Abstract
Maintaining appropriate levels of the global regulator FNR is critical to its function as an O2 sensor. In this study, we examined the mechanisms that control transcription of fnr to increase our understanding of how FNR protein levels are regulated. Under anaerobic conditions, one mechanism that controls fnr expression is negative autoregulation by the active [4Fe-4S] form of FNR. Through DNase I footprinting and in vitro transcription experiments, we observed that direct binding of [4Fe-4S]-FNR to the predicted downstream FNR binding site is sufficient for repression of the fnr promoter in vitro. In addition, the downstream FNR binding site was required for repression of transcription from fnr′-lacZ fusions in vivo. No repression of fnr was observed in vivo or in vitro with the apoprotein form of FNR, indicating that repression requires the dimeric, Fe-S cluster-containing protein. Furthermore, our in vitro and in vivo data suggest that [4Fe-4S]-FNR does not bind to the predicted upstream FNR binding site within the fnr promoter. Rather, we provide evidence that integration host factor binds to this upstream region and increases in vivo expression of Pfnr under both aerobic and anaerobic conditions.
The ability of Escherichia coli to efficiently sense and respond to O2 is primarily controlled by the global regulatory protein FNR (22, 42, 58). FNR is selectively active as a transcription factor under anaerobic growth conditions, where it has been shown to control the transcription of hundreds of genes, many of which are necessary for adaptation to O2-limiting growth conditions (10, 16, 25, 46). The large number of genes whose expression is regulated by changes in O2 and the dramatic reprogramming of metabolic pathways have made the study of FNR and its regulon ideal for a system level approach. The primary mechanism of regulation is the direct inactivation of FNR via the O2-dependent destruction of its [4Fe-4S] cluster, which is required for its activity (20, 28, 33). Recent studies indicate that this inactivation mechanism is optimized for normal cellular levels of FNR protein (2,600 to 4,100 molecules per cell) (56) since excess FNR protein (even twofold) escapes O2 inactivation (4, 37, 38, 53). Defining the mechanisms that control FNR protein levels is important in understanding the global response to O2.
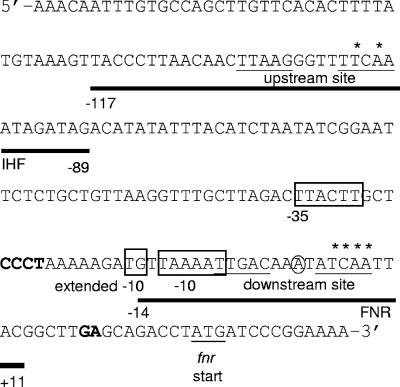
Both transcriptional and posttranscriptional control of FNR protein levels has been observed, providing a foundation for understanding how FNR levels are regulated. Under aerobic growth conditions, proteolysis decreases FNR protein levels (12, 38), while under anaerobic conditions, FNR represses its own transcription (24, 41, 44, 47, 54). While the mechanism that regulates FNR proteolysis has been elucidated (12, 38), a detailed analysis of fnr transcription has not been carried out. In vivo studies using either fnr::lacZ transcriptional or translational fusions demonstrated that the fnr promoter was repressed approximately two- to threefold in an FNR-dependent manner under anaerobic conditions (24, 41, 44, 47, 54). Surprisingly, repression of fnr::lacZ was shown to be further enhanced when fnr was expressed from a multicopy plasmid (44, 54), suggesting that repression is limited by FNR levels under anaerobic conditions. Two sequence elements that showed similarity to the FNR consensus binding site (TTGAT-N4-ATCAA) (22) were identified within the fnr promoter region (Fig. 1). The predicted upstream FNR binding site (TTAAG-N4-TTCAA) is centered at bp −103.5 relative to the transcription start site, whereas the predicted downstream FNR binding site (TTGAC-N4-ATCAA; underlined nucleotide match the consensus) is centered at bp −0.5 and overlaps the transcription start site (22). While binding of reconstituted [4Fe-4S]-FNR to the predicted downstream site has been reported in vitro (21), the contribution of this site or the predicted upstream site to Pfnr repression was not examined.
FIG. 1.
fnr promoter region. Shown are nucleotides −163 to +40 relative to the transcription start site (circled). The predicted upstream and downstream FNR binding sites are underlined, and the asterisks indicate the bases in both of the sites that were mutated in this study. The fnr start codon is also underlined, and the −35, −10, and extended −10 promoter elements are boxed. The bold horizontal lines mark the areas of protection by FNR or IHF from DNase I cleavage as determined in this study. Positions of enhanced DNase I cleavage are in bold.
In this study, we determined the roles of the two predicted FNR binding sites in the regulation of fnr transcription using both in vitro and in vivo approaches. DNase I footprinting and in vitro transcription experiments were used to determine whether direct binding of [4Fe-4S]-FNR to either the upstream or the downstream site was sufficient for Pfnr repression. β-Galactosidase activities from wild-type or mutant fnr::lacZ fusions in which base substitutions were made within the upstream or downstream binding sites were monitored in both anaerobically and aerobically grown strains. In addition, the involvement of other transcription factors, in addition to FNR, in the regulation of fnr transcription was investigated.
MATERIALS AND METHODS
FNR protein purification.
Isolation of [4Fe-4S]-FNR was carried out as described previously (38, 55), using a Pharmacia fast-performance liquid chromatography system equipped with a BioRex-70 cation-exchange column (Bio-Rad Laboratories) in a Coy anaerobic chamber with an atmosphere of 80% N2, 10% CO2, and 10% H2. To further enrich for the dimeric, cluster-containing form of FNR, the [4Fe-4S]-FNR preparation was subject to size exclusion chromatography under anaerobic conditions as previously described (40) and the pooled dimeric FNR protein fraction was analyzed for protein, iron, and sulfide content (2, 26, 27). The isolated [4Fe-4S]-FNR was ∼100% occupied with [4Fe-4S] clusters, calculated on the basis of the sulfide content (27). Apo-FNR was purified as described previously (38), using a Pharmacia fast-performance liquid chromatography system equipped with a 5-ml Hi-Trap heparin column (Amersham), followed by concentration with a 1-ml Hi-Trap heparin column.
DNase I footprinting.
