Abstract
Secretins are oligomeric proteins that mediate the export of macromolecules across the bacterial outer membrane. The members of the secretin superfamily possess a C-terminal homology domain that is important for oligomerization and channel formation, while their N-terminal halves are thought to be involved in system-specific interactions. The XcpQ secretin of Pseudomonas spp. is a component of the type II secretion pathway. XcpQ from Pseudomonas alcaligenes is not able to functionally replace the secretin of the closely related species Pseudomonas aeruginosa. By analysis of chimeric XcpQ proteins, a region important for species-specific functioning was mapped between amino acid residues 344 and 478 in the C-terminal homology domain. Two chromosomal suppressor mutations were obtained that resulted in the proper functioning in P. aeruginosa of P. alcaligenes XcpQ and inactive hybrids. These mutations caused a defect in the synthesis of the lipopolysaccharide (LPS) outer core region. Subsequent analysis of different LPS mutants showed that changes in the outer core and not the loss of O antigen caused the suppressor phenotype. High concentrations of divalent cations in the growth medium also allowed P. alcaligenes XcpQ and inactive hybrids to function properly in P. aeruginosa. Since divalent cations are known to affect the structure of LPS, this observation supports the hypothesis that LPS has a role in the functioning of secretins.
Pseudomonas aeruginosa is able to secrete proteins into the extracellular medium via at least five different pathways (16). Most of the exoproteins are secreted via the type II secretion system. This pathway, which is widely distributed among the Proteobacteria, involves two separate translocation steps (8, 26). First, the secreted proteins are directed to the Sec translocon in the cytoplasmic membrane by the presence of a signal sequence. Upon translocation across the inner membrane, the signal sequence is removed, and the mature protein is released in the periplasm. In this compartment, the exoproteins acquire a folded conformation with the help of specific chaperones and of general folding catalysts, before they are translocated across the outer membrane. Some type II substrates use the Tat system rather than the Sec system to cross the inner membrane (46), and in these cases folding may occur in the cytoplasm. The second step, transport of the folded exoproteins across the outer membrane, is mediated by the type II secretion machinery, which in P. aeruginosa consists of at least 12 different proteins, designated XcpA and XcpP to XcpZ (15, 16).
Since the Xcp secretion machinery transports folded exoproteins across the outer membrane, the secretion channel has to be large. Consistently, the only outer membrane component of the machinery, XcpQ, has been shown to form large channels (6). XcpQ belongs to a family of proteins generically designated the secretins. This family also includes members that are involved in type III protein secretion, in the formation of type IV pili, in the production of filamentous phages, and in the uptake of DNA (20). Therefore, secretins constitute a very important group of macromolecule transporters in the outer membrane of gram-negative bacteria. Members of this family have been shown by electron microscopy to form large, ring-shaped, oligomeric complexes with a central cavity, which could represent the protein-conducting channel (2). The homology between members of the family is in their C-terminal halves (20). Therefore, this part of the secretins has been implicated in the formation of the ring-shaped structure and the transport channel, which was confirmed by limited proteolysis of XcpQ (6) and PulD of Klebsiella oxytoca (34). The N-terminal domains exhibit homology only in secretins that are involved in similar transport processes and could therefore play a role in the interaction with other components of the transport system or with the substrates. Studies performed with chimeric secretins have supported this notion, since they showed that the N termini of OutD of Erwinia chrysanthemi (3) and of the phage-encoded pIV secretin (10) are important for substrate specificity. Moreover, the EpsD secretin of Vibrio cholerae was shown to interact with the XcpP homolog EpsC via the N-terminal domain (27).
Pseudomonas alcaligenes is closely related to P. aeruginosa and also contains a complete xcp gene cluster, the products of which are involved in secretion of a lipase (21). Generally, type II secretion systems are highly specific, and secretion of heterologous exoproteins is rarely observed, even when they are expressed in closely related species. However, P. aeruginosa is able to secrete exoproteins of P. alcaligenes and vice versa (11), which indicates that the two secretion systems are closely related. Although most xcp genes of these two species can be functionally exchanged, expression of the xcpQ gene from P. alcaligenes does not efficiently complement the secretion defect of an xcpQ mutant of P. aeruginosa (11). In this study, we constructed chimeric XcpQ proteins to identify the domain that is involved in species-specific functioning of the XcpQ protein. Furthermore, to identify host factors important for the lack of exchangeability, we isolated and characterized suppressor mutations that allowed proper functioning of P. alcaligenes XcpQ in P. aeruginosa.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and plasmids.
Escherichia coli strains DH5α and MC1061 were obtained from The Netherlands Culture Collection of Bacteria (NCCB). DH5α was used for general cloning purposes, and MC1061 was used for isolation of chimeric xcpQ genes. The Pseudomonas strains used in this study are listed in Table 1. The P. aeruginosa and E. coli strains were grown at 37°C in a modified Luria-Bertani (LB) broth (43). P. alcaligenes was grown as previously described (21). Plasmids used in this study are listed in Table 1 and Fig. 1. Genes derived from P. aeruginosa and P. alcaligenes are indicated by the subscripts aer and alc, respectively, in the gene designations. The P. aeruginosa-infecting bacteriophages F116L and E79 were obtained from the NCCB and Bengt Wretlind, respectively. Bacteriophage PAF3 was isolated from sewage water obtained at the wastewater purification system in Houten, The Netherlands. Phage-resistant mutants were isolated by plating 0.1 ml of an overnight culture together with 0.1 ml of a phage stock solution (1010 to 1011 PFU/ml) in a 5-ml agar top layer. Subsequently, the plates were grown for 30 h at 37°C, potential phage-resistant mutants were streaked twice on fresh LB medium plates, and the lipopolysaccharide (LPS) patterns were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Transduction experiments were performed with phage F116L. Chromosomal xcpQ mutations were obtained by using the suicide plasmid pKNWQ1, which contains part of the xcpQ gene and a gentamicin resistance cassette, as described previously (1). Plasmids were introduced by the CaCl2 procedure into E. coli (38) and by triparental mating into Pseudomonas strains using the conjugative helper plasmid pRK2013. For plasmid maintenance or selection, the following antibiotics were used: for E. coli, ampicillin (50 μg/ml), kanamycin (25 μg/ml), tetracycline (15 μg/ml), and gentamicin (15 μg/ml); and for P. aeruginosa, piperacillin (40 μg/ml), kanamycin (50 μg/ml), tetracycline (80 μg/ml), and gentamicin (40 μg/ml). Sensitivity to antibiotics and EDTA was determined as described previously (45).
TABLE 1.
