Abstract
YycI and YycH are two membrane-anchored periplasmic proteins that regulate the essential Bacillus subtilis YycG histidine kinase through direct interaction. Here we present the crystal structure of YycI at a 2.9-Å resolution. YycI forms a dimer, and remarkably the structure resembles that of the two C-terminal domains of YycH despite nearly undetectable sequence homology (10%) between the two proteins.
Two-component signaling systems (TCS) are widespread among bacteria, archaea, and lower eukaryotes. A typical bacterium employs 30 to 40 such signaling systems (10) for a wide variety of signal-responsive gene regulons. To date, only a small number of TCS have been shown to be essential for viability. Whereas no essential TCS has been found for Escherichia coli or its close relatives, low-G+C-content gram-positive bacteria express one TCS that is highly conserved and essential (9, 16, 18), generically referred to here as the YycFG system, where YycF is the response regulator and YycG is the histidine kinase (9). Orthologs in other organisms include VicRK in Streptococcus pneumoniae and RR07/HK07 in Enterococcus faecalis (2, 8, 29).
A majority of yyc operons (except in streptococci) include two other genes, yycH and yycI (19, 26). Their protein products share no detectable homology with any other proteins in the databases, as evidenced from a BLAST search. Deletion of the genes coding for either of these proteins in B. subtilis gives identical phenotypes, with a 10-fold upregulation of the YycFG system. Furthermore, YycG, YycH, and YycI have been shown to form a ternary complex. Both YycH and YycI localize to the periplasm and are anchored to the membrane via a single N-terminal transmembrane helix (25, 26). Therefore, YycH and YycI appear to act together to negatively regulate the YycG histidine kinase.
In contrast to the high sequence conservation of the YycG kinase and the YycF response regulator, YycH and YycI orthologs are poorly conserved. We recently solved the crystal structure of YycH, which revealed a three-domain architecture and a novel fold (27) but gave no clues to the function of the protein. Here we present the crystal structure of YycI at a 2.9-Å resolution and discuss insights into its biochemical function derived from the structure.
Cloning, expression, and purification of YycI.
The B. subtilis yycI gene encoding amino acids 31 to 280 was PCR amplified utilizing two oligonucleotides (restriction sites in boldface): yycIf (5′-GTTTTTTCAAAAACATATGGCTACCGGCAAAG-3′) and yycIr (5′-TTGCAAGCGGATCCTTGTCTCACTCACTCC-3′).
The resulting PCR fragment was digested with NdeI and EcoRI restriction enzymes and cloned into the same sites of vector pET28a (Novagen), generating pJS32. This vector expresses the yycI gene devoid of its N-terminal transmembrane helix and translationally fused to an N-terminal thrombin-cleavable His6 tag. Plasmid pJS32 was transformed into BL21(DE3) (Stratagene) for production of native YycI and into the methionine auxotroph B834(DE3) (Stratagene) for production of selenomethionine-labeled YycI.
The overexpression and purification protocol for unlabeled and selenomethionine-labeled YycI was identical to that previously described for YycH (27). Following purification of His-tagged YycI, thrombin was added at 1 U/mg YycI, and the mixture was dialyzed against three changes of buffer B (5 mM Tris-HCl [pH 8], 50 mM NaCl, 20 mM dithiothreitol) over a 14-h period. Thrombin, cleaved His tag, and undigested protein were removed by addition of p-amino-benzamidine agarose (Sigma) and Ni-nitrilotriacetic acid resin, respectively. This procedure yielded more than 15 mg/liter of pure YycI protein.
Crystallization and structure solution of YycI(31-280).
YycI(31-280) was crystallized at 22°C by the hanging drop vapor diffusion method using selenomethionine-labeled protein concentrated to 15 to 30 mg/ml in 5 mM Tris-HCl buffer (pH 8.0) plus 50 mM NaCl and a reservoir solution consisting of 18% polyethylene glycol 1500, 100 mM citrate (pH 3.2), and 200 mM LiSO4. Crystals grew to a size of 0.2 by 0.2 by 0.4 mm3 in 7 to 14 days, and adopted space group P21 with cell dimensions a = 60.8 Å, b = 161.9 Å, c = 180.2 Å, and β = 90.9°. Twenty percent glycerol was used as a cryoprotectant for data collection.