DNA fragments containing the fnr promoter region were isolated from plasmid pPK7665 (bp −155 to +25 relative to the transcription start site) or pPK8221 (bp −214 to +25) (Table 1) with either HindIII and BamHI or EcoRI and BamHI. A Klenow fragment (New England Biolabs) was used to 3′ radiolabel the HindIII or EcoRI end of the DNA fragment with [α-32P]dATP (∼3,000 Ci mmol−1, i.e., ∼110 TBq mmol−1) (GE Healthcare). Labeled DNA fragments were isolated from a nondenaturing 5% acrylamide gel and were subsequently purified with elutip-d columns (Schleicher and Schuell). DNase I footprinting was carried out in a Coy anaerobic chamber in a total volume of 20 μl by incubating 6 nM DNA and either isolated [4Fe-4S]-FNR (100 to 400 nM), apo-FNR (200 to 400 nM), integration host factor (IHF) (250 to 750 nM), or cyclic AMP receptor protein (CRP) (0.5 to 4 μM) proteins for 30 min at 37°C in 40 mM Tris (pH 7.9), 70 mM KCl, 100 μg ml−1 bovine serum albumin, and 1 mM dithiothreitol. Cyclic AMP (cAMP) was added to a final concentration of 0.2 mM where indicated. DNase I (2 μg ml−1) and MgCl2 (10 mM) were added, and after 30 s, the reaction was terminated by the addition of 300 mM sodium acetate and 20 mM EDTA. The reaction mixtures were then ethanol precipitated, resuspended in loading dye (8 M urea, 0.5× TBE [Tris-borate-EDTA], 0.05% bromophenol blue, 0.05% xylene cyanol), heated for 30 s at 90°C, and loaded onto a 7 M urea-8% polyacrylamide gel in 0.5× TBE buffer. A+G sequencing ladders were generated as previously described (35). The reaction products were visualized by phosphorimaging and ImageQuant software.
TABLE 1.
E. coli strains and plasmids used in this work
| Construct | Relevant genotype | Reference or source |
|---|---|---|
| Strains | ||
| PK22 | BL21 (DE153) Δcrp-bs990 rpsL ΔfnrΩSpr/Smrzcj-3061::Tn10 | 32 |
| BW25993 | lacIq ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 11 |
| DBP102 | himA452::mini-tet Δ(lac pro) rpsL | 3 |
| DM0068 | crp::cat | 30 |
| BW25113 | lacIqhsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 11 |
| PK7510 | BW25113 arcA::cat | This laboratory |
| MG1655 | F− λ−rph-1 | This laboratory |
| PK4811 | MG1655 but ΔfnrΩSpr Smr | This laboratory |
| PK6988 | MG1655 (−155 to +25) Pfnr′-lacZ | This study |
| PK6997 | PK4811 (−155 to +25) Pfnr′-lacZ | This study |
| PK6990 | Same as PK6988 but bp +4 to +7 of Pfnr changed to 5′-CTGG-3′ | This study |
| PK6999 | Same as PK6997 but bp +4 to +7 of Pfnr changed to 5′-CTGG-3′ | This study |
| PK7608 | Same as PK6988 but arcA::cat | This study |
| PK7609 | Same as PK6997 but arcA::cat | This study |
| PK7610 | Same as PK6988 but himA::tet | This study |
| PK7611 | Same as PK6997 but himA::tet | This study |
| PK8208 | Same as PK6988 but crp::cat | This study |
| PK8209 | Same as PK6997 but crp::cat | This study |
| PK6989 | Same as PK6988 but bp −100 to −98 of Pfnr changed to 5′-CCG-3′ | This study |
| PK6998 | Same as PK6997 but bp −100 to −98 of Pfnr changed to 5′-CCG-3′ | This study |
| PK7602 | Same PK6989 but bp +4 to +7 of Pfnr changed to 5′-CTGG-3′ | This study |
| PK7603 | Same as PK6998 but bp +4 to +7 of Pfnr changed to 5′-CTGG-3′ | This study |
| PK7616 | Same as PK6989 but himA::tet | This study |
| PK7617 | Same as PK6998 but himA::tet | This study |
| PK8293 | MG1655 (bp −214 to +25) Pfnr′-lacZ | This study |
| PK8294 | PK4811 (−214 to +25) Pfnr′-lacZ | This study |
| PK8295 | Same as PK8293 but crp::cat | This study |
| PK8296 | Same as PK8294 but crp::cat | This study |
| PK8434 | Same as PK8293 but bp −142 to −138 of Pfnr changed to 5′-CTGAG-3′ | This study |
| PK8435 | Same as PK8294 but bp −142 to −138 of Pfnr changed to 5′-CTGAG-3′ | This study |
| PK7667 | PK6997 but with pRZ7411 | This study |
| PK7668 | PK6997 but with pACYC184 | This study |
| PK7669 | PK6997 but with pPK852 | This study |
| PK7670 | PK6997 but with pPK436 | This study |
| PK7671 | PK6997 but with pPK853 | This study |
| PK7672 | PK6997 but with pPK438 | This study |
| PK7673 | PK6997 but with pPK6928 | This study |
| PK7674 | PK6997 but with pPK6929 | This study |
| RZ7416 | λ (bp −481 to +55) Pfnr′-lacZ | This laboratory |
| RZ7426 | Same as RZ7416 but ΔfnrΩSpr Smr | This laboratory |
| PK8242 | Same as RZ7416 but arcA::cat | This study |
| PK8243 | Same as RZ7426 but arcA::cat | This study |
| Plasmids | ||
| pPK823 | Apr; bp +1 to +1115 of fnr in NdeI and BamHI sites of pET-11a | 32 |
| pPK7179 | Apr; pUC19-spf′ with XhoI site replacing SalI site | 25 |
| pPK7665 | bp −155 to +25 of Pfnr in XhoI and BamHI sites of pPK7179 | This study |
| pPK8221 | bp −214 to +25 of Pfnr in XhoI and BamHI sites of pPK7179 | This study |
| pACYC184 | Cmr | 8 |
| pRZ7411 | Cmr; HindIII-BamHI of fnr; bp −521 to +1115 of fnr in pACYC184 | 32 |
| pPK6979 | Same as pRZ7411 but bp +4 to +7 of Pfnr changed to 5′-CTGG-3′ | This study |
| pPK7035 | Knr gene from pHP45Ω and BamHI-NdeI fragment from pRS1553 into pBR322 | 25 |
| pPK6978 | bp −155 to +25 of Pfnr in XhoI and BamHI sites of pPK7035 | This study |
| pPK6980 | Same as pPK6978 but bp −100 to −98 of Pfnr changed to 5′-CCG-3′ | This study |
| pPK6981 | Same as pPK6978 but bp +4 to +7 of Pfnr changed to 5′-CTGG-3′ | This study |
| pPK7000 | Same as pPK6981 but bp −100 to −98 of Pfnr changed to 5′-CCG-3′ | This study |
| pPK8429 | Same as pPK8278 but bp −142 to −138 of Pfnr changed to 5′-CTGAG-3′ | This study |
| pKD46 | Phage λ gam-bet-exo genes under ParaB control | B. Wanner |
| pPK852 | Same as pRZ7411 but fnr-CS20 | This laboratory |
| pPK436 | Same as pRZ7411 but fnr-CS23 | 32 |
| pPK853 | Same as pRZ7411 but fnr-CS29 | This laboratory |
| pPK438 | Same as pRZ7411 but fnr-CS122 | 32 |
| pPK6928 | Same as pRZ7411 but fnr-CA23 | 38 |
| pPK6929 | Same as pRZ7411 but fnr-CA122 | 38 |
In vitro transcription assays.