Pseudomonas strains and plasmids used
| Strain or plasmida | Relevant characteristicsb | Reference or source |
|---|---|---|
| P. aeruginosa strains | ||
| PAO25 | leu arg | 22 |
| PAN1 | PAO25 xcpQ::Gm | 1 |
| PAN15 | PAN1, suppressor, LPS core deficient | This study |
| PAN16 | PAN1, suppressor, LPS core deficient | This study |
| AK1012 | PAO1, LPS core deficient | 24 |
| AK1401 | PAO1, LPS B-band deficient | 24 |
| PAN38 | AK1012 xcpQ::Gm | This study |
| PAN39 | AK1401 xcpQ::Gm | This study |
| P. alcaligenes 93A | 21 | |
| Plasmids | ||
| pUC18 | Ampr, ColE1 | NCCB |
| pMMB67EH | Ampr, IncQ | 18 |
| pUR6500HE | pMMB67HE derivative, Kmr | 17 |
| pVDZ′2 | Tetr, IncP | 12 |
| pRK2013 | Kmr Tra+ Mob+ | 14 |
| pLAF600SB | xcpPQalc in PLAFR3 | 21 |
| pB28 | xcpQaer in pMMB67EH | 1 |
| pCK25 | xcpQaer in pUR6500EH | This study |
| pCK27 | xcpPalc in pMMB67EH | This study |
| pCK28 | xcpQalc in pMMB67EH | 1 |
| PCK29 | xcpPalc in pVDZ′2 | This study |
| pCK31 | xcpPQalc in pMMB67EH | This study |
| pCK32 | xcpQaer and xcpQalc in pMMB67EH | This study |
| pUWL6 | xcpQaer in pUC19 | 1 |
| pUWL15 | xcpQF497 in pUC18 | This study |
| pUWL20 | xcpQH13 in pUC18 | This study |
| pKNWQ1 | xcpQaer::Gm in suicide vector pKNG101, Smr | 1 |
P. aeruginosa strains PAN15, PAN16, PAN38, and PAN39 have been deposited in NCCB and are available as strains PC4313, PC4314, PC4319, and PC4320, respectively.
Gm, gentamicin; Amp, ampicillin; Km, kanamycin; Tet, tetracycline; Sm, streptomycin.
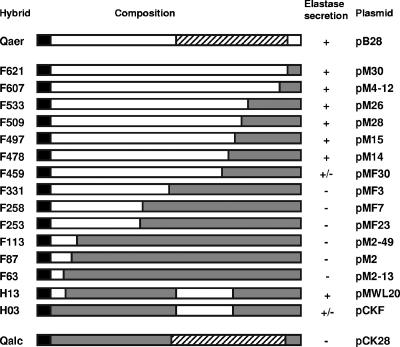
FIG. 1.
Schematic diagrams of the different (chimeric) XcpQ proteins. Open bars, XcpQaer; shaded bars, XcpQalc; solid bars, signal sequences; cross-hatched bars, C-terminal domain that is conserved in the various members of the secretin family. The designations of the proteins are indicated on the left; in each case the number refers to the position of the amino acid residue at the fusion site (the numbering includes the signal sequence). The corresponding plasmids are indicated on the right. The functionality of XcpQ was based on the ability of the xcpQ mutant PAN1 supplemented with the different xcpQ genes to form haloes on elastin-containing plates after 24 h of growth. +, formation of haloes whose sizes were similar to the sizes of haloes formed by the wild-type strain; +/−, formation of smaller haloes; −, no halo formation.
Isolation and characterization of chimeric xcpQ genes.
Plasmid isolation and DNA manipulations were performed using standard procedures (38). Plasmid pCK27 was constructed by cloning the EcoRI/SalI fragment of pLAF600SB containing the xcpPalc gene in the EcoRI/SalI-digested cloning vector pMMB67EH. The xcpPalc gene was subsequently recloned into the vector pVDZ′2 using the EcoRI/HindIII sites, resulting in plasmid pCK29. Plasmid pCK31, carrying the xcpPQalc genes, was obtained by cloning the EcoRI/SphI fragment of pCK27 and the EcoRI/HindIII fragment of pCK28 in SphI/HindIII-digested pMMB67EH by three-point ligation. The xcpQaer gene was obtained by digesting pUWL6 with HindIII/SacI and cloning of the fragment with xcpQaer in the same sites of the pUR6500HE vector, resulting in plasmid pCK25.
An in vivo recombination strategy (42) was used to construct hybrid genes. The xcpQaer and xcpQalc genes were cloned in tandem in the pMMB67EH vector with a unique restriction site between the two genes. First, the vector was digested with EcoRI/BamHI and religated after the overhanging ends were filled in, which resulted in a cloning vector lacking most of the multiple cloning site. The xcpQalc gene was obtained from plasmid pCK28 with KpnI/HindIII, and the xcpQaer gene was obtained from plasmid pCK25 using PstI and KpnI. The two genes were introduced by three-point ligation into the altered pMMB67EH vector digested with PstI and HindIII. The resulting plasmid, pCK32, contained the two genes in tandem with a unique KpnI site between them. To obtain xcpQ chimeras in vivo, plasmid pCK32 was linearized with KpnI. After inactivation of the restriction enzyme, the overhanging ends were made blunt with T4 DNA polymerase. The linear plasmid was used for electroporation of E. coli strain MC1061. Alternatively, pCK32 was linearized with SmaI and used for electroporation. Linear plasmids cannot be replicated in vivo, but the linear plasmids can become circular by recombination at homologous sites in the plasmids, resulting in the desired chimeric genes (42). The plasmids were isolated from transformants and subjected to restriction enzyme analysis to identify clones in which a recombination event had occurred. The nucleotide sequences of the relevant fragments subsequently showed the exact fusion sites. The chimeras obtained are shown schematically in Fig. 1.
The chimeric xcpQH13 gene was created by a modified version of the QuikChange mutagenesis procedure (Stratagene). First, xcpQF497 was subcloned in pUC18, resulting in pUWL15. Subsequently, an internal fragment of xcpQalc was amplified by PCR, using primers WILQF1 (5′-TCCGCGAATTCATCGACCAG-3′) and WILQF3 (5′-CAGCAGGACCAGGGCATTG-3′), both of which were designed to hybridize with xcpQaer as well as with xcpQalc. The resulting PCR product was extracted with phenol and purified from an agarose gel by using Jetsorb (Genomed). Subsequently, this material was used as a megaprimer in a QuikChange mutagenesis protocol with pUWL15 as the template. This protocol was performed with Pwo polymerase (Boehringer) using 18 10-min amplification steps. The final PCR product was treated with DpnI and used to transform DH5α cells. The clone that was isolated, which contained xcpQH13, was subjected to restriction enzyme analysis and DNA sequencing and was designated pUWL20. Subsequently, the xcpQH13 gene was introduced into pMMB67EH by using restriction enzymes XbaI and HindIII, resulting in plasmid pMWL20. This plasmid was used to construct pCKF (carrying xcpQH03) by replacing the SmaI/HindIII fragment, encoding the N-terminal domain of xcpQH13, with the corresponding fragment of pCK28 (xcpQalc).
Isolation of suppressor mutations.
Suppressor mutants were selected on the basis of their ability to secrete the staphylolytic enzyme LasA. Cells of strain PAN1 carrying plasmid pM2 (Fig. 1) were treated for 90 min with ethyl methanesulfonate. Subsequently, the bacteria were washed with 5% NaS2O3 and 0.9% NaCl, incubated in LB medium for 2 h at 37°C, and plated on selection plates. These plates contained 25 ml of 10-fold-diluted LB medium in M9 medium (without an additional carbon source), piperacillin, 1.5% agar, and heat-killed Staphylococcus aureus cells from a 10-ml overnight culture, which were pelleted and washed. After overnight growth, colonies of LasA-secreting bacteria could be identified by their larger sizes and by the small clear halo surrounding each colony. Putative suppressor mutants were subsequently analyzed for the secretion of elastase on elastin plates as described previously (5).
