tn21Km was inserted in one of the binding sites for the regulation protein KorB (insertion site and consensus sequence, TTTA↓GCGGCTAAA). This may inhibit transcription of the kfr operon downstream, which was recently shown to be involved in plasmid stability (1). This finding is also compatible with our previous observation that IncP-1β plasmids have transposons in regions where they do not disrupt transcription of genes and operons (44). Finally, our data also showed to some extent how minimizing the plasmid cost by acquisition of a transposon at a specific site can be the third mechanism that allows the IncP-1 plasmids with the typical structure to persist longer than other cognate plasmids and thus accumulate in the currently known pool of structurally similar plasmids (Table 2).
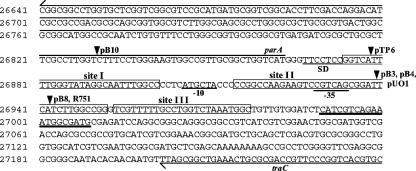
Although most IncP-1 plasmids, regardless of their subgroups, have a second “hot spot” for insertion of transposons in the tra-trb region (2, 44, 52), we detected only a few Tn21Km insertions in this region of pBP136 (Fig. 1). We have previously reported that a different type of transposons, the Tn402/5053 group, are predominantly located in this region of IncP-1β plasmids (44). Moreover, the resolution (res) sites of resolvases have been shown to be a specific target for these transposons (27, 31), and many IncP-1 plasmids have a resolvase gene, parA, between the tra and trb operons (4, 23, 24, 26, 33, 37, 42, 46, 48). The reports for RP4 ParA and other resolvases (16, 22, 32) helped us to identify the putative res site for the ParA protein of pBP136 (Fig. 4). This identification subsequently clarified the insertion sites of Tn402/5053-like transposons in IncP-1β plasmids in or around the putative res site (Fig. 4). Such Tn402/5053-like insertions are also found in the res site for the ParA resolvase of the IncP-1α plasmid pTB11 (48) and upstream of the resolvase genes of the IncP-1γ plasmid pQKH54 (21) and the recently described IncP-1 plasmid pKJK5 (4). These observations suggest that the specific insertion of the Tn402/5053-like transposons in the tra-trb region of IncP-1 plasmids is most likely due to the presence of the res site and ParA resolvase. This idea is consistent with the reports that (i) the target specificity of Tn402 (=Tn5090) to the res site of the IncP-1α plasmid RP1 (=RP4) depends on the presence of both ParA and the res site (27) and (ii) the IncN plasmid R46 carries a Tn402-like insertion just upstream of the resP resolvase gene (AY046276). Since several IncP-1 plasmids have transposons that are not related to Tn402/5053-like transposons in the tra-trb regions (30, 54), the res site for the parA gene might form a unique DNA structure that also attracts other transposons. This hypothesis is compatible with the fact that other parA homologue-carrying plasmids, pK245 (accession no. DQ449578) and pSC138 (9), also have transposons just upstream of their parA genes (data not shown).
FIG. 4.
Nucleotide sequence between the parA and traC genes of pBP136. The first nucleotide of the parA gene was redefined in this study and thus differs from the previously defined location (26). The numbers shown on the left of the sequence indicate nucleotide positions of pBP136. The insertion sites (inverted triangles) of Tn402/5053-like transposons in various IncP-1β plasmids are shown at homologous locations of the pBP136 sequence. Boxed sequences indicate three resolvase-binding regions (sites I to III) in the putative res site of the ParA resolvase of pBP136. Putative −10 and −35 sequences for parA are underlined. The sequence in bold and underlined represents the 20-bp IR. Sequence data for pB3, pB4, pB8, pB10, R751, pTP6, and pUO1 are compiled from the DDBJ/EMBL/GenBank databases: pB3, accession no. AJ639924; pB4, AJ431260; pB8; AJ863570; pB10, AJ564903; R751, U67194; pTP6, AM048832; and pUO1, AB063332.
It should be noted that IncP-1β plasmids have one more region where a transposon has been found, i.e., between traM and kfrC. Plasmid pB4 is at present the only example carrying an insertion, Tn5393c, in this region (44, 46). Although the long-term stability of pB4 has not yet been investigated, our stability experiment using pBP136Km, which has a Kmr determinant between traM and kfrC, demonstrated that this plasmid was as stable as the plasmids with transposons in the oriV-trfA and the tra-trb regions (Fig. 2).