The fnr promoter regions (bp −155 to +25 or −214 to +25 relative to the transcription start site) were PCR amplified using pRZ7411 (Table 1) (32) as a template and primers containing XhoI and BamHI sites, digested with XhoI and BamHI, and cloned into pPK7179, a pUC19-spf′ derivative containing an XhoI site (25). The resulting plasmids (pPK7665 or pPK8221) were purified with a QIAfilter Maxi kit (QIAGEN). Assays were carried out in a Coy anaerobic chamber using isolated [4Fe-4S]-FNR, IHF, and CRP proteins. Supercoiled plasmid DNA (2 nM) (pPK7179 or pPK8221) was incubated with the indicated protein(s), 5 μCi (185,000 Bq) of [α-32P]UTP (∼3,000 Ci/mmol, i.e., ∼110 TBq mmol−1), unlabeled UTP (50 μM), and 500 μM final concentrations each of ATP, CTP, and GTP (GE Healthcare) for 30 min at 37°C in 40 mM Tris (pH 7.9), 70 mM KCl, 100 μg ml−1 bovine serum albumin, 1 mM dithiothreitol, and 10 mM MgCl2. cAMP (0.2 mM) was present where indicated. Eσ70 RNA polymerase (50 nM) (Epicentre) was added, and each reaction (in a 20-μl total volume) was terminated after 5 min by adding 10 μl of 95% (vol/vol) formamide, 20 mM EDTA, 0.05% (wt/vol) bromophenol blue, and 0.05% (wt/vol) xylene cyanol FF (USB Corporation). After the mixture was heated to 90°C for 30 seconds, 5 μl was loaded onto an 8% polyacrylamide-7 M urea gel (0.5× TBE). Upon exposure to a PhosphorImager screen, transcripts were quantified using Molecular Dynamics ImageQuant software and fnr transcription was normalized to the amount of RNA-1 transcript (13). Each assay was repeated at least three times.
Construction of strains and plasmids.
The construction of fnr promoter-lacZ fusions involved two steps. First, pPK7035 (Table 1) (25) plasmid derivatives containing base substitutions within the downstream FNR binding site in the fnr promoter region were made via site-directed mutagenesis of pRZ7411 to create pPK6979 (Table 1). DNA fragments containing the wild-type or downstream mutant fnr promoter region (bp −155 to +25 or −214 to +25 relative to the transcription start site) were PCR amplified using pRZ7411 or pPK6979 as a template and primers containing XhoI and BamHI sites, digested with XhoI and BamHI, and cloned into pPK7035 to create pPK6978 (bp −115 to +25 of Pfnr), pPK6981 (bp −155 to +25 of downstream mutant Pfnr), or pPK8278 (bp −214 to +25 of Pfnr) (Table 1). Base substitutions within the upstream FNR binding site were made via site-directed mutagenesis of pPK6978 and pPK6981 to create pPK6980 (bp −155 to +25 of upstream mutant Pfnr) and pPK7000 (bp −155 to +25 of downstream and upstream mutant Pfnr), respectively. Base substitutions within the predicted CRP binding site were made via site-directed mutagenesis of pPK8278 to create pPK8429 (bp −214 to +25 of Pfnr with a mutation in the CRP binding site).
The second step involved PCR amplification of the lacI-Kn promoter-lacZ fragment from the pPK7035 plasmid derivatives and recombination into the chromosome as previously described (14). Kn promoter-lacZ fusions were introduced into MG1655 and its FNR− derivative, PK4811, via P1 transduction and selection for kanamycin resistance. Transduction with P1 was also used to introduce himA::tet, crp::cat, and arcA::cat from strains DPB102, DM0068, and PK7510 (Table 1), respectively, into strains containing wild-type or mutant fnr promoter-lacZ fusions.
β-Galactosidase assays.
β-Galactosidase activity was measured in strains containing wild-type or mutant fnr promoter-lacZ fusions as described previously (39). Cells were grown aerobically or anaerobically to an optical density at 600 nm of ∼0.2 in either M9 minimal medium with 0.2% (wt/vol) glucose (or 0.2% [wt/vol] fructose where indicated), 10 μM ferric ammonium citrate, and 0.2 μM ammonium molybdate or LB as previously described (55). Casamino Acids or chloramphenicol was added to the medium where indicated. To terminate cell growth and any further protein synthesis, either chloramphenicol (final concentration, 20 μg ml−1) or tetracycline (final concentration, 10 μg ml−1) was added and cells were placed on ice until assayed for β-galactosidase activity (39). β-Galactosidase assays were repeated at least three times. β-Galactosidase activity was normalized to account for the difference in cell numbers per ml of culture for aerobically and anaerobically grown cells as determined via viable plating assays (56). At an optical density at 600 nm of 0.4, aerobic and anaerobic cultures contained (2.6 ± 0.2) × 108 and (4 ± 0.3) × 108 cells ml−1, respectively. Therefore, β-galactosidase activity was normalized by multiplying the aerobic values by a factor of 1.5.
RESULTS
In vivo negative autoregulation requires the presence of [4Fe-4S]-FNR.
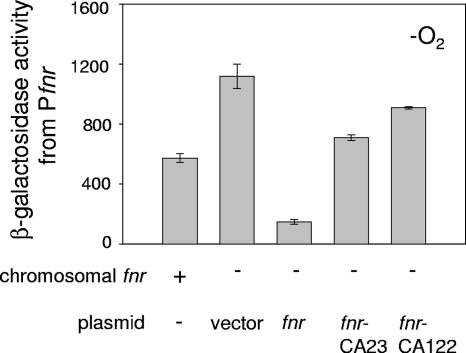
To determine if negative autoregulation requires the [4Fe-4S] form of FNR, expression of Pfnr′-lacZ (bp −155 to +25 of Pfnr relative to the transcription start site) was monitored in anaerobically grown strains expressing either wild-type FNR or FNR mutants (FNR-CA23 and FNR-CA122) that were previously shown to not contain [4Fe-4S] clusters (29, 32, 34, 37, 50, 53). Consistent with previous studies (24, 41, 44, 47, 54), expression from Pfnr was repressed approximately twofold when the chromosomal copy of fnr was present (Fig. 2). In addition, Pfnr expression was further decreased approximately fourfold in the presence of plasmid-derived wild-type FNR. In contrast, repression of Pfnr was approximately five- to sixfold less efficient in strains expressing the FNR-CA23 and FNR-CA122 mutants than Pfnr repression by plasmid-derived wild-type FNR. These data support the notion that negative autoregulation in vivo requires the presence of active [4Fe-4S]-FNR.
FIG. 2.
Effect of plasmid-derived FNR protein levels on fnr′-lacZ expression. Δfnr strains expressing wild-type FNR or FNR mutants from pACYC184 were grown in M9 minimal glucose medium containing a final concentration of 20 μg ml−1 of chloramphenicol under anaerobic growth conditions. β-Galactosidase activity from fnr ′-lacZ (bp −155 to +25) is also shown for the wild-type strain, containing fnr present in a single copy. FNR-CA23 and FNR-CA122 are mutant proteins that do not ligate [4Fe-4S] clusters (29, 32, 34, 37, 50, 53). Error bars represent the standard errors for three independent experiments.