SDS-PAGE and immunoblotting.
Proteins were separated by SDS-PAGE (29), with 0.2% SDS in the running gel. The proteins were stained in the gel with Coomassie brilliant blue, or they were transferred onto nitrocellulose membranes by semidry electroblotting. To improve immunodetection, the oligomeric XcpQ complexes were denatured in situ in the gel prior to the blotting procedure. To do this, the gels were incubated for 30 min in 10% trichloroacetic acid (TCA) and washed four times in distilled water and then in SDS-PAGE electrode buffer until the pH of the electrode buffer was reached. After blotting, the different XcpQ variants were visualized by immunodetection with antibodies directed against an N-terminal fragment of XcpQ (amino acid residues 12 to 100 of the mature protein) (1). Peroxidase activity on blots was detected by staining with 4-chloro-1-naphthol and H2O2, unless indicated otherwise.
Cell fractionation.
Secreted proteins were isolated by precipitation from the supernatant fractions of cell cultures with 5% TCA. Cell envelopes were harvested by centrifugation (20 min, 4°C, 20,000 × g) after ultrasonic disruption of P. aeruginosa cells in the presence of 1% NP-40 (Igepal CA-630) (32) and the Complete protease inhibitor cocktail (Boehringer Mannheim) to keep endogenous elastase inactive. For separation of inner and outer membranes, cells grown overnight in LB medium were pelleted by centrifugation and resuspended in a solution containing 0.25 M sucrose, 0.5 mM EDTA, 100 mM Tris-HCl (pH 8), and 60 μg/ml lysozyme (Merck). The mixture was incubated for 1 h at room temperature. After addition of dithiothreitol (DTT) and phenylmethylsulfonyl fluoride, both to a final concentration of 2 mM, the spheroplasts were collected by centrifugation for 75 min at 150,000 × g at 4°C, resuspended in H2O with 2 mM DTT and 1 mM phenylmethylsulfonyl fluoride, and disrupted by 20 s of ultrasonication. The membranes were pelleted by centrifugation for 75 min at 150,000 × g at 4°C, resuspended in 5 mM EDTA (pH 7.5)-2 mM DTT, and layered on a discontinuous sucrose gradient containing 30 to 55% (wt/wt) sucrose in 5 mM EDTA (pH 7.5)-2 mM DTT. The gradients were centrifuged at 35,000 rpm in an SW41 rotor (Beckman) for 20 h at 4°C and fractionated. NADH oxidase activity was determined as described previously (35). The presence of OprF in the different fractions was evaluated by SDS-PAGE.
Purification and analysis of LPS.
Routinely, LPS patterns in proteinase K-treated cell envelopes were analyzed by SDS-PAGE on 16% acrylamide gels. After separation, LPS was visualized in the gels by silver staining (44). For detailed analysis of the LPS composition, bacteria were grown in 10-liter cultures. Cell pellets were collected and washed with 0.9% NaCl and then with distilled water prior to lyophilization. The lyophilized cells were washed overnight with ethanol and then twice with acetone and finally with diethyl ether. Subsequently, the extracted cell debris was dried overnight in air. The dried material (10 to 15 g) was resuspended in 250 ml distilled water supplemented with DNase (10 mg) and RNase (10 mg) and stirred overnight at room temperature. Subsequently, proteinase K (10 mg) was added, and this was followed by incubation for 16 h at room temperature. The suspension was dialyzed against distilled water for 5 days at 4°C, and the insoluble material was collected by centrifugation, washed twice with acetone (250 ml), and dried in air. P. alcaligenes LPS was extracted with 45% hot phenol (47), and P. aeruginosa LPS was isolated by a modified phenol-chloroform-petroleum ether extraction method (19).
Chemical analysis of LPS.
Gas-liquid chromatography (GLC) was performed with a Hewlett-Packard model 5890 Series II instrument equipped with a 30-m SPB-5 capillary column (Supelco, Germany) using a temperature gradient which increased from 150 to 320°C at a rate of 5°C min−1. Monosaccharides were analyzed by GLC as the alditol acetates after hydrolysis of LPS with 0.1 M HCl for 48 h at 100°C for neutral sugars (39) or with 10 M HCl for 30 min at 80°C for amino sugars (13). Following hydrolysis, sugars were dried by evaporation in a vacuum, reduced with NaBH4, and acetylated with acetic anhydride in pyridine (30 min, 85°C). Fatty acids were analyzed by GLC as their methyl esters, liberated after strong methanolysis (2 M HCl-methanol, 120°C, 24 h) and extraction with chloroform. Total hexosamine contents were determined after acid hydrolysis (4 M HCl, 16 h, 100°C) using the Morgan-Elson reaction as described previously (41). Amino acids and amino sugars were identified as their phenylisothiocyanate derivatives by reversed-phase high-performance liquid chromatography using a Waters PICO-TAG system (Waters, Eschborn, Germany) (7, 9). 3-Deoxy-d-manno-octulosonic acid contents were determined by the thiobarbituric acid assay using the modified method (4). Total phosphate was quantified by the ascorbic acid method (31).
RESULTS
Construction and functionality of chimeric XcpQ secretins.
Although P. alcaligenes XcpQ (XcpQalc) is highly homologous to its counterpart in P. aeruginosa, exhibiting 69% identity (21), expression of xcpQalc in an xcpQ mutant of P. aeruginosa does not restore protein secretion via the type II secretion pathway (11). In order to identify the amino acid residues responsible for the species-specific functioning, hybrids between xcpQalc and xcpQaer were constructed. Since we expected that the more variable N-terminal domain would be involved in species specificity, we constructed chimeric genes that encoded different N-terminal fragments of XcpQaer and the complementing C-terminal domain of XcpQalc. The hybrid genes were obtained by in vivo recombination (see Materials and Methods), and the fusion sites were localized by restriction enzyme mapping and nucleotide sequencing (Fig. 1). The numbers in the designations of the hybrid proteins indicate the positions in the amino acid sequence of the XcpQ precursor where the fusions occurred. All the xcpQ genes were under control of the tac promoter.