We conclude that the conserved architecture of IncP-1 plasmids can be attributed to region-specific insertion of transposons, especially those of the Tn21 and Tn402/5053 groups, in combination with selection for plasmids that are most stable and transferable and least costly. More comparative genome analyses of IncP-1 plasmids will further improve our insight into the structural similarity and evolutionary history of these plasmids.
Acknowledgments
We thank K. Kamachi (National Institute for Infectious Diseases, Japan) for his kind gift of pBP136. We are also grateful to N. Packer, S. Bassler, J. Williams, and S. Poler for their assistance in the experiments.
This project was supported by NIH grant P20 RR16448 from the Idaho COBRE Program of the National Center for Research Resources (NCRR). The work at the lab of M.T. was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Agriculture, Forestry, and Fisheries, Japan.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Adamczyk, M., P. Dolowy, M. Jonczyk, C. M. Thomas, and G. Jagura-Burdzy. 2006. The kfrA gene is the first in a tricistronic operon required for survival of IncP-1 plasmid R751. Microbiology 152:1621-1637. [DOI] [PubMed] [Google Scholar]
- 2.Adamczyk, M., and G. Jagura-Burdzy. 2003. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim. Pol. 50:425-453. [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1988. Current protocols in molecular biology. Wiley, New York, NY.
- 4.Bahl, M. I., L. H. Hansen, A. Goesmann, and S. J. Sørensen. The multiple antibiotic resistance IncP-1 plasmid KJK5 isolated from a soil environment is phylogenetically divergent from members of the previously established α, β, and γ sub-groups. Plasmid, in press. [DOI] [PubMed]
- 5.Barth, P. T., N. J. Grinter, and D. E. Bradley. 1978. Conjugal transfer system of plasmid RP4: analysis by transposon 7 insertion. J. Bacteriol. 133:43-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergstrom, C. T., M. Lipsitch, and B. R. Levin. 2000. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155:1505-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouma, J. E., and R. E. Lenski. 1988. Evolution of a bacteria/plasmid association. Nature 335:351-352. [DOI] [PubMed] [Google Scholar]
- 8.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, C. H., P. Tang, C. Chu, S. Hu, Q. Bao, J. Yu, Y. Y. Chou, H. S. Wang, and Y. S. Lee. 2005. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 33:1690-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahlberg, C., and L. Chao. 2003. Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics 165:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, C. J., and C. A. Hutchison, 3rd. 1995. Insertion site specificity of the transposon Tn3. Nucleic Acids Res. 23:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Gelder, L., J. M. Ponciano, P. Joyce, and E. M. Top. 2007. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 153:452-463. [DOI] [PubMed] [Google Scholar]
- 13.de la Cruz, F., and J. Grinsted. 1982. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J. Bacteriol. 151:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennis, J. J. 2005. The evolution of IncP catabolic plasmids. Curr. Opin. Biotechnol. 16:291-298. [DOI] [PubMed] [Google Scholar]
- 15.Dunnett, C. W. 1964. New tables for multiple comparisons with a control. Biometrics 20:482-491. [Google Scholar]
- 16.Eberl, L., C. S. Kristensen, M. Givskov, E. Grohmann, M. Gerlitz, and H. Schwab. 1994. Analysis of the multimer resolution system encoded by the parCBA operon of broad-host-range plasmid RP4. Mol. Microbiol. 12:131-141. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Lopez, R., M. P. Garcillan-Barcia, C. Revilla, M. Lazaro, L. Vielva, and F. de la Cruz. 2006. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30:942-966. [DOI] [PubMed] [Google Scholar]
- 18.Frost, L. S., R. Leplae, A. O. Summers, and A. Toussaint. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722-732. [DOI] [PubMed] [Google Scholar]
- 19.Geurts, A. M., C. S. Hackett, J. B. Bell, T. L. Bergemann, L. S. Collier, C. M. Carlson, D. A. Largaespada, and P. B. Hackett. 2006. Structure-based prediction of insertion-site preferences of transposons into chromosomes. Nucleic Acids Res. 34:2803-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gogarten, J. P., and J. P. Townsend. 2005. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 3:679-687. [DOI] [PubMed] [Google Scholar]
- 21.Haines, A. S., P. Akhtar, E. R. Stephens, K. Jones, C. M. Thomas, C. D. Perkins, J. R. Williams, M. J. Day, and J. C. Fry. 2006. Plasmids from freshwater environments capable of IncQ retrotransfer are diverse and include pQKH54, a new IncP-1 subgroup archetype. Microbiology 152:2689-2701. [DOI] [PubMed] [Google Scholar]
- 22.Hallet, B., V. Vanhooff, and F. Cornet. 2004. DNA site-specific resolution systems, p. 145-180. In G. Phillips and B. Funnel (ed.), Plasmid biology. ASM Press, Washington, DC.