FNR directly binds to the predicted downstream site but not the upstream site in vitro.
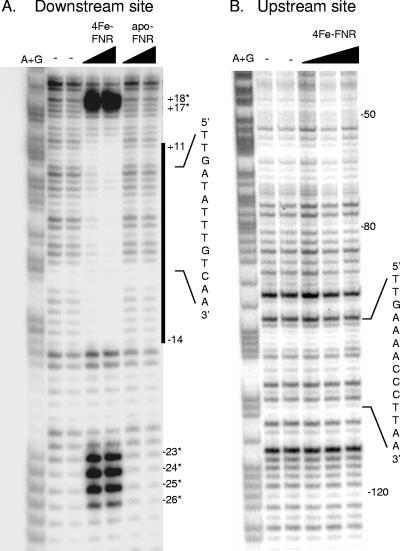
To address whether FNR directly binds to both of the predicted FNR binding sites within the fnr promoter region, in vitro DNase I footprinting experiments with [4Fe-4S]-FNR were performed. At the proposed downstream FNR binding site, a clear pattern of protection was observed from bp −14 to +11 relative to the fnr transcription start site by [4Fe-4S]-FNR (200 to 400 nM) (Fig. 3A). In addition, strong enhancements of DNase I cleavage were present at positions −26 through −23 and at positions +17 and +18. The presence of the [4Fe-4S] cluster was required for FNR to bind to the downstream site, since no enhancements or regions of protection were detected when equivalent amounts of apo-FNR were used in the assay (Fig. 3A). In contrast, no enhancements or regions of protection were detected for the predicted upstream FNR binding site by either [4Fe-4S]-FNR (Fig. 3B) or apo-FNR (data not shown). These data indicate that [4Fe-4S]-FNR binds only to the downstream site under the in vitro solution conditions used in this study.
FIG. 3.
Footprinting of FNR at the predicted downstream (A) and upstream (B) FNR binding sites within the fnr promoter region. The DNA sequences for the predicted FNR binding sites are indicated at the right of each panel. Numbers indicate the distances relative to the transcription start site. Samples were electrophoresed with Maxam-Gilbert (A+G) ladders made using the same DNA. (A) DNase I cleavage of the EcoRI-BamHI DNA fragment from pPK7665 (Table 1), in which the 3′ EcoRI end was radiolabeled. FNR protein concentrations are given from left to right in terms of nM total protein: [4Fe-4S]-FNR (4Fe-FNR), 200 and 400; apo-FNR, 200 and 400. The area of protection from DNase I by [4Fe-4S]-FNR is indicated with a vertical line. Asterisks indicate the positions of enhanced DNase I cleavage. (B) DNase I cleavage of the HindIII-BamHI DNA fragment from pPK7665, in which the 3′ HindIII end was radiolabeled. [4Fe-4S]-FNR (4Fe-FNR) concentrations are given from left to right in terms of nM total protein: 100, 200, and 400.
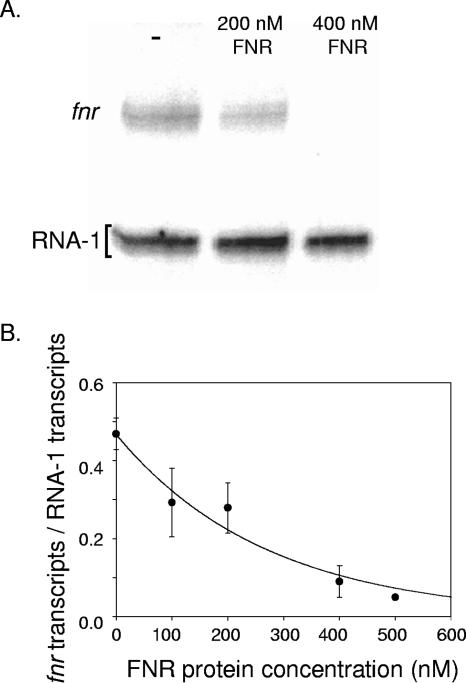
To test whether binding of this downstream site is sufficient for repression of Pfnr, in vitro transcription assays were carried out with purified RNA polymerase and a plasmid template containing bp −155 to +25 of the fnr promoter region. In the absence of FNR protein, distinct transcripts that initiated from the fnr promoter (expected sizes of 135 and 134 nucleotides) and the control RNA-1 promoter (13) were detected (Fig. 4). In the presence of increasing amounts of [4Fe-4S]-FNR protein (0 to 500 nM), the amount of fnr transcript decreased in a concentration-dependent manner, whereas no effect on the amount of RNA-1 transcript was observed. Since the in vitro transcription assays were carried out under reaction conditions similar to those for the DNase I footprinting experiments, these data suggest that binding of [4Fe-4S]-FNR to the downstream site is sufficient to repress Pfnr transcription in vitro.
FIG. 4.
Effect of [4Fe-4S]-FNR on Pfnr transcription in vitro. (A) Assay mixtures contained 2 nM plasmid DNA containing bp −155 to +25 of the fnr promoter region relative to the transcription start site, 50 nM Eσ70 RNA polymerase, and, where indicated, 200 nM or 400 nM [4Fe-4S]-FNR protein. (B) Quantified data showing the amounts of in vitro transcription from Pfnr in the presence of increasing concentrations of [4Fe-4S]-FNR. Pfnr transcription was normalized by dividing the amount of the fnr transcript by the amount of the RNA-1 control transcript. Error bars represent the standard errors for three independent experiments.
Repression of Pfnr transcription in vivo also requires the downstream FNR binding site but not the upstream binding site.
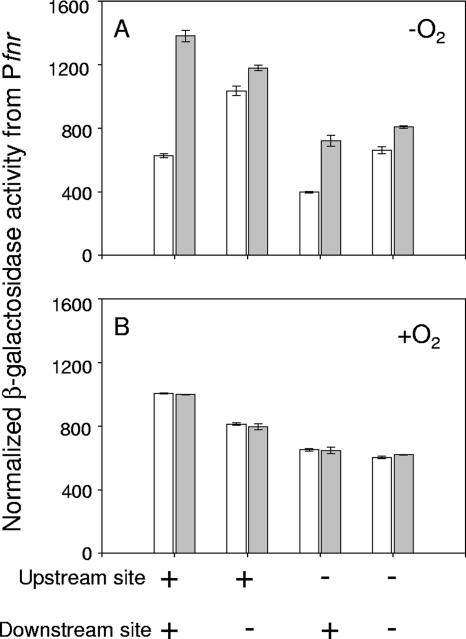
To determine if either of these sequence elements plays a role in regulating transcription of Pfnr in vivo, expression from wild-type or mutant fnr promoters (bp −155 to +25 relative to the transcription start site) fused to lacZ was measured (Fig. 5). Analysis of mutations within the upstream (−110TTAAG-N4-TCCGA−97) or downstream (−7TTGAC-N4-ACTGG+7) (base substitutions are underlined) site showed that only the downstream site was required for repression of Pfnr, in agreement with the in vitro results. Surprisingly, the mutation in the upstream site caused an approximately twofold decrease in expression from the mutant fnr promoter in both FNR+ and FNR− strains. Furthermore, the mutations in the downstream and upstream sites appear to act independently, since substitutions in both of the sites resulted in both a loss of negative autoregulation and an overall approximately twofold decrease in fnr expression. Collectively, these results indicate that only the downstream FNR binding site is required for negative autoregulation, while the upstream site may play a role in transcription activation of Pfnr by an FNR-independent mechanism.