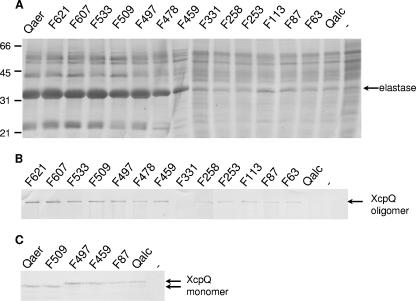
Subsequently, the constructs containing the hybrid genes were mobilized into P. aeruginosa strain PAN1, in which the chromosomal xcpQ gene is inactivated by insertion of a gentamicin resistance gene, and they were tested to determine the abilities to complement the secretion defect of this strain. Secretion of the type II substrate elastase was examined on elastin-containing plates, on which secretion-proficient strains formed haloes around the colonies (Fig. 1), and by SDS-PAGE analysis of the extracellular protein profiles, in which elastase was the most prominent band (Fig. 2A). The hybrids containing an N-terminal pre-XcpQaer fragment consisting of 478 or more amino acid residues were able to restore the secretion of elastase very efficiently. The XcpQF459 hybrid exhibited partial complementation, whereas all the fusions with an N-terminal pre-XcpQaer fragment consisting of less than 331 amino acid residues were not functional. Thus, the protein segment between residues 331 and 478 of XcpQaer appears to contain an important determinant for species-specific functioning.
FIG. 2.
Functionality and complex formation for the different XcpQ secretins. (A) Extracellular protein profiles of P. aeruginosa strain PAN1 with constructs encoding the different XcpQ variants or with the empty vector (lane −). Secreted proteins were precipitated with 5% TCA and separated by SDS-PAGE. The position of the major protein secreted by the type II pathway, elastase, is indicated on the right, and the positions of the molecular mass standard proteins (in kDa) are indicated on the left. (B and C) Cell envelopes (B) or whole-cell preparations (C) of strain PAN1 with constructs encoding different XcpQ variants or with the empty vector (lane −) were subjected to SDS-PAGE. Proteins were transferred to nitrocellulose membranes and visualized by immunodetection with polyclonal antibodies directed against XcpQ. The positions of the XcpQ oligomers and monomers are indicated on the right.
To investigate whether additional specificity determinants are present in the N-terminal domain of XcpQ, we constructed two additional chimeric genes, encoding the H13 and H03 hybrids (Fig. 1), in vitro. The resulting hybrids contained the same C-terminal segment of XcpQalc that was present in hybrid F497 and another XcpQalc segment extending from either amino acid residue 1 (in H03) or residue 68 (in H13) up to residue 344. Both genes were able to complement the secretion defect of strain PAN1 (Fig. 1), although the gene encoding H03 did not restore secretion completely. We concluded that the region between amino acid residues 344 and 478 plays an important role in preventing XcpQalc from functioning in P. aeruginosa. All of this region is located in the C-terminal secretin homology domain, which encompasses amino acid residues 326 to 606 of the XcpQaer precursor.
Characteristics of the oligomers formed by the chimeric XcpQ secretins.
To study the production of the chimeric proteins and their abilities to form oligomers, cell envelope preparations were analyzed by SDS-PAGE, followed by immunoblotting. Most of the chimeric proteins were detected as high-molecular-weight oligomeric complexes; the only exception was the hybrid protein XcpQF331, which did not seem to form any oligomers (Fig. 2B). However, the amounts of oligomeric complexes formed by the nonfunctional fusions were smaller than the amounts formed by the functional fusions; in particular, the complexes formed by XcpQalc were hardly visible after immunodetection (Fig. 2B). Analysis of whole-cell preparations revealed that a small proportion of XcpQ was present in the monomeric form. Interestingly, the amounts of monomers were the same for all the XcpQ variants, as shown in Fig. 2C for a number of representative proteins, indicating that a constant pool of stable XcpQ monomers was present in the cells, which may have represented assembly intermediates.
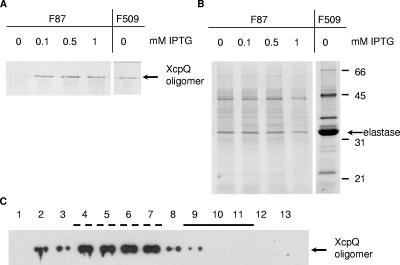
To determine whether there is a direct correlation between functionality and the amount of oligomer, PAN1 producing the inactive fusion protein XcpQF87 from pM2 was grown in the presence of various concentrations of isopropyl-β-d-thiogalactopyranoside (IPTG). Addition of IPTG to the growth medium resulted in an increase in the amount of the XcpQ complex (Fig. 3A); however, this did not lead to restoration of elastase secretion (Fig. 3B). In contrast, addition of IPTG was not required to obtain type II-mediated secretion in the case of the active fusions (shown for fusion protein XcpQF509 in Fig. 3A and B). These results show that the failure of the hybrids to function was not correlated with a quantifiable defect in oligomer formation.
FIG. 3.
Characterization of the nonfunctional hybrid protein XcpQF87. The xcpQ mutant PAN1 with pM2 encoding the nonfunctional hybrid protein XcpQF87 was grown to an optical density at 660 nm of 2.3, after which IPTG was added at the concentrations indicated above the lanes, and growth was continued for 2 h. PAN1(pM28) producing the active fusion protein XcpQF509 was grown without IPTG. To determine the production of XcpQ oligomers, cell envelopes were isolated and subjected to SDS-PAGE and Western blotting with polyclonal antibodies directed against XcpQ (A). To determine the functionality of XcpQ hybrids, secreted proteins were precipitated with 5% TCA and separated by SDS-PAGE (B). The position of the major protein secreted via the type II pathway, elastase, and the positions of the molecular mass standard proteins (in kDa) are indicated on the right. (C) Cellular localization of XcpQF87. A cell envelope preparation of P. aeruginosa PAN1(pM2) expressing XcpQF87 was applied to a 30 to 55% sucrose gradient, which was centrifuged and fractionated. Samples from all fractions were analyzed by SDS-PAGE, and proteins were transferred to a nitrocellulose membrane and visualized by immunodetection using antibodies against XcpQ and enhanced chemiluminescence for detection. The fractions that represented the inner membrane (solid line) and outer membrane (dashed line) were identified based on NADH oxidase activity and the presence of OprF, respectively. The position of the XcpQ oligomers is indicated on the right.
The inability of the inactive fusions to function in xcpQ mutant strain PAN1 could have been due to incorrect targeting to the outer membrane. To examine this possibility, the subcellular localizations of the oligomers of the functional hybrid protein XcpQF509 and the nonfunctional protein XcpQF87 were determined. Neither XcpQ protein could be detected in the soluble fraction (data not shown). The membrane fractions were analyzed further by sucrose gradient centrifugation to separate the inner and outer membranes. As expected, the functional XcpQF509 oligomeric complexes were found in the fractions containing the outer membrane proteins (data not shown). However, the oligomers of the inactive hybrid XcpQF87 protein also fractionated with the outer membranes (Fig. 3C). These results show that the oligomers formed by this chimeric protein were targeted to the outer membrane.
Isolation of suppressor mutations.
The malfunctioning of the XcpQalc protein in P. aeruginosa is probably due to aberrant interactions with components of the heterologous host. We first examined whether XcpP might be such a component, since several reports have indicated that XcpP and its homologs in other bacterial species interact with their cognate secretins (26, 27). Therefore, we introduced a plasmid, pCK29, containing P. alcaligenes xcpP into strain PAN1 producing in trans one of the various inactive XcpQ hybrids and examined the protein secretion profiles by SDS-PAGE. However, no restoration of protein secretion was observed (results not shown).