- 23.Harada, K. M., Y. Aso, W. Hashimoto, B. Mikami, and K. Murata. 2006. Sequence and analysis of the 46.6-kb plasmid pA1 from Sphingomonas sp. A1 that corresponds to the typical IncP-1β plasmid backbone without any accessory gene. Plasmid 56:11-23. [DOI] [PubMed] [Google Scholar]
- 24.Heuer, H., R. Szczepanowski, S. Schneiker, A. Pühler, E. M. Top, and A. Schlüter. 2004. The complete sequences of plasmids pB2 and pB3 provide evidence for a recent ancestor of the IncP-1β group without any accessory genes. Microbiology 150:3591-3599. [DOI] [PubMed] [Google Scholar]
- 25.Jung, S. H., S. H. Kang, and C. Ahn. 2003. χ-square test for R × C contingency tables with clustered data. J. Biopharm. Stat. 13:241-251. [DOI] [PubMed] [Google Scholar]
- 26.Kamachi, K., M. Sota, Y. Tamai, N. Nagata, T. Konda, T. Inoue, E. M. Top, and Y. Arakawa. 2006. Plasmid pBP136 from Bordetella pertussis represents an ancestral form of IncP-1β plasmids without accessory mobile elements. Microbiology 152:3477-3484. [DOI] [PubMed] [Google Scholar]
- 27.Kamali-Moghaddam, M., and L. Sundstrom. 2000. Transposon targeting determined by resolvase. FEMS Microbiol. Lett. 186:55-59. [DOI] [PubMed] [Google Scholar]
- 28.Kostelidou, K., and C. M. Thomas. 2000. The hierarchy of KorB binding at its 12 binding sites on the broad-host-range plasmid RK2 and modulation of this binding by IncC1 protein. J. Mol. Biol. 295:411-422. [DOI] [PubMed] [Google Scholar]
- 29.Lenski, R. E., and J. E. Bouma. 1987. Effects of segregation and selection on instability of plasmid pACYC184 in Escherichia coli B. J. Bacteriol. 169:5314-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minakhina, S., G. Kholodii, S. Mindlin, O. Yurieva, and V. Nikiforov. 1999. Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol. Microbiol. 33:1059-1068. [DOI] [PubMed] [Google Scholar]
- 32.Nash, H. A. 1996. Site-specific recombination: integration, excision, resolution, and inversion of defined DNA segments, p. 2363-2376. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella, 2nd ed., vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 33.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncPα plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 34.Parke, D. 1990. Construction of mobilizable vectors derived from plasmids RP4, pUC18 and pUC19. Gene 93:135-137. [DOI] [PubMed] [Google Scholar]
- 35.Rawlings, D. E., and E. Tietze. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65:481-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schilf, W., and V. Krishnapillai. 1986. Genetic analysis of insertion mutations of the promiscuous IncP-1 plasmid R18 mapping near oriT which affect its host range. Plasmid 15:48-56. [DOI] [PubMed] [Google Scholar]
- 37.Schlüter, A., H. Heuer, R. Szczepanowski, S. M. Poler, S. Schneiker, A. Pühler, and E. M. Top. 2005. Plasmid pB8 is closely related to the prototype IncP-1β plasmid R751 but transfers poorly to Escherichia coli and carries a new transposon encoding a small multidrug resistance efflux protein. Plasmid 54:135-148. [DOI] [PubMed] [Google Scholar]
- 38.Sherratt, D. 1989. Tn3 and related transposable elements: site-specific recombination and transposition, p. 163-184. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, DC.