FIG. 5.
In vivo expression from wild-type or mutant fnr′-lacZ (−155 to +25) promoter fusions from anaerobically (A) or aerobically (B) grown cells measured using β-galactosidase assays. FNR+ and FNR− strains are represented by the white and gray bars, respectively. “−” indicates that base substitutions (Fig. 1) were made within the predicted upstream FNR binding site, the predicted downstream FNR binding site, or both sites. “+” indicates that the wild-type sequence is present. β-Galactosidase activity from the fnr′-lacZ promoter fusions was normalized by correcting for the difference in cell numbers ml−1 of culture for aerobically and anaerobically grown cells as explained in Materials and Methods. Error bars represent the standard errors for three independent experiments.
Activation of Pfnr occurs under aerobic and anaerobic growth conditions.
The effect of the mutation of the upstream DNA element indicated that an unknown factor may function to weakly activate fnr expression under anaerobic conditions. To determine whether disruption of the upstream element also affected Pfnr expression under aerobic growth conditions, β-galactosidase activity from the wild-type and mutant fnr promoters was measured in aerobically grown cells (Fig. 5B). As expected (24, 41, 44, 47, 54), no repression of Pfnr by FNR was observed under aerobic growth conditions. However, mutation of the upstream element caused the same approximately twofold decrease in expression in aerobically grown cells as that observed under anaerobic growth conditions. The possibility that mutation of this upstream sequence decreased the function of a second fnr promoter was eliminated since in vivo mapping of transcription start sites within the wild-type fnr promoter region indicates that there is a single transcription start site similar to that observed in vitro (data not shown). Rather, these data are consistent with the notion that another transcription factor may be recruited by the upstream site to activate Pfnr expression under both aerobic and anaerobic growth conditions.
IHF increases transcription from the fnr promoter in vivo.
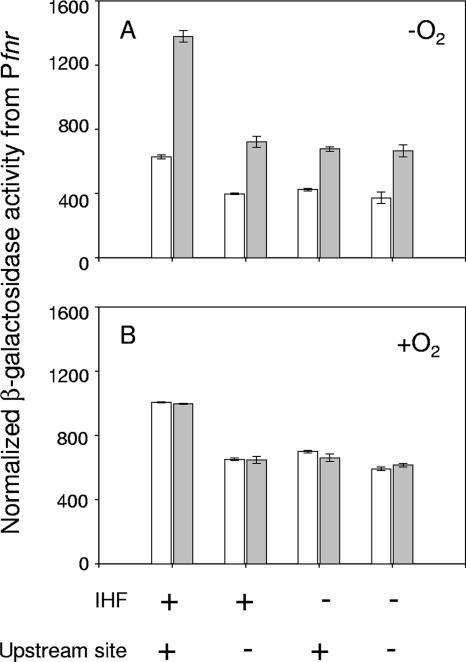
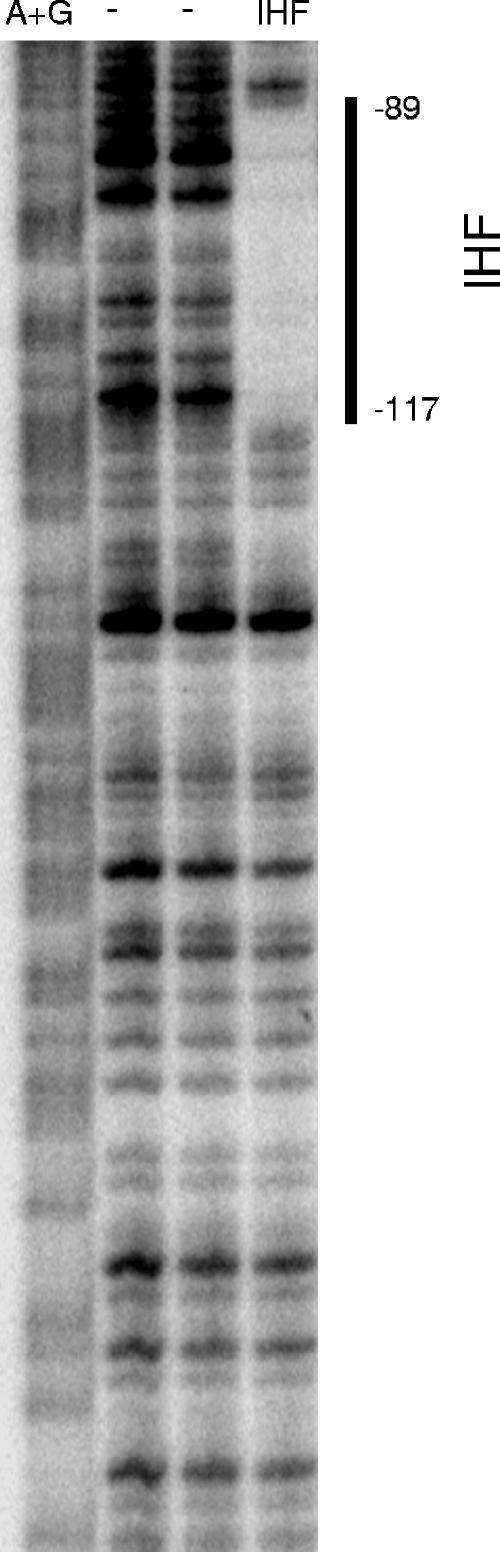
To examine whether other transcription factors regulate Pfnr, we tested whether the DNA-bending protein IHF had an effect on fnr transcription, since it has been shown to be involved in the regulation of several FNR-dependent promoters, such as narG, nir, nrfA, dmsA, ubiC, ndh, pfl, sodA, and narK (5-7, 9, 19, 31, 36, 43, 48, 51, 52). In addition, the DNA sequences that IHF binds to (consensus site YAANNNNTTGAW, where W is A or T, Y is T or C, and N is any nucleotide) (15, 23, 60) show some similarity to the sequence within this upstream region of the fnr promoter (−110TTAAGGGTTTTCAA−97). An approximately twofold decrease in Pfnr expression was observed in both aerobic and anaerobic cells lacking IHF (Fig. 6). Furthermore, the IHF-dependent increase in fnr expression was not observed in the construct containing the mutant upstream site (Fig. 6), suggesting that IHF binds to the upstream element. In support of this notion, the results of DNase I footprinting experiments revealed that the region of DNA from bp −117 to −89 relative to the fnr transcription start site was protected in the presence of 0.5 μM IHF (Fig. 7) and includes the sequence of DNA that was originally predicted to be the upstream FNR binding site centered at bp −103.5. These data suggest that binding of IHF to the fnr promoter enhances transcription.
