Abstract
We performed a comprehensive study of the distribution and function of an insertion sequence (IS) element, IS1237, in the genome of Leifsonia xyli subsp. cynodontis, a useful genetic carrier for expressing beneficial foreign genes in plants. Two shorter IS1237 isoforms, IS1237d1 and IS1237d2 resulting from precise deletion between two nonperfect repeats, were found in the bacterial genome at a level that was one-fifth the level of wild-type IS1237. Both the genome and native plasmid pCXC100 harbor a truncated toxin-antitoxin cassette that is precisely fused with a 5′-truncated IS1237 sequence at one nonperfect repeat, indicating that it is a hot site for DNA rearrangement. Nevertheless, no transposition activity was detected when the putative transposase of IS1237 was overexpressed in Escherichia coli. Using thermal asymmetric interlaced PCR, we identified 13 upstream and 10 downstream unique flanking sequences, and two pairs of these sequences were from the same loci, suggesting that IS1237 has up to 65 unique loci in the L. xyli subsp. cynodontis chromosome. The presence of TAA or TTA direct repeat sequences at most insertion sites indicated that IS1237 inserts into the loci by active transposition. IS1237 showed a high propensity for insertion into other IS elements, such as ISLxc1 and ISLxc2, which could offer IS1237 a nonautonomous transposition pathway through the host IS elements. Interestingly, we showed that IS1237 has a strong promoter at the 3′ end and a weak promoter at the 5′ end, and both promoters promote the transcription of adjacent genes in different gram-positive bacteria. The high-copy-number nature of IS1237 and its promoter activity may contribute to bacterial fitness.
Insertion sequence (IS) elements are mobile genetic elements that generally encode a transposase required for transposition. The sizes of these elements vary from 0.7 to 3.5 kb, and the elements have imperfect terminal inverted repeat (IR) sequences that are 10 to 60 bp long (30, 32, 34). IS elements are found in the genomes of a wide range of bacteria. They cause insertion mutations and genome rearrangements and help mediate the spread of resistance and virulence determinants within and among species. In other cases, however, IS insertion also leads to activation or alteration of the expression of adjacent genes (3, 4, 6, 17, 18, 19, 30, 36, 38).
Leifsonia xyli subsp. cynodontis is a gram-positive coryneform bacterium that was originally isolated from the xylem of Bermuda grass (Cynodontis dactylon L. Per.) (8, 10, 27). L. xyli subsp. cynodontis colonization does not cause Bermuda grass decline or stunt symptoms (7). L. xyli subsp. cynodontis also colonizes and grows in the xylem vessels to a high titer in maize (Zea mays L.) when it is artificially inoculated (24). We recently found that colonization of rice with L. xyli subsp. cynodontis increases the growth of some rice strains (26). It may be possible to use L. xyli subsp. cynodontis as a genetic carrier to express agriculturally useful genes in crops of interest or in turf grass. Progress in this area has been made in the past few years, and the advances have included successful transformation of L. xyli subsp. cynodontis by electroporation (31), expression of the genes of interest from an integrative vector (15, 24), discovery of strong promoters in L. xyli subsp. cynodontis (14), and characterization of the native plasmid of L. xyli subsp. cynodontis and construction of a stable expression vector (25, 26, 40).
An IS element belonging to the IS5 family, IS1237 (0.9 kb), was identified in the genome of L. xyli subsp. cynodontis by Laine et al. (23). Southern blot analysis showed that at least 16 copies of IS1237 are present in the genome. Sequence analysis showed that production of the full-length transposase encoded by IS1237 requires a +1 frameshift between ORF2 and ORF1. It is not known if this multicopy IS element is active in transposition. Moreover, the distribution of IS1237 in the L. xyli subsp. cynodontis genome and its function are not clear either. In this study, we first analyzed variants of IS1237 in the genome and native plasmid pCXC100 of L. xyli subsp. cynodontis and found two shorter IS1237 isoforms, IS1237d1 and IS1237d2, in addition to the wild-type IS1237 in the genome and a toxin-antitoxin fusion variant, TA-3′IS1237, on the plasmid. No active transposase activity was detected in Escherichia coli. However, thermal asymmetric interlaced PCR (TAIL-PCR) (28) analysis of the upstream and downstream sequences of IS1237 revealed that this transposon has about 65 unique loci in the L. xyli subsp. cynodontis genome and that it has a high propensity to transpose into other IS elements, such as the newly identified elements ISLxc1 and ISLxc2, suggesting that this IS element may nonautonomously transpose with other active IS elements. Interestingly, we found that IS1237 harbors a strong promoter at the 3′ end and a weak promoter at the 5′ end and that both promoters promote transcription of the adjacent genes, suggesting that the high-copy-number nature of IS1237 may be a result of the contribution of its promoter to bacterial fitness.