- 39.Shintani, M., H. Yano, H. Habe, T. Omori, H. Yamane, M. Tsuda, and H. Nojiri. 2006. Characterization of the replication, maintenance, and transfer features of the IncP-7 plasmid pCAR1, which carries genes involved in carbazole and dioxin degradation. Appl. Environ. Microbiol. 72:3206-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 41.Simonsen, L. 1991. The exsistence conditions for bacterial plasmids: theory and reality. Microb. Ecol. 22:187-205. [DOI] [PubMed] [Google Scholar]
- 42.Smalla, K., A. S. Haines, K. Jones, E. Krögerrecklenfort, H. Heuer, M. Schloter, and C. M. Thomas. 2006. Increased abundance of IncP-1β plasmids and mercury resistance genes in mercury-polluted river sediments: first discovery of IncP-1β plasmids with a complex mer transposon as the sole accessory element. Appl. Environ. Microbiol. 72:7253-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sota, M., M. Endo, K. Nitta, H. Kawasaki, and M. Tsuda. 2002. Characterization of a class II defective transposon carrying two haloacetate dehalogenase genes from Delftia acidovorans plasmid pUO1. Appl. Environ. Microbiol. 68:2307-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sota, M., H. Kawasaki, and M. Tsuda. 2003. Structure of haloacetate-catabolic IncP-1β plasmid pUO1 and genetic mobility of its residing haloacetate-catabolic transposon. J. Bacteriol. 185:6741-6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sota, M., H. Yano, A. Ono, R. Miyazaki, H. Ishii, H. Genka, E. M. Top, and M. Tsuda. 2006. Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J. Bacteriol. 188:4057-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tauch, A., A. Schlüter, N. Bischoff, A. Goesmann, F. Meyer, and A. Pühler. 2003. The 79,370-bp conjugative plasmid pB4 consists of an IncP-1β backbone loaded with a chromate resistance transposon, the strA-strB streptomycin resistance gene pair, the oxacillinase gene blaNPS-1, and a tripartite antibiotic efflux system of the resistance-nodulation-division family. Mol. Gen. Genomics 268:570-584. [DOI] [PubMed] [Google Scholar]
- 47.Taylor, L. A., and R. E. Rose. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tennstedt, T., R. Szczepanowski, I. Krahn, A. Pühler, and A. Schlüter. 2005. Sequence of the 68,869 bp IncP-1α plasmid pTB11 from a waste-water treatment plant reveals a highly conserved backbone, a Tn402-like integron and other transposable elements. Plasmid 53:218-238. [DOI] [PubMed] [Google Scholar]
- 49.Thomas, C. M. 2000. Paradigms of plasmid organization. Mol. Microbiol. 37:485-491. [DOI] [PubMed] [Google Scholar]
- 50.Thomas, C. M., and K. M. Nielsen. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711-721. [DOI] [PubMed] [Google Scholar]
- 51.Thomas, C. M., and C. A. Smith. 1987. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu. Rev. Microbiol. 41:77-101. [DOI] [PubMed] [Google Scholar]
- 52.Thorsted, P. B., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. M. Thomas. 1998. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 53.Top, E. M., and D. Springael. 2003. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14:262-269. [DOI] [PubMed] [Google Scholar]
- 54.Tralau, T., A. M. Cook, and J. Ruff. 2001. Map of the IncP1β plasmid pTSA encoding the widespread genes (tsa) for p-toluenesulfonate degradation in Comamonas testosteroni T-2. Appl. Environ. Microbiol. 67:1508-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsuda, M., and T. Iino. 1988. Identification and characterization of Tn4653, a transposon covering the toluene transposon Tn4651 on TOL plasmid pWW0. Mol. Gen. Genet. 213:72-77. [DOI] [PubMed] [Google Scholar]
- 56.Tsuda, M., K. Minegishi, and T. Iino. 1989. Toluene transposons Tn4651 and Tn4653 are class II transposons. J. Bacteriol. 171:1386-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsuda, M., H. M. Tan, A. Nishi, and K. Furukawa. 1999. Mobile catabolic genes in bacteria. J. Biosci. Bioeng. 87:401-410. [DOI] [PubMed] [Google Scholar]
- 58.van der Meer, J. R., and V. Sentchilo. 2003. Genomic islands and the evolution of catabolic pathways in bacteria. Curr. Opin. Biotechnol. 14:1-7. [DOI] [PubMed] [Google Scholar]
- 59.Ward, J. M., and J. Grinsted. 1982. Physical and genetic analysis of the Inc-W group plasmids R388, Sa, and R7K. Plasmid 7:239-250. [DOI] [PubMed] [Google Scholar]
- 60.Wolkow, C. A., R. T. DeBoy, and N. L. Craig. 1996. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 10:2145-2157. [DOI] [PubMed] [Google Scholar]
- 61.Zatyka, M., L. Bingle, A. C. Jones, and C. M. Thomas. 2001. Cooperativity between KorB and TrbA repressors of broad-host-range plasmid RK2. J. Bacteriol. 183:1022-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 2000. Conjugative-DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool. Harwood Academic Publishers, Amsterdam, The Netherlands.