FIG. 6.
In vivo expression from wild-type or upstream mutant fnr′-lacZ promoter fusions in anaerobically (A) or aerobically (B) grown cells lacking IHF. FNR+ and FNR− strains are represented by the white and gray bars, respectively. “−” indicates that the strains are IHF−, whereas “+” indicates that the strains are IHF+. For the predicted upstream FNR binding site, “−” indicates that base substitutions (Fig. 1) were made within the upstream sequence, whereas “+” indicates that the wild-type sequence is present. β-Galactosidase activity from the fnr′-lacZ promoter fusions was normalized by correcting for the difference in cell numbers ml−1 of culture for aerobically and anaerobically grown cells as explained in Materials and Methods. Error bars represent the standard errors for three independent experiments.
FIG. 7.
Footprinting of the fnr promoter region by IHF. Samples contained the HindIII-BamHI DNA fragment from pPK8221 (Table 1), in which the 3′ HindIII end was radiolabeled. The area of protection from DNase I by 0.5 μM IHF is indicated with a vertical line, and numbers indicate the distances relative to the transcription start site. Samples were electrophoresed with Maxam-Gilbert ladders (A+G) made using the same DNA.
In vitro transcription experiments revealed that IHF did not alter transcription from the fnr promoter (data not shown), raising the possibility that IHF works in conjunction with another transcription factor to regulate transcription of Pfnr as has been previously observed for several FNR-dependent promoters (5-7, 9, 19, 31, 36, 43, 48, 51, 52). A recent study, which evaluated the distribution of CRP binding sites along the E. coli chromosome, identified a potential CRP binding site within the fnr promoter region centered at bp −145.5 relative to the transcription start site (18). Although DNase I footprinting revealed a weak binding site in this position, no increase in Pfnr transcription was observed in vitro in the presence of purified CRP-cAMP (0.5 to 1 μM), either in the presence or in the absence of IHF (0.5 μM) (data not shown). Furthermore, no effect of CRP was found in vivo by either mutation of the CRP binding site or use of strains that lacked CRP (data not shown), indicating that under the conditions tested, CRP does not regulate the FNR promoter.
We also tested whether the anaerobic regulator ArcA plays a role in regulating fnr transcription since recent reverse transcription-PCR studies have shown that levels of fnr transcript are slightly higher in cells lacking ArcA than in wild-type cells (49). However, we found that expression from Pfnr (bp −155 to +25 or −418 to +55) in ArcA− cells was similar to that in wild-type cells under both aerobic and anaerobic growth conditions (data not shown). Thus, it is unclear how IHF increases the expression of the fnr promoter.
DISCUSSION
In this study, we have defined the role of the two predicted FNR binding sites in fnr transcription. In addition to defining one sequence element required for negative autoregulation by FNR, we found a second element for IHF binding, which, through an unknown mechanism, enhances Pfnr expression. Thus, these studies have expanded our knowledge of how FNR is regulated at the transcriptional level and have provided new insights into how FNR protein levels are achieved for the global response to O2.
Regulation of fnr repression.
While our data indicate that [4Fe-4S]-FNR represses its own synthesis by binding to the predicted downstream FNR binding site within the fnr promoter region, the mechanism by which [4Fe-4S]-FNR prevents RNA polymerase from transcribing the fnr gene is not known. Given that the downstream FNR binding site is centered at bp −0.5 relative to the transcription start site, these results may suggest that [4Fe-4S]-FNR represses Pfnr by blocking RNA polymerase from binding to Pfnr through steric hindrance. However, further investigations are needed to examine this hypothesis. Although the downstream site contains 9 out of 10 bp which match the consensus FNR binding site, previous findings (44, 54) and data presented in this study suggest that the fnr promoter is not fully saturated by endogenous levels of [4Fe-4S]-FNR. For example, repression of Pfnr was increased approximately fourfold in vivo when FNR was expressed from a multicopy plasmid and was larger (approximately fivefold) in vitro than in vivo (approximately twofold) when FNR was expressed from the chromosome. Whether this is due to competition between FNR and RNA polymerase for binding to Pfnr and/or reflects additional sequence requirements for FNR binding in vivo is not known. Furthermore, it appears that FNR binding sites cannot be predicted based on bioinformatic data alone. For example, FNR did not bind to the predicted upstream site, which contains 7 out of 10 bp that match the consensus site. In contrast, FNR has been shown to directly bind to two sites within the FNR-repressed ndh promoter (19), one of which contains only 6 out of 10 bp that match the consensus site. Perhaps the efficiency of FNR binding is also influenced by differences in the architecture of FNR-dependent promoters.
Regulation of fnr activation.
Although our studies indicate that the predicted upstream site is not an FNR binding site, we found that this sequence binds IHF and increases expression of fnr under both aerobic and anaerobic conditions. This finding is also in agreement with a previous study which indicated that DNA sequences upstream of bp position −41 relative to the transcription start site are important for maximal fnr expression (47). While IHF alone had no effect on Pfnr transcription in vitro, it is possible that conditions of the assay may have bypassed a role for IHF or that another transcription factor, along with IHF, is required to activate the fnr promoter. Alternatively, IHF may function by preventing another transcription factor from repressing Pfnr. Indeed, this appears to be the case for the nir promoter, in which binding of IHF to the IHF II site decreases the repression of nir mediated by IHF and Fis binding at other sites (7). Our studies suggest that neither CRP nor ArcA is this transcription factor even though a previous study indicated that expression of fnr is slightly higher (approximately twofold) in ArcA− cells under microaerobic growth conditions (49). However, a recent study, which mapped the distribution of Fis binding sites across the E. coli genome, identified a potential Fis binding site within the fnr promoter region (17). Thus, further investigation is necessary to determine the role of Fis and other transcription factors in regulating fnr transcription.
Relevance of negative autoregulation in O2 sensing.
Negative autoregulation is not an uncommon regulatory mechanism found in E. coli. In fact, it has been reported that over 40% of known E. coli transcriptional factors are subject to negative autoregulation (45). Mathematical modeling and studies with synthetic gene circuits have indicated that negative autoregulation decreases the response times of transcription networks because the steady-state concentration of the transcription factor is achieved faster (1, 45, 57, 59). Since E. coli lives in environments with regular changes in O2 tension, rapidly achieving a new steady-state level of active FNR may provide an advantage during adaptation to various growth conditions by quickly allowing a new transcription rate for the FNR regulon.