MATERIALS AND METHODS
Strains, media, and growth conditions.
E. coli strains DH5α and BL21 were used to construct plasmids and to express transposases in E. coli, respectively. L. xyli subsp. cynodontis strain #3 (8, 10, 31), Corynebacterium glutamicum ATCC 13032, and Bacillus subtilis 168 (from the China General Microbiological Culture Collection Center) were used to assay the promoter activity of IS1237. E. coli and B. subtilis were cultured in Luria broth at 37°C. C. glutamicum was cultured in Luria broth containing 0.5% glucose at 30°C. L. xyli subsp. cynodontis was cultured on DM agar at 30°C (31). The concentrations of antibiotics used in this study were 60 μg/ml of ampicillin, 2.5 μg/ml of tetracycline, 25 to 50 μg/ml of kanamycin, and 10 μg/ml of chloramphenicol.
Plasmids, DNA manipulation, and plasmid construction.
Plasmids used in this study are shown in Table 1. Plasmids were extracted from B. subtilis and C. glutamicum by the alkaline lysis method, except that 10 mg/ml lysozyme was added to the lysis solution and the solution was incubated at 37°C for 1 h (31, 33). A plasmid was released from L. xyli subsp. cynodotis by the STE method (5) and then transformed into E. coli for further characterization and identification. Transformation of L. xyli subsp. cynodotis, B. subtilis, and C. glutamicum was performed by electroporation as previously described (2, 31, 41).
TABLE 1.
Plasmids used in this study
| Plasmid | Phenotype | Marker(s) | Source or reference |
|---|---|---|---|
| Vectors | |||
| pMD18-T | E. coli TA cloning vector | Ampr | Takara |
| pBR325 | E. coli cloning vector | Ampr Tcr | Lab collection |
| pCXC100 | L. xyli subsp. cynodotis native 51-kb plasmid | 25 | |
| pBE2 | E. coli-B. subtilis shuttle vector | Kmr | 12 |
| pEC-K18mob2 | E. coli-C. glutamicum shuttle vector | Kmr | 20 |
| pBL121 | E. coli cloning vector containing GUS gene | Kmr | Lab collection |
| pET28a | E. coli expression vector | Kmr | Lab collection |
| pBAD33 | E. coli cloning vector | Cmr | 13 |
| Clones | |||
| pLC011 | E. coli-L. xyli subsp. cynodotis shuttle vector, pBR325 containing the 51-kb replicon of pCXC100 | Ampr | This study |
| pUCISD | pMD18-T vector containing IS1237 in the HindIII-to-EcoRI direction | Ampr | This study |
| pUCISR | pMD18-T vector containing IS1237 in the EcoRI-to-HindIII direction | Ampr | This study |
| pUC-GUS | pMD18-T vector containing GUS gene | Ampr | This study |
| pUCISD-GUS | pUCISD containing GUS gene at the EcoRI-XbaI site; IS1237 and GUS gene have the same orientation | Ampr | This study |
| pUCISR-GUS | pUCISR containing GUS gene at the EcoRI-XbaI site; IS1237 and GUS gene have opposite orientations | Ampr | This study |
| pUC9-GUS | pUC-GUS vector containing 850-bp L. xyli subsp. cynodotis chromosome fragment before GUS gene | Ampr | This study |
| pLCISD-GUS | pLC011 vector containing the IS1237-GUS cassette from pUCISD-GUS | Ampr | This study |
| pLCISR-GUS | pLC011 vector containing the reverse IS1237-GUS cassette from pUCISR-GUS | Ampr | This study |
| pLC9-GUS | pLC011 vector containing 850-bp L. xyli subsp. cynodotis chromosome fragment before GUS gene | Ampr | This study |
| pLCP-GUS | pLC011 vector containing the L. xyli subsp. cynodotis strong promoter PIII before GUS gene | This study | |