A three-wavelength data set to a 2.9-Å resolution suitable for MAD (multiwavelength anomalous dispersion) phasing was collected at ∼100 K at beamline 12.3.1 of the Advanced Light Source and processed with HKL2000 (20). A heavy-atom search was performed with SHELX-D (28), and the top 72 sites were used as input to autoSHARP (3) for phasing and density modification with SOLOMON (1) and DM (6). Mean figures of merit before and after density modification were 0.42 and 0.82 for reflections between 50 and 2.9 Å. Data reduction and phasing statistics are presented in Table 1.
TABLE 1.
Data reduction and phasing statistics
| Parameter | Result fora:
|
||
|---|---|---|---|
| λ1 (remote) | λ2 (inflection) | λ3 (peak) | |
| Resolution (Å) | 50-2.89 (3.0-2.89) | 50-2.89 (3.0-2.89) | 50-2.89 (3.0-2.89) |
| Wavelength | 0.94645 | 0.97969 | 0.97955 |
| Completeness (%) | 92.7 (53.9) | 88.3 (33.0) | 93.2 (56.7) |
| Average redundancy | 3.8 (2.1) | 3.8 (1.6) | 2.1 (1.4) |
| No. of unique reflections | 71,733 | 68,606 | 72,213 |
| Rmerge | 0.084 (0.334) | 0.095 (0.395) | 0.098 (0.325) |
| I/σ(I) | 12.4 (1.4) | 10.7 (1.1) | 13.4 (1.5) |
| Phasing power | |||
| Anomalous | 0.72 | 1.33 | |
| Isomorphous | 1.34 | 0.66 | |
| Cullis R | |||
| Anomalous | 0.90 | 0.75 | |
| Isomorphous | 0.48 | 0.51 | |
Numbers in parentheses refer to the highest-resolution shell.
Native Patterson maps revealed strong off-origin peaks, ∼15% (2.9-Å data) or 25% (5-Å data) of the origin height, consistent with noncrystallographic translational symmetry elements (0,0,1/3) and (0,0,2/3). The ∼90° β angle further suggested that the monoclinic lattice could be derived by distortion of the orthorhombic lattice observed for the wild-type crystals. We therefore predicted that the asymmetric unit would comprise six noncrystallographic symmetry-related copies of a repeating molecular unit (a dimer, for a total of 12 proteins per asymmetric unit, assuming a solvent content of 51%).
The clearest region of the initial electron density map was selected for manual building using O (14) of one protein subunit, allowing 85% of expected residues to be built as a polyalanine model. Eleven further copies of this model were manually placed into the map, and one round of energy minimization was performed with Refmac5 (17). The arrangement of the 12 polypeptide chains in three rows of four, each generated by noncrystallographic translation and containing two dimers related by a two-fold axis nearly coincident with the y axis, confirmed the prediction from the Patterson analysis.
The amino acid sequence was assigned to the polyalanine models by utilizing the six selenomethionine sites per chain and several clearly visible aromatic side chains as guides. Refinement was carried out using Refmac5 with phase restraints using medium and loose ncs restraints for molecules related by ncs translation or two-fold rotation, respectively, with further model building in O as necessary. Simulated annealing with tight ncs restraints in CNS was performed for the next two cycles of refinement (4). The free set of reflection for cross-validation was created from a pseudo-orthorhombic data set expanded to monoclinic (i.e., h,k,l and −h,k,l).
The final model contains 12 protein chains, each comprising residues from 34 to 276 to 280, with up to 9 missing residues in the loop between strands β20 and β21 and 6 putative Cl− ions, one at each dimer interface. The root mean square deviation (RMSD) for Cα atoms when each subunit was superposed onto a reference molecule (chain A) varied between 0.27 and 1.16 Å. No water molecules were included as none could be found that obeyed the symmetry in more than four ncs-related copies (using ARP/wARP (22). The final R values are Rwork = 0.212 and Rfree = 0.262. The stereochemical quality of the model was excellent, as judged by PROCHECK (15) (Table 2). All but 2 of the 21 outliers in the Ramachandran plot are located in loops with strong but ambiguous electron density, suggesting the presence of alternative main-chain conformations. Surface area calculations were performed in CNS with a probe radius of 1.4 Å. Refinement statistics are summarized in Table 2.