In addition to providing a means for rapidly reaching steady-state levels of [4Fe-4S]-FNR, the amount of [4Fe-4S]-FNR protein produced by the negative autoregulation mechanism also seems optimal for the efficient inactivation of FNR by O2. Even though under standard aerobic growth conditions the O2 concentration in the medium can never exceed ∼220 μM at 37°C, it is in excess relative to the cellular concentration of FNR (∼7 μM) (56). Previous kinetic studies suggest that the rate of FNR inactivation is moderately fast at 220 μM O2 (half-life of ∼30 seconds at 25°C) (56). Despite this, it has been shown that even small increases in FNR protein result in increased FNR activity under aerobic growth conditions, indicating that excess FNR is not efficiently inactivated, presumably as a result of insufficient time to inactivate the additional protein (4, 37, 38, 53). Thus, we hypothesize that under anaerobic conditions, negative autoregulation also prevents [4Fe-4S]-FNR from exceeding a critical level beyond which it can be efficiently inactivated. Taken together, negative autoregulation appears to provide an optimal balance of FNR protein levels, directing a new rate of synthesis of the FNR regulon under anaerobic conditions and allowing the efficient inactivation of [4Fe-4S]-FNR upon exposure to O2. Future studies involving construction of mathematical models will be necessary to test these predictions.
Acknowledgments
We thank Barry Wanner for providing plasmid pKD46 and E. coli strains BW25113 and BW25993, Richard Gourse for purified CRP protein and strain DM0068, and Marcin Filutowicz for purified IHF and strain DBP102. We thank Jamie Visintainer and Andrea Nans for their generous help in constructing strains and performing preliminary β-galactosidase assays. In addition, we thank Helmut Beinert for evaluating the iron and sulfide content of isolated FNR protein.
This work was supported by NIH grant GM045844 (to P.J.K.).
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Becskei, A., and L. Serrano. 2000. Engineering stability in gene networks by autoregulation. Nature 405:590-593. [DOI] [PubMed] [Google Scholar]
- 2.Beinert, H. 1983. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 131:373-378. [DOI] [PubMed] [Google Scholar]
- 3.Biek, D. P., and S. N. Cohen. 1989. Involvement of integration host factor (IHF) in maintenance of plasmid pSC101 in Escherichia coli: mutations in the topA gene allow pSC101 replication in the absence of IHF. J. Bacteriol. 171:2066-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnefoy, V., M. Fons, J. Ratouchniak, M. C. Pascal, and M. Chippaux. 1988. Aerobic expression of the nar operon of Escherichia coli in a fnr mutant. Mol. Microbiol. 2:419-425. [DOI] [PubMed] [Google Scholar]
- 5.Browning, D. F., C. M. Beatty, A. J. Wolfe, J. A. Cole, and S. J. Busby. 2002. Independent regulation of the divergent Escherichia coli nrfA and acsP1 promoters by a nucleoprotein assembly at a shared regulatory region. Mol. Microbiol. 43:687-701. [DOI] [PubMed] [Google Scholar]
- 6.Browning, D. F., J. A. Cole, and S. J. Busby. 2000. Suppression of FNR-dependent transcription activation at the Escherichia coli nir promoter by Fis, IHF and H-NS: modulation of transcription initiation by a complex nucleo-protein assembly. Mol. Microbiol. 37:1258-1269. [DOI] [PubMed] [Google Scholar]
- 7.Browning, D. F., J. A. Cole, and S. J. Busby. 2004. Transcription activation by remodelling of a nucleoprotein assembly: the role of NarL at the FNR-dependent Escherichia coli nir promoter. Mol. Microbiol. 53:203-215. [DOI] [PubMed] [Google Scholar]
- 8.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Compan, I., and D. Touati. 1993. Interaction of six global transcription regulators in expression of manganese superoxide dismutase in Escherichia coli K-12. J. Bacteriol. 175:1687-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Constantinidou, C., J. L. Hobman, L. Griffiths, M. D. Patel, C. W. Penn, J. A. Cole, and T. W. Overton. 2006. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 281:4802-4815. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dibden, D. P., and J. Green. 2005. In vivo cycling of the Escherichia coli transcription factor FNR between active and inactive states. Microbiology 151:4063-4070. [DOI] [PubMed] [Google Scholar]
- 13.Erickson, J. W., and C. A. Gross. 1989. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 3:1462-1471. [DOI] [PubMed] [Google Scholar]
- 14.Giel, J. L., D. Rodionov, M. Liu, F. R. Blattner, and P. J. Kiley. 2006. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 60:1058-1075. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich, J. A., M. L. Schwartz, and W. R. McClure. 1990. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 18:4993-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grainger, D. C., H. Aiba, D. Hurd, D. F. Browning, and S. J. Busby. 2007. Transcription factor distribution in Escherichia coli: studies with FNR protein. Nucleic Acids Res. 35:269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grainger, D. C., D. Hurd, M. D. Goldberg, and S. J. Busby. 2006. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 34:4642-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grainger, D. C., D. Hurd, M. Harrison, J. Holdstock, and S. J. Busby. 2005. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. USA 102:17693-17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, J., M. F. Anjum, and J. R. Guest. 1997. Regulation of the ndh gene of Escherichia coli by integration host factor and a novel regulator, Arr. Microbiology 143:2865-2875. [DOI] [PubMed] [Google Scholar]
- 20.Green, J., B. Bennett, P. Jordan, E. T. Ralph, A. J. Thomson, and J. R. Guest. 1996. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem. J. 316:887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green, J., A. S. Irvine, W. Meng, and J. R. Guest. 1996. FNR-DNA interactions at natural and semi-synthetic promoters. Mol. Microbiol. 19:125-137. [DOI] [PubMed] [Google Scholar]
- 22.Guest, J. R., J. Green, A. S. Irvine, and S. Spiro. 1996. The FNR modulon and FNR-regulated gene expression, p. 317-342. In E. C. C. Lin and A. S. Lynch (ed.), Regulation of gene expression in Escherichia coli. R. G. Landes Company, Austin, TX.