| pBEISD-GUS | pBE2 vector containing the direct IS1237-GUS cassette from pUCISD-GUS | Kmr | This study |
| pBEISR-GUS | pBE2 vector containing the reverse IS1237-GUS cassette from pUCISR-GUS | Kmr | This study |
| pBE9-GUS | pBE2 vector containing 850-bp L. xyli subsp. cynodotis chromosome fragment before GUS gene | Kmr | This study |
| pBE2-GUS | pBE2 vector containing GUS gene after the SP6 promoter | Kmr | This study |
| pECISD-GUS | pEC-K18mob2 vector containing direct IS1237-GUS cassette from pUCISD-GUS | Kmr | This study |
| pECISR-GUS | pEC-K18mob2 vector containing reverse IS1237-GUS cassette from pUCISR-GUS | Kmr | This study |
| pEC-GUS | pEC-K18mob2 vector containing GUS gene | Kmr | This study |
| pETIS1 | pET28a vector containing the wild-type IS1237 transposase gene | Kmr | This study |
| pETIS2 | pET28a vector containing 4-bp insertion at position 230 of IS1237 transposase gene | Kmr | This study |
| pETIS3 | pET28a vector containing 1-bp insertion at position 239 of IS1237 transposase gene | Kmr | This study |
| pETIS1-IR | pETIS1 containing IS1237-like element | Kmr | This study |
| pETIS2-IR | pETIS2 containing IS1237-like element | Kmr | This study |
| pETIS3-IR | pETIS3 containing IS1237-like element | Kmr | This study |
| pBADIS | pBAD33 vector containing whole IS1237 sequence | Cmr | This study |
To construct plasmids for the purpose of testing the transposition activity of IS1237 in E. coli (Fig. 1 and Table 1), the wild-type transposase gene was PCR amplified with primers d12-5′ and d12-3′ (Table 2) and inserted into the NcoI and HindIII sites of pET28a, resulting in plasmid pETIS1. Insertion of 4 bp at position 230 of the transposase gene by filling in the stick end of the BamHI site and by inserting a G residue at position 239 by mutagenesis PCR created two frameshifted mutants, pETIS2 and pETIS3, respectively. These mutants are predicted to produce the full-length active transposase without frameshifting. The whole IS1237 sequence was cloned into pBAD33 at XbaI-HindIII sites to obtain pBADIS. Construction of plasmids pETIS1-IR, pETIS3-IR, and pETIS3-IR is described in the Results and Discussion.
FIG. 1.
Structure of the plasmids used in this study. (A) IS1237 transposition activity detection system I. The IS1237 transposase gene cloned into pET28a between HindIII and NcoI sites is indicated by open boxes above the plasmid map. The solid arrows indicate the ORFs of the transposase gene. The mutation sites in IS1237 mutants are indicated by asterisks. The designations of plasmids having different transposase genes are indicated to the left of the corresponding transposase genes. (B) IS1237 transposition activity detection system II. An IS1237-like element (IS1237::Amp) was inserted into pETIS1, pETIS2, and pETIS3 at the SmaI site. The solid triangles indicate the IR sequence of IS1237 (IRR and IRL). The gray box indicates the inserted ampicillin resistance gene. (C) IS1237 promoter activity assay system. The shaded boxes indicate the GUS gene, and the open boxes indicate the upstream sequences flanking the GUS gene. The designations of the GUS-containing plasmids containing different upstream sequences are indicated to the left of the corresponding cloned elements.
TABLE 2.