As an alternative approach, we isolated suppressor mutations in strain PAN1 that allowed functioning of an inactive XcpQ hybrid protein. The selection procedure was based on the ability of the type II-secreted LasA protein, also known as staphylolytic enzyme, to lyse S. aureus cells. LasA-secreting cells exhibit increased growth on plates containing S. aureus cells and clearing zones around the colonies. Cells of strain PAN1 containing plasmid pM2, which encodes the inactive hybrid protein XcpQF87, were mutagenized with ethyl methanesulfonate and plated on plates containing S. aureus cells. Twelve fast-growing colonies were analyzed on elastin-containing plates. Two of these colonies exhibited extracellular elastase activity, and they were subsequently cured of pM2. The cured strains, designated PAN15 and PAN16, were secretion defective, whereas reintroduction of pM2 into both mutants, but not reintroduction of pM2 into PAN1, resulted in elastase secretion, as revealed by halo formation on elastin plates (results not shown) and by SDS-PAGE analysis of the extracellular protein profiles (shown for PAN16 in Fig. 4A). These results show that the suppressor mutations were located on the chromosome.
FIG. 4.
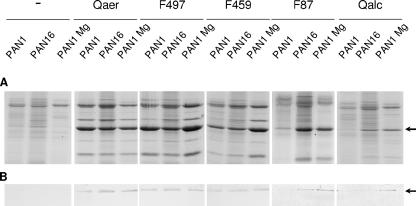
Effects of the suppressor mutation and divalent cations on the functionality and production of various XcpQ proteins. P. aeruginosa strains PAN1 and PAN16 containing the empty vector (−) or xcpQaer, xcpQF497, xcpQF459, xcpQF87, or xcpQalc on plasmids were grown in normal LB medium or LB medium supplemented with 5 mM MgCl2 (Mg). Extracellular proteins were precipitated with 5% TCA and separated by SDS-PAGE (A). The position of the major protein secreted via the type II pathway, elastase, is indicated by an arrow. (B) Amounts of XcpQ oligomers produced by the strains. Cell envelopes were subjected to SDS-PAGE followed by Western blotting with polyclonal antibodies directed against XcpQ. The position of the XcpQ oligomers is indicated by an arrow.
All the constructs containing different xcpQ genes were introduced into PAN16, and the extracellular protein patterns of the transconjugants were analyzed. Most of the fusions that were inactive in PAN1 were able to restore, at least to some extent, protein secretion in the suppressor mutant PAN16 (shown for XcpQF87 in Fig. 4A); the only exception was XcpQF331, which, like the results obtained for PAN1 (Fig. 2B), did not form any detectable oligomers in this strain (data not shown). Even the wild-type XcpQalc protein expressed from plasmid pCK28 was functional in PAN16 (Fig. 4A), although the efficiency of secretion was relatively low and the XcpQalc oligomers formed were hardly visible (Fig. 4B). The plasmids encoding the intact XcpQaer protein, the functional XcpQF497 fusion protein, or the partially functional XcpQF459 fusion protein restored secretion in the suppressor mutant to levels similar to those in the PAN1 strain (Fig. 4A). In contrast, introduction of the empty vector pMMB67HE into strain PAN1 or into the suppressor strain PAN16 did not result in secretion of elastase and other type II-dependent exoproteins (Fig. 4A).
Analysis of the cell envelopes showed that the yield of oligomers formed by XcpQalc or XcpQF87 in the suppressor strain was slightly greater than the yield in PAN1, whereas the amounts of XcpQaer, XcpQFF497, and XcpQF459 oligomers were the same in both strains (Fig. 4B).
LPS biosynthesis is affected in suppressor strains PAN15 and PAN16.
To identify the chromosomal mutations responsible for the suppressor phenotype, the obvious candidates (i.e., the genes encoding components of the type II secretion machinery) were evaluated first. Since the xcpP and xcpRSTUVWXYZ genes are adjacent to xcpQ on the chromosome, transduction experiments were performed. However, the suppressor phenotype could not be cotransduced with the xcpQ::Gm allele, which indicated that the suppressor mutation is not located in the xcp gene cluster.
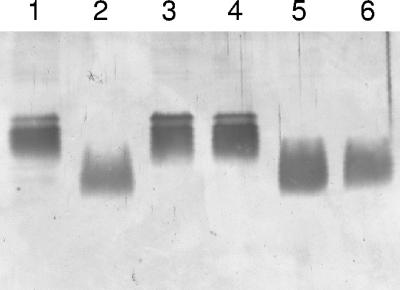
Analysis of the cell envelope protein profiles of PAN15 and PAN16 by SDS-PAGE revealed no significant differences compared with the profile of PAN1 (data not shown). Also, the growth characteristics of the mutants were not altered. In addition, the sensitivities of the two suppressor strains to various antibiotics (ampicillin, polymyxin B, and kanamycin) and EDTA did not differ from the sensitivities of PAN1, which indicates that the integrity of the outer membrane was not grossly affected. Subsequently, the LPS patterns of the different strains were analyzed by SDS-PAGE, using the well-characterized LPS mutants AK1012, which produces LPS molecules consisting of the lipid A part and the inner core domain, and AK1401, which is deficient only in the synthesis of the B-band O antigen, as references (Fig. 5, lanes 2 and 3, respectively). Both wild-type strain PAO1 and the original xcpQ mutant PAN1 produced intact lipid A-core molecules (Fig. 5, lanes 1 and 4, respectively) and O-antigen subunits (results not shown). In contrast, LPS isolated from strains PAN15 and PAN16 clearly exhibited increased mobility on SDS-PAGE gels (Fig. 5, lanes 5 and 6), similar to the mobility observed for the LPS of strain AK1012, which indicated that there was a defect in the synthesis of the LPS core region. Consistently, PAN15 and PAN16 were resistant to the P. aeruginosa-infecting phage E79, which uses LPS as its receptor (25).
FIG. 5.
LPS patterns of P. aeruginosa strains. Partially purified LPS of P. aeruginosa PAO1 (wild type) (lane 1), AK1012 (core mutant) (lane 2), AK1401 (B-antigen mutant) (lane 3), PAN1 (lane 4), and suppressor strains PAN15 (lane 5) and PAN16 (lane 6) were separated by SDS-PAGE and visualized by silver staining. LPS molecules containing multiple repeating units of the O antigens were not detected due to the small amounts of LPS loaded.
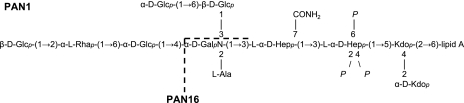
To obtain more insight into the LPS structure required for XcpQalc to function, the LPS of P. aeruginosa strains PAN1 and PAN16 were isolated, and their chemical compositions were determined (Table 2). Differences between PAN1 and the suppressor mutant were found in the content of glucose and rhamnose, which are components of the outer core. The composition of PAN16 LPS is consistent with the proposed LPS structure of the outer core mutant AK1012 (37) and is shown in Fig. 6. The chemical composition of P. alcaligenes LPS was also determined (Table 2). Interestingly, the LPS of this bacterium is characterized by a lower glucose content. Since glucose residues are found in the outer core and are not present in the suppressor mutant, their presence in wild-type P. aeruginosa LPS could be responsible for the malfunctioning of XcpQalc.