TABLE 2.
Refinement statistics
| Statistic | Result for statistic |
|---|---|
| Rwork | |
| Overall | 21.2 |
| Last shell (limits) | 32.7 (2.97-2.89) |
| Rfree | |
| Overall | 26.2 |
| Last shell (limits) | 38.5 (2.97-2.89) |
| σ cutoff | None |
| Nonhydrogen atoms | |
| Protein | 23,399 |
| Ion | 6 |
| Avg B factors (Å2) | |
| Main chain | 29.1a |
| Side chains | 31.7a |
| Ion | 21.3 |
| Wilson | 63.6 |
| RMSD | |
| Bond lengths (Å) | 0.10 |
| Bond angles (°) | 1.17 |
| Ramachandran plot: no. of residues (%) | |
| Most favored | 2,324 (87.1) |
| Additionally allowed | 323 (12.1) |
| Generously allowed | 15 (0.6) |
| Disallowed | 6 (0.2) |
Average B factors are residual values after calculation of total least-squares parameters.
Overall structure of YycI(31-280).
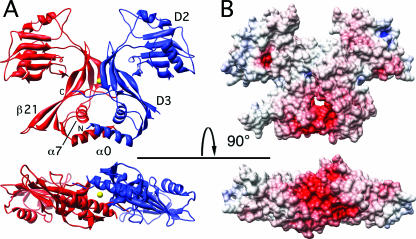
The structure of the periplasmic domain of YycI was determined at a 2.9-Å resolution and is shown in a ribbon representation and as a solvent-accessible surface (Fig. 1A and B). YycI is a mostly β-protein that crystallizes as a dimer, consistent with its behavior in solution as observed by native gel electrophoresis (11; data not shown) and bacterial two-hybrid data indicating a homotypic interaction in vivo (25). Each subunit is comprised of two domains plus an N-terminal α-helix (Fig. 1A).
FIG. 1.
YycI dimer in two orthogonal views. (A) Subunits are shown as red and blue in the ribbon representation, and the chloride ion is shown as a yellow sphere. N and C termini, domains 2 and 3 (D2 and D3), and structural elements mentioned in the text are labeled on one subunit. The β20-β21 hairpin is fully visible in the red subunit (see text). Two orthogonal views related by a horizontal axis are shown, from the side and from the bottom of the molecule along the pseudo-two-fold symmetry axis. Molecular graphics objects were created with Chimera (23). (B) Accessible surface of the YycI dimer in the same two views as panel A, colored according to electrostatic potential on a scale from −20 kT/e (red) to 20 kT/e (blue). The electrostatic potential map was calculated with Spock (5).
YycI shares a common fold with domains 2 and 3 of YycH.
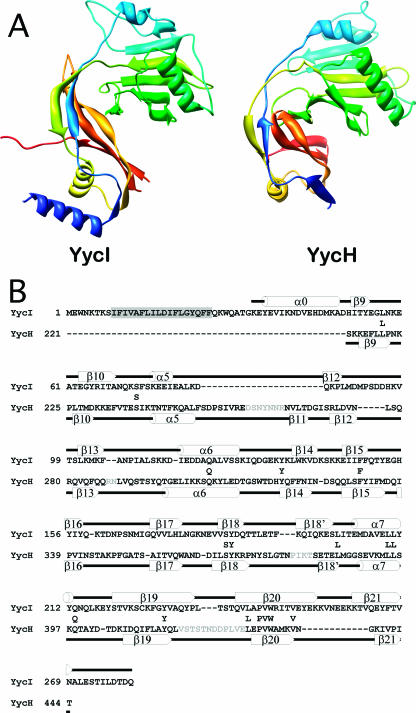
The topology of YycI is nearly identical to that of domains 2 and 3 of YycH, with differences only in the length and positioning of loops, helices, and strands (Fig. 2A). For consistency with YycH, we named the two domains in YycI as domain 2 (amino acids 71 to 190) and domain 3 (amino acids 51 to 70 and 191 to 280) and use the same nomenclature for the homologous secondary structure elements, as shown in Fig. 2B. According to the DALI algorithm (12), the structures can be superposed with an RMSD of 4.6 Å over 177 Cα atoms (or 3.1 Å and 2.7 Å, respectively, when domains 2 and 3 are aligned separately) and with 10% sequence identity.