Primers used in this study
| Primer | Sequence | Description |
|---|---|---|
| d3 | GGTCACTTGTTGGCGGCG | Amplification of partial sequences of IS1237 |
| d4 | CGAGCGCGGTGATCGTTG | Amplification of partial sequences of IS1237 |
| d5 | GAGGTGGTTTCAGTAGTC | Amplification of IS1237 from L. xyli subsp. cynodotis chromosome |
| d12-5′ | GTCCCATGGCGACGCTGGCTG | Amplification of the IS1237 transposase gene |
| d12-3′ | CCCAAGCTTGTTTACCCAGTC | Amplification of the IS1237 transposase gene |
| d13 | GGACCTAACCCCACGGATC | Amplification of downstream flanking sequence of IS1237 in L. xyli subsp. cynodotis chromosome |
| d14 | CTCGTGGAACCTGTGTTAGAC | Amplification of downstream flanking sequence of IS1237 in L. xyli subsp. cynodotis chromosome |
| d15 | CTCGTCGCTCTAGCAATCG | Amplification of downstream flanking sequence of IS1237 in L. xyli subsp. cynodotis chromosome |
| d16 | CACTTACGGAGTCCAGGCAG | Amplification of upstream flanking sequence of IS1237 in L. xyli subsp. cynodotis chromosome |
| d17 | CCGCTTCGGACCATTCACGC | Amplification of upstream flanking sequence of IS1237 in L. xyli subsp. cynodotis chromosome |
| d18 | CACAACATATCCGTAATGAACCG | Amplification of upstream flanking sequence of IS1237 in L. xyli subsp. cynodotis chromosome |
| d48 | CGGCCATGGTGTGGGAGC | Construction of the mutant transposase gene |
| d49 | CGTGGAGAGAGCTCCC | Construction of the mutant transposase gene |
| d50 | GGGAGCTCTCTCCACG | Construction of the mutant transposase gene |
| d51 | TAAAAGCTTGTTTCAGTAGTCGTT | Construction of the mutant transposase gene |
| AMP1 | AAGGGATCCGTGATACGCC | Amplification of the ampicillin resistance gene |
| AMP2 | GAAGGATCCTCACCTAGATC | Amplification of the ampicillin resistance gene |
| Tn5564-p1 | ACATCAACGGCCCGGAATAC | Amplification of the ISLxc2 sequence |
| Tn5564-p2 | GCAACAGCCTCTCGTGGGAC | Amplification of the ISLxc2 sequence |
| Tn5564-p3 | ACTGGAACAAGGCGAGAGGGAC | Amplification of the ISLxc2 sequence |
| Tn5564-p4 | CATAAGGCCCGGCAACTACA | Amplification of the ISLxc2 sequence |
| Tn5564-p5 | CAGGGTGCTCTCCGACAACG | Amplification of the ISLxc2 sequence |
| Tn5564-p6 | CAGGGAGAGCGGAACAAGGTC | Amplification of the ISLxc2 sequence |
| AD1 | NTCGA(G/T)T(A/T)T(G/C)G(A/T)GTT | Arbitrary degenerate primer |
| AD2 | NGTCGA(G/C)(A/T)TGANA(A/T)GAA | Arbitrary degenerate primer |
| AD3 | (A/T)GTGNAG(A/T)ANCANAGA | Arbitrary degenerate primer |
| AD4 | AG(A/T)GNAG(A/T)ANCA(A/T)AGG | Arbitrary degenerate primer |
| GUS7 | CCACACTTTGCCGTAATG | RT-PCR primer for the GUS gene |
| GUS8 | CACAAACGGTGATACGTAC | RT primer for the GUS gene |
| AAP | GGCCACACATCGACTAGTACGGGIIGGGIIGGGII | 5′ RACE abridged anchor primer |
| AUAP | GGCCACGCGTCGACTAGTAC | Abridged universal amplification primer |
| Pu1 | ACAACCACCGCCACGC | RT-PCR primer for Pu |
| Pu2 | CCGCTAACCAGCCACAA | RT-PCR primer for Pu |
| Pd1 | CTGAAACCACCTCATCTC | RT-PCR primer for Pd |
| Pd2 | CAAAAACGACTACTGAAAC | RT-PCR primer for Pd |
The E. coli-L. xyli subsp. cynodontis shutter vector pLC011 was constructed as follows: first the 5-kb NcoI-EcoRI fragment of pCXC100 that contained the replication origin (25) was cloned into the corresponding restriction sites of the pET28a vector, and then the NcoI-HindIII fragment containing the NcoI-EcoRI fragment of pCXC100 from the intermediate construct was cloned into the corresponding restriction sites of pBR325. Note that the original EcoRI site of pBR325 was eliminated prior to use of the vector in this cloning procedure.