TABLE 2.
Composition of LPS isolated from P. aeruginosa PAN1 and PAN16 and P. alcaligenes 93A
| Componenta | Concn (nmol/mg) in:
|
||
|---|---|---|---|
| Strain PAN1 | Strain PAN16 | Strain 93A | |
| Total HexN | 631 | 663 | 445 |
| Total GlcN | 427 | 553 | 321 |
| GalN | 225 | 290 | 161 |
| Ala | 199 | 278 | 214 |
| Kdo | 231 | 303 | 237 |
| P | 1,612 | 2,176 | 1,261 |
| Glc | 703 | 117 | 345 |
| Hep | 118 | 195 | 109 |
| Rha | 527 | 90 | 1438 |
| 10:0(3-OH) | 161 | 239 | 170 |
| 12:0 | 115 | 159 | 247 |
| 12:0(2-OH) | 200 | 292 | |
| 12:0(3-OH) | 265 | 380 | 241 |
| 16:0 | 31 | 39 | |
HexN, hexosamine; GlcN, glucosamine; GalN, galactosamine; Kdo, 3-deoxy-d-manno-octulosonic acid; Glc, glucose; Hep, l-glycero-d-manno-heptose; Rha, rhamnose.
FIG. 6.
Structural model of the LPS alteration in suppressor mutant PAN16. The structures of PAN1 and PAN16 LPS are based on data obtained previously (37) and data reported here. Kdo, 3-deoxy-d-manno-octulosonic acid.
Functionality of XcpQalc in P. aeruginosa is dependent on LPS core mutations or the presence of divalent cations.
To verify that the suppressor phenotype of strains PAN15 and PAN16 correlates with the defect in the synthesis of the LPS core region, xcpQ deletion mutant derivatives of the LPS mutants AK1012 and AK1401 were constructed. As expected, the resulting strains, designated PAN38 and PAN39, respectively, were secretion deficient (Table 3). Subsequently, plasmids carrying different xcpQ genes were introduced into PAN38 and PAN39, and the secretion profiles were evaluated. Introduction of pB28 encoding XcpQaer restored elastase secretion in both strains, although the levels of secretion were slightly lower than those of strain PAO25 (Table 3). Introduction of plasmids encoding either XcpQalc or the inactive XcpQF87 hybrid protein into PAN39 did not result in complementation of the xcpQ mutant phenotype. However, both plasmids restored the secretion defect in strain PAN38, which confirmed that mutations affecting LPS outer core biosynthesis facilitate functioning of the inactive XcpQ hybrids. To substantiate this conclusion, we selected mutants of strain PAN1 containing plasmid pM2 (xcpQF87) that were resistant to the LPS-specific phage PAF3. Nine of 12 mutants isolated had an altered LPS core, whereas the other mutants only had a defect in O-antigen biosynthesis, as revealed by SDS-PAGE analysis of proteinase K-treated cell envelope preparations (results not shown). In contrast to the three O-antigen mutants, all nine mutants with a defect in outer core biosynthesis had a suppression phenotype, as determined by halo formation on elastin plates. Together, these results show that defects in the LPS outer core, rather than the loss of O antigen, allow the nonactive hybrid XcpQ proteins to become functional.
TABLE 3.
Complementation of LPS mutant strains with xcpQ wild-type genes and chimerasa
| Strain | Mutationb | Secretion with different xcpQ genes
|
||||
|---|---|---|---|---|---|---|
| None | xcpQaer | xcpQalc | xcpQF87 | xcpQF497 | ||
| PAO25 | None | ++ | ++ | ++ | ++ | ++ |
| PAN1 | xcpQ | − | ++ | − | − | ++ |
| PAN15 | xcpQ, suppressor | − | ++ | + | ++ | ++ |
| PAN16 | xcpQ, suppressor | − | ++ | + | ++ | ++ |
| PAN38 | xcpQ, core | − | + | + | + | + |
| PAN39 | xcpQ, O antigen | − | + | − | − | + |
Secretion was analyzed on elastin plates by evaluating halo formation after 24 h of incubation and by analyzing extracellular protein profiles by SDS-PAGE. ++, secretion comparable to that of wild-type PAO25 cells; +, decreased but significant secretion; −, no significant secretion.
core, mutation causing a defective LPS outer core; O antigen, mutation causing a lack of O antigen.
Since the structure of LPS can also be influenced by the growth conditions, the activity of the nonfunctional hybrid protein XcpQF87 in the xcpQ mutant PAN1 was determined at different temperatures and in medium with various concentrations of salt or divalent cations. The temperature or different salt concentrations did not influence the functionality of XcpQF87. However, addition of Mg2+ or Ca2+ to the growth medium increased the ability of XcpQF87 to restore elastase secretion in PAN1 (Fig. 4A) and to form oligomers (Fig. 4B). The effect of the divalent cations on the functionality of this hybrid was similar to the effect of the LPS mutations. Divalent cations also stimulated protein secretion in strain PAN1 producing the XcpQalc protein (Fig. 4A) or the other inactive chimeric hybrids (data not shown). The presence of divalent cations did not alter the extracellular protein profiles of PAN1 producing XcpQaer or the active fusion protein XcpQF497 (Fig. 4A). In contrast, the functionality of the partially functional fusion protein XcpQF459 was improved by the presence of Mg2+ (Fig. 4A).
DISCUSSION
P. aeruginosa and P. alcaligenes are closely related organisms that both secrete proteins via the type II secretion pathway. Previously, it has been shown that most of the components of the Xcp machinery of P. alcaligenes can replace their P. aeruginosa counterparts; the exceptions are XcpP and XcpQ. Only when the genes encoding XcpP and XcpQ were expressed simultaneously was some complementation observed (11). However, this complementation was observed only when the cells were grown on plates and not when the cells were grown in liquid cultures. In this study, we determined the region of the XcpQalc protein that is responsible for species-specific functioning of the secretin and isolated suppressor mutations on the chromosome of P. aeruginosa that allowed XcpQalc to be functional.
Isolation of the chimeric proteins XcpQaer and XcpQalc enabled us to pinpoint a domain determining the nonfunctionality of XcpQalc in P. aeruginosa. Interestingly, this region was found to be in the C-terminal homology domain. This result was not expected, since it is the N-terminal half of secretins that is thought to be involved in system-specific interactions. The region identified is 74% identical in XcpQalc and XcpQaer. Moreover, the experiments also revealed that the chimeric secretin XcpQF478 was completely functional, whereas with XcpQF459 there was only partial restoration of secretion in the P. aeruginosa xcpQ mutant. These two XcpQ variants differ at only a single amino acid residue. Whereas XcpQF478 contains a lysine residue at position 460 (the numbering includes the signal sequence), XcpQF459 contains a glutamate residue at the equivalent position. Therefore, this lysine appears to be important for proper functioning of the secretin in P. aeruginosa.