FIG. 2.
Comparison of a YycI subunit and a YycH monomer. (A) The two structures are shown in the same orientation after structural alignment colored in blue-green-yellow-orange-red so that corresponding structural elements are in similar colors. Only the fragment of YycH that aligns with YycI is shown for clarity. (B) A structure-based sequence alignment of YycI and YycH was generated using the DALI server. For accuracy, individual domains were aligned separately and then assembled into a single alignment. Residues conserved between YycH and YycI are shown between the two sequences. Secondary structure elements for YycI and YycH are shown above and below each sequence, respectively, and were labeled as previously defined for YycH (27). One strand in YycH located between β18 and β19 was not previously labeled and was therefore named β18′. The N-terminal helix in YycI does not have a counterpart in YycH and was hence named α0. The putative transmembrane domain in YycI is highlighted in gray. Sequences in YycH that were not resolved are in gray.
The N-terminal helix (amino acids 34 to 50) of YycI, which packs perpendicular to α7, has no counterpart in YycH and was therefore named α0. Dimerization is mediated by this helix as well as domain 3, including a contribution from an extended C terminus contacting helix α7 and strand β21 in the apposing subunit, burying a total surface area of 3,000 to 3,500 Å2 per dimer. Electron density for the C terminus is visible to a varied extent in each of the 12 chains. The pair of helices made from α0 from each subunit forms the base of the dimer and is part of a central dimerization moiety, while domain 2 points away from the center.
The Fold and Function Assignment System (FFAS) server predicts a common fold for YycH and YycI.
A regular BLAST or iterative Psi-BLAST search was unable to predict any similarity between YycH and YycI; thus, the common protein fold revealed by crystallography was surprising at first. We subsequently used the FFAS server at the Burnham Institute for Medical Research (http://ffas.ljcrf.edu) (13) to further analyze this discrepancy. The algorithm uses profile-profile alignments to take advantage of information present in sequences of homologous proteins to amplify the sequence pattern defining the family (it does not directly use 3-dimensional structural information for the homology search). As a result, it can detect remote homologies beyond the limits of other methods. When the YycH structure was included in the database, the FFAS server correctly predicted the fold of YycI to be homologous to that of YycH, with a confidence level of >97% (Z score = −16.1; sequence identity, 10%).
Strong negative charge is present at the base of the dimer.
The periplasmic domain of YycI has a calculated isoelectric point of 5.5, with a net charge of −6 at neutral pH. Notable positive and negative electrostatic potentials occur on the outer surface at the top of the molecule and the “lower” surface of the N-terminal helix pair, respectively (Fig. 1B). The latter may be relevant to the signaling mechanism, as this surface is predicted to appose the membrane in vivo. A net negative charge is generally, though not universally, observed at this site in orthologs (not shown). In the crystal structure, there are only seven periplasmic residues unresolved between the transmembrane helices and the ordered N termini, which begin at Lys34. Given that the two ordered N termini within the dimer are ∼26 Å apart, it is unlikely that the transmembrane helices can interact within the homodimer without some geometric constraints. This suggests that the periplasmic domain is held rather rigidly vis-à-vis the membrane, either through electrostatic repulsion or through divalent cation-mediated interactions. In either case, we predict that the short periplasmic domain-transmembrane linker severely restricts the translational freedom of the transmembrane domains. That the latter play a role in the interaction between YycI, YycH, and YycG is suggested by the good sequence conservation between orthologs, in contrast to the low conservation of the periplasmic domains. We previously described several conserved hydrophilic residues in the transmembrane helix of YycH, including an SXXX(T/S) motif, that could confer hetero- or homotypic interactions (7, 27). Similarly, conserved hydrophilic residues are found in the YycI and YycG transmembrane helices, despite somewhat lower conservation. Therefore, one function of the periplasmic domain of YycI could be the regulation of the behavior of its transmembrane helix. We further note that the flat shape of the YycI dimer may allow juxtaposition of the transmembrane domains of YycI, YycH, and YycG by minimizing steric clashes between their periplasmic domains and therefore facilitate their interactions.