A series of plasmids were also constructed to test whether IS1237 has promoter functions that affect the expression of the flanking genes (Table 1 and Fig. 1). The full-length IS1237 element was PCR amplified using primer d5 and then cloned into the pMD18-T vector, and this was followed by sequence confirmation. Clones pUCISD and pUCISR with IS1237 inserted in opposite orientations were obtained. Then the beta-glucuronidase (GUS) gene was cleaved with the XbaI and EcoRI restriction enzymes and inserted into the corresponding sites in pUCISD and pUCISR, resulting in plasmids pUCISD-GUS and pUCISR-GUS. In pUCISD-GUS, the open reading frames (ORFs) of IS1237 were in the same orientation as the GUS gene, while they were in the opposite orientation in pUCISR-GUS. The GUS gene was also cloned into the XbaI-EcoRI sites of pMD18-T to construct pUC-GUS. An 850-bp unrelated fragment amplified from the L. xyli subsp. cynodontis genome was cloned into pUC-GUS to obtain pUC9-GUS. The latter two plasmids served as negative controls.
The IS1237-GUS gene cassettes were subsequently subcloned into pLC011, E. coli-B. subtilis shuttle vector pBE2 (12), and E. coli-C. glutamicum shuttle vector pEC-K18mob2 (20) at the restriction sites indicated in Fig. 1 to determine the IS1237 promoter activities in L. xyli subsp. cynodontis, B. subtilis, and C. glutamicum, respectively (Table 1).
All of the sequences of primers used in plasmid construction are shown in Table 2.
TAIL-PCR to obtain the upstream and downstream flanking sequences of IS1237.
TAIL-PCR (28) was used to amplify DNA sequences flanking IS1237 integration sites on the L. xyli subsp. cynodontis genome. In TAIL-PCR three nested specific primers are utilized in consecutive reactions together with arbitrary degenerate (AD) primers to enhance the amplification efficiency of specific products. Specific primers d13, d14, and d15 and four AD primers (AD1, AD2, AD3, and AD4) were used to amplify the downstream flanking sequences of IS1237 in the L. xyli subsp. cynodontis chromosome. Three specific primers (d16, d17, and d18) and four AD primers were used to amplify the upstream sequences. The specific primers were designed to have a melting temperature (60 to 65°C) that was higher than that of the AD primers (45°C). The PCR systems and cycling conditions used have been described previously (28). The PCR products were cloned into pMD18-T vector and sequenced. Database searches were performed using the BLAST program at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/) covering the GenBank, EMBL, DDBL, and PDB databases (1, 35).
Promoter activity assay.
All B. subtilis and C. glutamicum strains containing corresponding plasmids were cultured overnight under appropriate conditions, while L. xyli subsp. cynodontis strains were grown on DM agar for 7 days. Cell pellets from 1-ml cultures having an optical density at 600 nm of 5 were collected by centrifugation at 10,000 rpm for 1 min at 4°C and resuspended in 500 μl of distilled water. Then the cells were broken by ultrasonication for 4 ms at 40 W 20 times. The GUS activity was measured as previously described (29). The reaction was performed for 6 h before a 12.5-μl aliquot was transferred to 237.5 μl of stop buffer (0.2 M Na2CO3) to stop the reaction. The light signals for all the bacterial strains tested were photographed.
5′ RACE RT-PCR.