With the exception of XcpQF331, all the fusions were capable of oligomerization. Although the amounts of the inactive XcpQ proteins were generically smaller than the amounts of the active proteins, we demonstrated that the reduced levels of expression were not responsible for the secretion defect. Furthermore, localization studies showed that the oligomers formed by the nonfunctional hybrid protein XcpQF87 colocalized with the outer membrane markers. The cells also contained a monomeric XcpQ pool, which was the same size for all the variants. This result suggests that the monomeric form of the inactive fusions is stable but that reduced efficiency during the subsequent stages in the biogenesis process makes the assembly intermediates susceptible to proteolytic degradation.
The nonfunctionality of XcpQalc and the inactive fusions in P. aeruginosa might be explained by inappropriate interactions with host factors. Interestingly, mutations affecting the biosynthesis of the outer core region of LPS enabled XcpQalc and inactive XcpQ hybrids to function in P. aeruginosa. It should be noted, however, that the functionality of XcpQalc in the suppressor strain was lower than that of the hybrid proteins, which indicates that the N-terminal 63 amino acid residues may also contribute to the species specificity. Consistently, fusion protein XcpQH03, which has the N terminus of XcpQalc, was not as functional as XcpQH13, which is similar except that the N-terminal 63 amino acid residues correspond to the residues in XcpQaer (Fig. 1). The XcpP protein is a good candidate to interact with the N terminus. However, we did not observe any restoration of secretion in the xcpQ mutant during coexpression of XcpPalc with its cognate secretin, which may have been due to competition between XcpPalc and the endogenous XcpP for binding to other Xcp components. Presumably, both the correct LPS and the cognate XcpP are required for optimal functioning.
The stimulatory effect of the LPS mutation on the functionality of inactive hybrids could be either direct or indirect. In the case of the latter possibility, the alterations in the outer core might have pleiotropic effects, resulting in stimulation of protein folding or reduced activity of proteases in the periplasm. Alternatively, improved secretion in the LPS mutants could be due to an altered interaction between LPS and XcpQ. The following findings support this hypothesis. (i) The inactive secretins form outer membrane-associated oligomers, indicating that they are correctly folded. (ii) The region involved in species specificity is present in the C-terminal homology domain of the secretin that is thought to be located in the outer membrane. Therefore, this part of the protein is probably in close contact with LPS. (iii) It has been shown previously that a different LPS mutation affects the functioning of XcpQaer negatively (33). Hence, mutations affecting the LPS structure can have opposite effects on the functionality of XcpQ in P. aeruginosa, namely, a negative effect on the endogenous secretin and a positive effect on the XcpQalc secretin, which indicates that there is a direct interaction between LPS and XcpQ. (iv) The addition of divalent cations also allows the XcpQalc secretin to function in P. aeruginosa. Divalent cations are known to affect LPS structure, either directly by influencing the charge and/or packaging of the LPS (40) or indirectly by downregulating LPS-modifying enzymes (36).
The ability of LPS to affect the functioning of outer membrane proteins has been reported previously. For instance, the outer membrane protease OmpT requires LPS for activity (28). The secretin-LPS interaction could also affect the interpretation of data obtained in several previous studies, in which secretion was studied in heterologous hosts. For example, the OutD secretin of Erwinia carotovora was shown to be unable to mediate the secretion of E. chrysanthemi exoproteins in a reconstituted pathway in E. coli, although the gene was expressed efficiently (30). However, the same protein did function correctly in an E. chrysanthemi background, indicating that an inappropriate interaction with LPS might play a role.
In conclusion, a part of the XcpQalc protein that is responsible for the inability of this secretin to function properly in the closely related host P. aeruginosa is located in the C-terminal secretin homology domain. This inability to function properly can be suppressed by alterations in the outer core of the LPS of P. aeruginosa or by the presence of divalent cations, indicating that changes in LPS structure influence the functioning of these protein transport channels. In future experiments we will try to identify the chromosomal mutations in the suppressor mutants and determine the precise role of LPS in the functioning of the XcpQ secretin.
Acknowledgments
We thank the NCCB, Gerard Michel, and Bengt Wretlind for providing strains, plasmids, and bacteriophages and Gijs Gerritse for providing the xcp gene cluster of P. alcaligenes. We are grateful to Hans de Cock and Mohamed El Khattabi for helpful discussions.
This work was supported by EU grants BIO4-CT96-0119 and QLRT-2001-02086 and by Sonderforschungsbereich (SFB) 470, project B4 (S.P.C. and U.Z.).
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Bitter, W., M. Koster, M. Latijnhouwers, H. de Cock, and J. Tommassen. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27:209-219. [DOI] [PubMed] [Google Scholar]
- 2.Bitter, W. 2003. Secretins of Pseudomonas aeruginosa: large holes in the outer membrane. Arch. Microbiol. 179:307-314. [DOI] [PubMed] [Google Scholar]
- 3.Bouley, J., G. Condemine, and V. E. Shevchik. 2001. The PDZ domain of OutC and the N-terminal region of OutD determine the secretion specificity of the Type II Out pathway of Erwinia chrysanthemi. J. Mol. Biol. 308:205-219. [DOI] [PubMed] [Google Scholar]
- 4.Brade, H., C. Galanos, and O. Lüderitz. 1983. Differential determination of the 3-deoxy-d-mannooctulosonic acid residues in lipopolysaccharides of Salmonella minnesota rough mutants. Eur. J. Biochem. 131:195-200. [DOI] [PubMed] [Google Scholar]
- 5.Braun, P., W. Bitter, W., and J. Tommassen. 2000. Activation of Pseudomonas aeruginosa elastase in Pseudomonas putida by triggering dissociation of the propeptide-enzyme complex. Microbiology 146:2565-2572. [DOI] [PubMed] [Google Scholar]
- 6.Brok, R., P. Van Gelder, M. Winterhalter, U. Ziese, A. J. Koster, H. de Cock, M. Koster, J. Tommassen, and W. Bitter. 1999. The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J. Mol. Biol. 294:1169-1179. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, P.-W. 1987. High-performance liquid chromatographic analysis of galactosamine, glucosamine, glucosaminitol and galactosaminitol. Anal. Biochem. 167:265-269. [DOI] [PubMed] [Google Scholar]
- 8.Cianciotto, N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13:581-588. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, S. A., and D. J. Strydom. 1988. Amino acid analysis utilizing phenyl-isothiocyanate derivatives. Anal. Biochem. 174:1-16. [DOI] [PubMed] [Google Scholar]
- 10.Daefler, S., M. Russel, and P. Model. 1997. Module swaps between related translocator proteins pIV(f1), pIV(IKe) and PulD: identification of a specificity domain. J. Mol. Biol. 266:978-992. [DOI] [PubMed] [Google Scholar]
- 11.de Groot, A., M. Koster, M. Gérard-Vincent, G. Gerritse, A. Lazdunski, J. Tommassen, and A. Filloux. 2001. Exchange of Xcp (Gsp) secretion machineries between Pseudomonas aeruginosa and Pseudomonas alcaligenes: species specificity unrelated to substrate recognition. J. Bacteriol. 183:959-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deretic, V., S. Chandrasekharappa, J. F. Gill, D. K. Chatterjee, and A. M. Chakrabarty. 1987. A set of cassettes and improved vectors for genetic and biochemical characterization of Pseudomonas genes. Gene 57:61-72. [DOI] [PubMed] [Google Scholar]
- 13.Di Fabio, J. L., M. B. Perry, and D. R. Bundle. 1987. Analysis of the lipopolysaccharide of Pseudomonas maltophilia 555. Biochem. Cell Biol. 65:968-977. [DOI] [PubMed] [Google Scholar]
- 14.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filloux, A., G. Michel, and M. Bally. 1998. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 22:177-198. [DOI] [PubMed] [Google Scholar]
- 16.Filloux, A., S. Bleves, P. van Ulsen, and J. Tommassen. 2004. Protein secretion mechanisms in Pseudomonas, p. 749-792. In J. Ramos (ed.), Pseudomonas, genomics, life style and molecular architecture. Kluwer Academic/ Plenum Publishers, New York, NY.