- 23.Hales, L. M., R. I. Gumport, and J. F. Gardner. 1996. Examining the contribution of a dA+dT element to the conformation of Escherichia coli integration host factor-DNA complexes. Nucleic Acids Res. 24:1780-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, H. M., and R. P. Gunsalus. 1987. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J. Bacteriol. 169:3340-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang, Y., K. D. Weber, Y. Qiu, P. J. Kiley, and F. R. Blattner. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 187:1135-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy, M. C., T. A. Kent, M. Emptage, H. Merkle, H. Beinert, and E. Münck. 1984. Evidence for the formation of a linear [3Fe-4S] cluster in partially unfolded aconitase. J. Biol. Chem. 259:14463-14471. [PubMed] [Google Scholar]
- 27.Khoroshilova, N., H. Beinert, and P. J. Kiley. 1995. Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA binding. Proc. Natl. Acad. Sci. USA 92:2499-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khoroshilova, N., C. Popescu, E. Münck, H. Beinert, and P. J. Kiley. 1997. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc. Natl. Acad. Sci. USA 94:6087-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiley, P. J., and H. Beinert. 1998. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341-352. [DOI] [PubMed] [Google Scholar]
- 30.Kim, J., S. Adhya, and S. Garges. 1992. Allosteric changes in the cAMP receptor protein of Escherichia coli: hinge reorientation. Proc. Natl. Acad. Sci. USA 89:9700-9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolesnikow, T., I. Schroder, and R. P. Gunsalus. 1992. Regulation of narK gene expression in Escherichia coli in response to anaerobiosis, nitrate, iron, and molybdenum. J. Bacteriol. 174:7104-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lazazzera, B. A., D. M. Bates, and P. J. Kiley. 1993. The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 7:1993-2005. [DOI] [PubMed] [Google Scholar]
- 33.Lazazzera, B. A., H. Beinert, N. Khoroshilova, M. C. Kennedy, and P. J. Kiley. 1996. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 271:2762-2768. [DOI] [PubMed] [Google Scholar]
- 34.Lonetto, M. A., V. Rhodius, K. Lamberg, P. Kiley, S. Busby, and C. Gross. 1998. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase sigma70 subunit. J. Mol. Biol. 284:1353-1365. [DOI] [PubMed] [Google Scholar]
- 35.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560. [DOI] [PubMed] [Google Scholar]
- 36.McNicholas, P. M., R. C. Chiang, and R. P. Gunsalus. 1998. Anaerobic regulation of the Escherichia coli dmsABC operon requires the molybdate-responsive regulator ModE. Mol. Microbiol. 27:197-208. [DOI] [PubMed] [Google Scholar]
- 37.Melville, S. B., and R. P. Gunsalus. 1990. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J. Biol. Chem. 265:18733-18736. [PubMed] [Google Scholar]
- 38.Mettert, E. L., and P. J. Kiley. 2005. ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe-4S] cluster. J. Mol. Biol. 354:220-232. [DOI] [PubMed] [Google Scholar]
- 39.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 40.Moore, L. J., and P. J. Kiley. 2001. Characterization of the dimerization domain in the FNR transcription factor. J. Biol. Chem. 276:45744-45750. [DOI] [PubMed] [Google Scholar]
- 41.Pascal, M. C., V. Bonnefoy, M. Fons, and M. Chippaux. 1986. Use of gene fusions to study the expression of fnr, the regulatory gene of anaerobic electron transfer in Escherichia coli. FEMS Microbiol. Lett. 36:35-39. [Google Scholar]
- 42.Patschkowski, T., D. M. Bates, and P. J. Kiley. 2000. Mechanisms for sensing and responding to oxygen deprivation, p. 61-78. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, DC.
- 43.Rabin, R. S., L. A. Collins, and V. Stewart. 1992. In vivo requirement of integration host factor for nar (nitrate reductase) operon expression in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 89:8701-8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reyes-Ramirez, F., and R. G. Sawers. 2006. Aerobic activation of transcription of the anaerobically inducible Escherichia coli focA-pfl operon by fumarate nitrate regulator. FEMS Microbiol. Lett. 255:262-267. [DOI] [PubMed] [Google Scholar]
- 45.Rosenfeld, N., M. B. Elowitz, and U. Alon. 2002. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 323:785-793. [DOI] [PubMed] [Google Scholar]
- 46.Salmon, K., S. P. Hung, K. Mekjian, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2003. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J. Biol. Chem. 278:29837-29855. [DOI] [PubMed] [Google Scholar]
- 47.Sawers, R. G. 2005. Expression of fnr is constrained by an upstream IS5 insertion in certain Escherichia coli K-12 strains. J. Bacteriol. 187:2609-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schroder, I., S. Darie, and R. P. Gunsalus. 1993. Activation of the Escherichia coli nitrate reductase (narGHJI) operon by NarL and Fnr requires integration host factor. J. Biol. Chem. 268:771-774. [PubMed] [Google Scholar]
- 49.Shalel-Levanon, S., K. Y. San, and G. N. Bennett. 2005. Effect of ArcA and FNR on the expression of genes related to the oxygen regulation and the glycolysis pathway in Escherichia coli under microaerobic growth conditions. Biotechnol. Bioeng. 92:147-159. [DOI] [PubMed] [Google Scholar]
- 50.Sharrocks, A. D., J. Green, and J. R. Guest. 1990. In vivo and in vitro mutants of FNR the anaerobic transcriptional regulator of E. coli. FEBS Lett. 270:119-122. [DOI] [PubMed] [Google Scholar]
- 51.Sirko, A., E. Zehelein, M. Freundlich, and G. Sawers. 1993. Integration host factor is required for anaerobic pyruvate induction of pfl operon expression in Escherichia coli. J. Bacteriol. 175:5769-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soballe, B., and R. K. Poole. 1997. Aerobic and anaerobic regulation of the ubiCA operon, encoding enzymes for the first two committed steps of ubiquinone biosynthesis in Escherichia coli. FEBS Lett. 414:373-376. [DOI] [PubMed] [Google Scholar]
- 53.Spiro, S., and J. R. Guest. 1988. Inactivation of the FNR protein of Escherichia coli by targeted mutagenesis in the N-terminal region. Mol. Microbiol. 2:701-707. [DOI] [PubMed] [Google Scholar]
- 54.Spiro, S., and J. R. Guest. 1987. Regulation and over-expression of the fnr gene of Escherichia coli. J. Gen. Microbiol. 133:3279-3288. [DOI] [PubMed] [Google Scholar]
- 55.Sutton, V. R., and P. J. Kiley. 2003. Techniques for studying the oxygen-sensitive transcription factor FNR from Escherichia coli. Methods Enzymol. 370:300-312. [DOI] [PubMed] [Google Scholar]
- 56.Sutton, V. R., E. L. Mettert, H. Beinert, and P. J. Kiley. 2004. Kinetic analysis of the oxidative conversion of the [4Fe-4S]2+ cluster of FNR to a [2Fe-2S]2+ cluster. J. Bacteriol. 186:8018-8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thieffry, D., A. M. Huerta, E. Perez-Rueda, and J. Collado-Vides. 1998. From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli. Bioessays 20:433-440. [DOI] [PubMed] [Google Scholar]
- 58.Unden, G., S. Achebach, G. Holighaus, H. G. Tran, B. Wackwitz, and Y. Zeuner. 2002. Control of FNR function of Escherichia coli by O2 and reducing conditions. J. Mol. Microbiol. Biotechnol. 4:263-268. [PubMed] [Google Scholar]
- 59.Wall, M. E., W. S. Hlavacek, and M. A. Savageau. 2003. Design principles for regulator gene expression in a repressible gene circuit. J. Mol. Biol. 332:861-876. [DOI] [PubMed] [Google Scholar]
- 60.Yang, C. C., and H. A. Nash. 1989. The interaction of E. coli IHF protein with its specific binding sites. Cell 57:869-880. [DOI] [PubMed] [Google Scholar]