The total RNAs of the strains containing corresponding plasmids were extract by the hot phenol method. Then 5′ rapid amplification of cDNA ends (RACE) reverse transcription-PCR (RT-PCR) was carried out to identify the 5′ ends of mRNA that initiated from the promoters. First, the cDNA of the GUS gene was produced by using reverse transcriptase with primer GUS8. Then the cDNA was isolated using a G-25 spin column (Roche), and poly(C) was added at the 5′ end of the cDNA by using terminal deoxynucleotidyl transferase (Takara, Dalian, China). The cDNA was then PCR amplified using primer GUS8 and the poly(G) primer AAP containing a stretch of the unique sequence 5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′. A nested PCR was performed with primers GUS7 and AUAP to enhance the amplification efficiency of specific products. The PCR products were cloned into the pMD18-T vector (Takara) and sequenced.
RESULTS AND DISCUSSION
Distribution of IS1237 and its variants in the L. xyli subsp. cynodontis genome.
In order to elucidate the transposition mechanism of IS1237, all the IS1237 homologous sequences located in the L. xyli subsp. cynodontis genome were cloned. Primer d5, consistent with the IS1237 IR sequence, was used to amplify IS1237-related sequences from the genomic DNA of plasmid-free L. xyli subsp. cynodontis strain #3 (Fig. 2A), which were then cloned into the pMD18-T vector. Restriction digestion revealed that IS1237 sequences that were different sizes were amplified from the L. xyli subsp. cynodontis genome. The majority of clones yielded restriction fragments corresponding to the size predicted based on the previously published IS1237 sequence (0.9 kb) (23), and the other clones contained a shorter IS1237 sequence (0.8 kb). Two clones of each IS1237 isoform were sequenced, which confirmed that the 0.9-kb IS1237 is identical to the previously published IS1237 and that the 0.8-kb IS1237 resulted from a 101-bp deletion inside IS1237 (Fig. 2A). The ratio of the number of 0.8-kb IS1237 sequences to the number of 0.9-kb IS1237 sequences in the L. xyli subsp. cynodontis genome was then determined by PCR amplification using primers d3 and d4, which yielded 466- and 567-bp DNA bands that could be easily separated on agarose gels (Fig. 3). Quantification of the two bands on agarose gels with the 1D 3.5 image analysis software (Kodak) revealed that the ratio of the shorter IS1237 species to the full-length IS1237 species in the L. xyli subsp. cynodotis genome is roughly 1:5.
FIG. 2.
IS1237 variants in L. xyli subsp. cynodontis. (A) IS1237 variants in the L. xyli subsp. cynodontis chromosome. The sequence and coding regions of IS1237 (orf2 and orf1) are shown. The positions of the d3 and d4 primers used to amplify the region around the deletion site of IS1237 and the position of the d5 primer used to amplify the entire IS1237 element are indicated. The scissor symbols indicate the deletion sites used to generate two shorter version of IS1237, and the shaded region is the deletion region of IS1237. The underlined sequence is the repeated sequence. IS1237d1-L and IS1237d1-R indicate the left and right sites at which IS1237 turns to IS1237d1; IS1237d2-L and IS1237d2-R indicate the left and right sites at which IS1237 turns to IS1237d2. (B) TA-IS1237 fusion variant in L. xyli subsp. cynodontis native plasmid pCXC100. The symbols used for the truncated IS1237 element are the same as those described above, and the truncated toxin-antitoxin cassette and the putative promoter upstream of the truncated IS1237 element are also shown. The positions of the d4 and d7 primers used to amplify the TA-IS1237 fusion variant from the chromosomal DNA of L. xyli subsp. cynodontis are indicated.
FIG. 3.