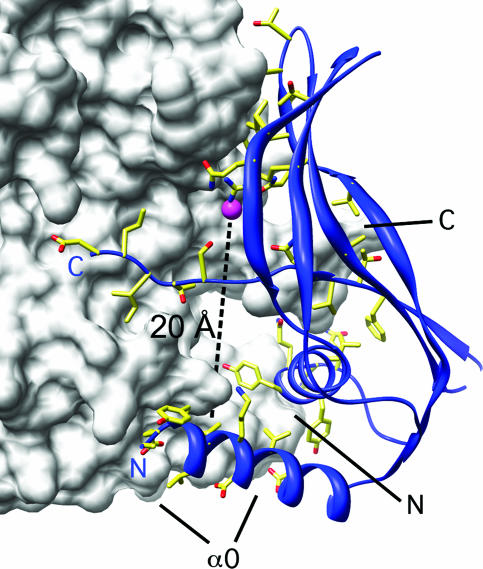
YycI displays a putative ligand binding site.
At the center of the dimer is a deep pocket lined by positively charged residues from domain 2. Strong residual electron density (5.5 to 6.0 σ) at the center of this pocket in the final 2Fo-Fc map (compared with 5.5 to 7.5 σ for the Se atoms) suggests the presence of a bound ion. We assigned the electron density to a chloride ion, based on electron count and the local chemistry, but we cannot be sure of its identity. The pocket brings basic residues from the two monomers into close apposition, suggesting that an anion is necessary at this site to promote dimer formation. From this pocket a narrow cavity extends all the way to the base of the dimer at helix α0, more than 20 Å away from the anion (Fig. 3). The presence of adjoining cavities suggests that the anion partially occupies a ligand-binding site that may serve the purpose of stabilizing or otherwise modulating the structure of the dimeric assembly in vivo. Our structure offers a rational approach through point mutagenesis to test this hypothesis.
FIG. 3.
Putative ligand binding site at the YycI dimeric interface. One subunit is shown as a white surface representation, and domain 3 of the other is shown as a blue ribbon. Side chains lining the dimerization interface are shown. The ion bound at the end of the putative ligand-binding cavity (positioned along the dashed line) is shown as a magenta sphere. N and C termini and α0 are labeled for reference in both subunits.
Conclusions.
YycI interacts with and shares a function with YycH in regulating the activity of the essential histidine kinase YycG (25). Remarkably, YycI also shares a common fold with YycH despite a lack of significant sequence similarity. This suggests that the two proteins were derived by gene duplication but have since diverged extensively. The few invariant residues are either localized to the N-terminal transmembrane helix or were found to serve a structural purpose. We conclude that the most likely function for YycI and YycH is the binding and integration of multiple periplasmic signals in order to modulate the activity of the YycG kinase. Interestingly YycG displays a periplasmic sensing domain predicted by the above-mentioned FFAS server to have a PAS-like fold similar to those found in the periplasmic sensing domains of the fumarate sensor DcuS of E. coli and the citrate sensor CitA of Klebsiella pneumoniae (21, 24). Additionally, YycG has a cytoplasmic PAS domain likely involved in signal sensing (19). Therefore, the ternary complex of YycG, YycH, and YycI displays at least four different ligand input domains potentially able to modulate YycG activity.
Protein structure accession number.
Structure factors and coordinates for YycI have been deposited in the Protein Data Bank (accession no. 2O3O).
Acknowledgments
This study was supported by grant GM019416 from the National Institute of General Medicine Sciences and grant AI055860 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, USPHS, to James A. Hoch and National Institutes of Health grant P01 AI055789 to Robert C. Liddington. The Stein Beneficial Trust supported in part oligonucleotide synthesis and DNA sequencing.
We thank Alexander Aleshin for synchrotron data collection and processing.
Footnotes
Published ahead of print on 16 February 2007.
Manuscript no. 18605 from The Scripps Research Institute.