- 17.Frenken, L. G., J. W. Bos, C. Visser, W. Müller, J. Tommassen, and C. T. Verrips. 1993. An accessory gene, lipB, required for the production of active Pseudomonas glumae lipase. Mol. Microbiol. 9:579-589. [DOI] [PubMed] [Google Scholar]
- 18.Fürste, J. P., W. Pansegrau, R. Frank, H. Blöcker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 19.Galanos, C., O. Lüderitz, and O. Westphal. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9:245-249. [DOI] [PubMed] [Google Scholar]
- 20.Genin, B., and C. A. Boucher. 1994. A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol. Gen. Genet. 243:112-118. [DOI] [PubMed] [Google Scholar]
- 21.Gerritse, G., R. Ure, F. Bizoullier, and W. J. Quax. 1998. The phenotype enhancement method identifies the Xcp outer membrane secretion machinery from Pseudomonas alcaligenes as a bottleneck for lipase production. J. Biotechnol. 64:23-38. [DOI] [PubMed] [Google Scholar]
- 22.Reference deleted.
- 23.Hancock, R. E. W., and H. Nikaido. 1978. Outer membranes of Gram-negative bacteria. J. Bacteriol. 136:381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarrell, K., and A. M. Kropinski. 1977. The chemical composition of the lipopolysaccharide from Pseudomonas aeruginosa strain PAO and a spontaneously derived rough mutant. Microbios 19:103-116. [PubMed] [Google Scholar]
- 25.Jarrell, K. F., and A. M. Kropinski. 1981. Isolation and characterization of a bacteriophage specific for the lipopolysaccharide of rough derivatives of Pseudomonas aeruginosa strain PAO. J. Virol. 38:529-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, T. L., J. Abendroth, W. G. Hol, and M. Sandkvist. 2006. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255:175-186. [DOI] [PubMed] [Google Scholar]
- 27.Korotkov, K. V., B. Krumm, M. Bagdasarian, and W. G. Hol. 2006. Structural and functional studies of EpsC, a crucial component of the type 2 secretion system from Vibrio cholerae. J. Mol. Biol. 363:311-321. [DOI] [PubMed] [Google Scholar]
- 28.Kramer, R. A., K. Brandenburg, L. Vandeputte-Rutten, M. Werkhoven, P. Gros, N. Dekker, and M. R. Egmond. 2002. Lipopolysaccharide regions involved in the activation of Escherichia coli outer membrane protease OmpT. Eur. J. Biochem. 269:1746-1752. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lindeberg, M., G. P. C. Salmond, and A. Collmer. 1996. Complementation of deletion mutants in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologues reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the type II pathway. Mol. Microbiol. 20:175-190. [DOI] [PubMed] [Google Scholar]
- 31.Lowry, O. H., N. R. Roberts, K. Y. Leiner, M. L. Wu, and A. L. Farr. 1954. The quantitative histochemistry of brain. J. Biol. Chem. 207:1-17. [PubMed] [Google Scholar]
- 32.McIver, K. S., E. Kessler, J. C. Olson, and D. E. Ohman. 1995. The elastase propeptide functions as an intramolecular chaperone required for the elastase activity and secretion in Pseudomonas aeruginosa. Mol. Microbiol. 18:877-889. [DOI] [PubMed] [Google Scholar]
- 33.Michel, G., G. Ball, J. B. Goldberg, and A. Lazdunski. 2000. Alteration of the lipopolysaccharide structure affects the functioning of the Xcp secretory system in Pseudomonas aeruginosa. J. Bacteriol. 182:696-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nouwen, N., H. Stahlberg, A. P. Pugsley, and A. Engel. 2000. Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J. 19:2229-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osborn, M. J., J. E. Gander, E. Parisi, and J. Carson. 1972. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 247:3962-3972. [PubMed] [Google Scholar]
- 36.Raetz, C. R. H. 2001. Regulated covalent modifications of lipid A. J. Endotoxin Res. 7:73-78. [PubMed] [Google Scholar]
- 37.Sadovskaya, I., J.-R. Brisson, J. S. Lam, J. C. Richards, and E. Altman. 1998. Structural elucidation of the lipopolysaccharide core regions of the wild-type strain PAO1 and O-chain-deficient mutant strains AK1401 and AK1012 from Pseudomonas aeruginosa serotype O5. Eur. J. Biochem. 225:673-684. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Sawardeker, J. S., J. H. Sloneker, and A. Jeanes. 1965. Quantitative determination of monosaccharides as their alditol acetates by gas liquid chromatography. Anal. Chem. 37:1602-1603. [Google Scholar]
- 40.Snyder, S., D. Kim, and T. J. McIntosh. 1999. Lipopolysaccharide bilayer structure: effect of chemotype, core mutations, divalent cations, and temperature. Biochemistry 38:10758-10767. [DOI] [PubMed] [Google Scholar]
- 41.Strominger, J. L., J. T. Park, and R. E. Thompson. 1959. Composition of the cell wall of Staphylococcus aureus: its relation to the mechanism of action of penicillin. J. Biol. Chem. 234:3263-3268. [PubMed] [Google Scholar]
- 42.Tommassen, J., P. van der Ley, M. van Zeijl, and M. Agterberg. 1985. Localization of functional domains in E. coli K-12 outer membrane porins. EMBO J. 4:1583-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tommassen, J., H. van Tol, and B. Lugtenberg. 1983. The ultimate localization of an outer membrane protein of Escherichia coli K-12 is not determined by the signal sequence. EMBO J. 2:1275-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 45.Van Gelder, P., N. Saint, R. van Boxtel, J. P. Rosenbusch, and J. Tommassen. 1997. Pore functioning of outer membrane protein PhoE of Escherichia coli: mutagenesis of the constriction loop L3. Protein Eng. 10:699-706. [DOI] [PubMed] [Google Scholar]
- 46.Voulhoux, R., G. Ball, B. Ize, M. L. Vasil, A. Lazdunski, L.-F. Wu, and A. Filloux. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20:6735-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westphal, O., and K. Jann. 1965. Extraction with phenol-water and further applications of the procedure. Carbohydr. Chem. 5:83-91. [Google Scholar]