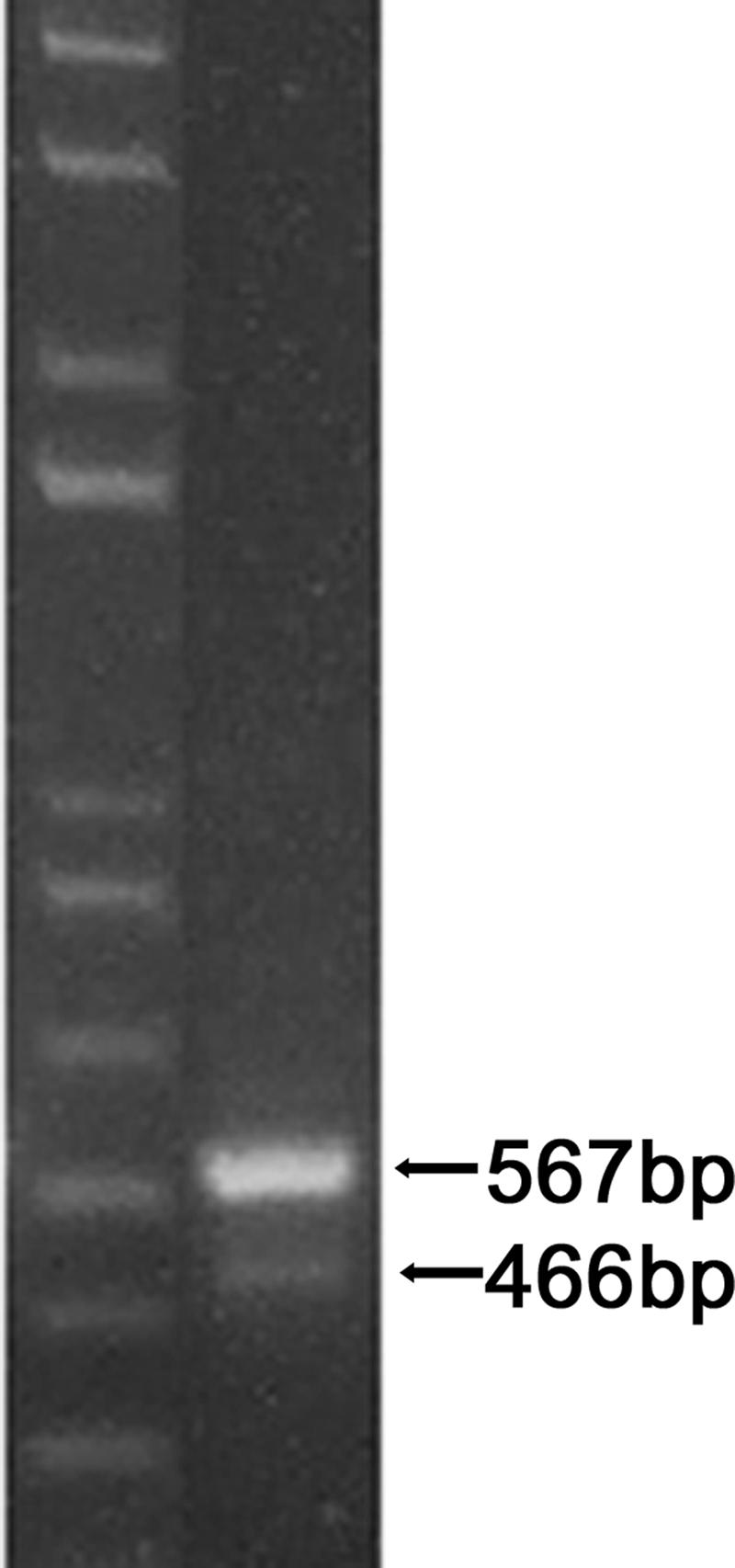
Determination of the ratio of IS1237 to IS1237d1 and IS1237d2. Left lane, 1-kb molecular weight marker; right lane, PCR product of genome of strain #3 obtained with primers d3 and d4. Gels were stained with ethidium bromide and then photographed using a Kodak 290 digital camera.
Eight additional clones containing the deletion form of IS1237 were sequenced to validate the sequence identity of the newly found IS1237 species, and this revealed the presence of two different variants of the shorter IS1237 species, namely IS1237d1 and IS1237d2. Interestingly, two nonperfect direct repeats were identified at the boundaries of deletion sites; the upstream repeat was 10 bp long, while the downstream repeat was 12 bp long. The 12-bp downstream repeat sequence and the 89-bp sequence between the repeats were deleted in the shorter variant, IS1237d1, while deletion of the same length of sequence was shifted 1 bp downstream in variant IS1237d2. We found that 5 of the 10 shorter IS1237 clones sequenced contained the IS1237d1 sequence and the other five contained the IS1237d2 sequence, indicating that roughly equal numbers of these two variants are present in L. xyli subsp. cynodnotis. The presence of 10- to 12-bp direct repeats at the boundaries of the deletion site of IS1237d1 and IS1237d2 underlines the importance of these direct repeats in creating the site-specific deletion variants of IS1237 (Fig. 2A).