REFERENCES
- 1.Abrahams, J. P., and A. G. Leslie. 1996. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. Sect. D Biol. Crystallogr. 52:30-42. [DOI] [PubMed] [Google Scholar]
- 2.Bent, C. J., N. W. Isaacs, T. J. Mitchell, and A. Riboldi-Tunnicliffe. 2004. Crystal structure of the response regulator 02 receiver domain, the essential YycF two-component system of Streptococcus pneumoniae in both complexed and native states. J. Bacteriol. 186:2872-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bricogne, G., C. Vonrhein, C. Flensburg, M. Schiltz, and W. Paciorek. 2003. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. Sect. D Biol. Crystallogr. 59:2023-2030. [DOI] [PubMed] [Google Scholar]
- 4.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 5.Christopher, J. A., R. Swanson, and T. O. Baldwin. 1996. Algorithms for finding the axis of a helix: fast rotational and parametric least-squares methods. Comput. Chem. 20:339-345. [DOI] [PubMed] [Google Scholar]
- 6.Cowtan, K., and P. Main. 1998. Miscellaneous algorithms for density modification. Acta Crystallogr. Sect. D Biol. Crystallogr. 54:487-493. [DOI] [PubMed] [Google Scholar]
- 7.Dawson, J. P., J. S. Weinger, and D. M. Engelman. 2002. Motifs of serine and threonine can drive association of transmembrane helices. J. Mol. Biol. 316:799-805. [DOI] [PubMed] [Google Scholar]
- 8.Echenique, J. R., and M.-C. Trombe. 2001. Competence repression under oxygen limitation through the two-component MicAB signal-transducing system in Streptococcus pneumoniae and involvement of the PAS domain of MicB. J. Bacteriol. 183:4599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabret, C., and J. A. Hoch. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J. Bacteriol. 180:6375-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galperin, M. Y. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedrick, J. L., and A. J. Smith. 1968. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch. Biochem. Biophys. 126:155-164. [DOI] [PubMed] [Google Scholar]
- 12.Holm, L., and C. Sander. 1995. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20:478-480. [DOI] [PubMed] [Google Scholar]
- 13.Jaroszewski, L., L. Rychlewski, Z. Li, W. Li, and A. Godzik. 2005. FFAS03: a server for profile-profile sequence alignments. Nucleic Acids Res. 33:W284-W288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjelgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 47:110-119. [DOI] [PubMed] [Google Scholar]
- 15.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK-A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 16.Martin, P. K., T. Li, D. Sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 18.Ng, W. L., G. T. Robertson, K. M. Kazmierczak, J. Zhao, R. Gilmour, and M. E. Winkler. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 50:1647-1663. [DOI] [PubMed] [Google Scholar]
- 19.Ng, W. L., and M. E. Winkler. 2004. Singular structures and operon organizations of essential two-component systems in species of Streptococcus. Microbiology 150:3096-3098. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 21.Pappalardo, L., I. G. Janausch, V. Vijayan, E. Zientz, J. Junker, W. Peti, M. Zweckstetter, G. Unden, and C. Griesinger. 2003. The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli. J. Biol. Chem. 278:39185-39188. [DOI] [PubMed] [Google Scholar]
- 22.Perrakis, A., R. Morris, and V. S. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458-463. [DOI] [PubMed] [Google Scholar]
- 23.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 24.Reinelt, S., E. Hofmann, T. Gerharz, M. Bott, and D. R. Madden. 2003. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 278:39189-39196. [DOI] [PubMed] [Google Scholar]
- 25.Szurmant, H., M. A. Mohan, P. M. Imus, and J. A. Hoch. 2007. YycH and Yycl interact to regulate the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 189:3280-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szurmant, H., K. Nelson, E.-J. Kim, M. Perego, and J. A. Hoch. 2005. YycH regulates the activity of the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 187:5419-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szurmant, H., H. Zhao, M. A. Mohan, J. A. Hoch, and K. I. Varughese. 2006. The crystal structure of YycH involved in the regulation of the essential YycFG two-component system in Bacillus subtilis reveals a novel tertiary structure. Protein Sci. 15:929-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uson, I., and G. M. Sheldrick. 1999. Advances in direct methods for protein crystallography. Curr. Opin. Struct. Biol. 9:643-648. [DOI] [PubMed] [Google Scholar]
- 29.Wagner, C., A. de Saizieu, H.-J. Schönfeld, M. Kamber, R. Lange, C. J. Thompson, and M. G. Page. 2002. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect. Immun. 70:6121-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]