Some previous studies revealed that spontaneous deletions are favored at repeated sequences in the lacI gene. Repeated sequences with five or eight bases were found at each deletion site, and the deletions removed one of the repeated sequences and 20- to 123-bp intervening fragments (11). It is also known that site-specific recombination between the direct repeats at the ends of a transposon results in transposon deletion (21). To the best of our knowledge, this is the first report demonstrating that site-specific deletion of a DNA segment occurs inside a putative bacterial transposon at a pair of direct repeats. However, it is not clear how such site-specific deletion occurs. The involvement of the recombination system of host bacteria in deletion should be tested. Furthermore, we noticed that the putative transposase of IS1237 contained the DDE motif (23). Recent research confirmed that a DDE family transposase is able to perform the functions of site-specific recombinases (22). Thus, it is possible that this transposase functions as a recombinase to catalyze the site-specific deletion of IS1237 in L. xyli subsp. cynodontis. Nevertheless, more experiments are necessary to elucidate the detailed deletion mechanism.
Presence of a truncated form of IS1237 in pCXC100, a native plasmid of L. xyli subsp. cynodontis.
The native plasmid of L. xyli subsp. cynodontis, pCXC100, has been reported to carry a DNA element capable of hybridizing to the chromosomal IS1237 element (23). Based on our previously reported restriction map of pCXC100 (25), restriction digestion of pCXC100 subclones and subsequent Southern blot analysis demonstrated that the plasmid copy of the IS1237-related sequence resides in a 2-kb HindIII fragment of pLXC102 (25; data not shown). This fragment was designated H2, and it was cloned and sequenced; the resulting sequence was deposited in the GenBank database (GenBank accession no. DQ287925). Searching the H2 sequence against the GenBank database showed that it contains two different cassettes that are fused together, forming TA-3′IS1237 (Fig. 2B). The downstream cassette is a 198-bp 3′ sequence of IS1237 containing the downstream IR sequence, and this truncated IS1237 sequence starts precisely at the position of IS1237d1-L (Fig. 2A and B), indicating that a precise deletion occurs at IS1237d1-L to generate the 198-bp truncated version of IS1237. Surprisingly, immediately upstream of this IS1237 segment there is a 499-nucleotide cassette exhibiting 55% (toxin) and 60% (antitoxin) amino acid homology to the toxin-antitoxin proteins encoded by Magnetospirillum magnetotacticum (GenBank accession no. ZP-00209851.1) (Fig. 2B). A −1 frameshift is required for in-frame translation of the toxin protein, and the toxin-antitoxin cassette is apparently incomplete because of the lack of a stop codon.
To examine the possible origin of TA-3′IS1237 in pCXC100, PCR amplification using genomic DNA from plasmid-free L. xyli subsp. cynodontis strain #3 as the template and primers d7 and d4 located in the antitoxin-encoding region and IS1237 region of H2 (Fig. 2B), respectively, was performed to determine whether there was a chromosomal copy of TA-3′IS1237. An expected 360-bp product was amplified; the sequence of this PCR product was identical to the sequence of the plasmid copy of TA-3′IS1237, which confirmed the presence of the TA-3′IS1237 fusion cassette in L. xyli subsp. cynodontis. TAIL-PCR using primers d4, d10, and d11 further revealed that the full TA-3′IS1237 fusion cassette, including an 84-bp upstream promoter region of the toxin-antitoxin cassette, is present in the L. xyli subsp. cynodontis genome. These results collectively suggested that the plasmid TA-3′IS1237 fusion cassette could have originated from the chromosome DNA.
The lack of six amino acids at the carboxyl terminus and the stop codon of the toxin protein in the TA-3′IS1237 fusion cassette indicates that the TA cassette in pCXC100 might not function in supporting plasmid stability in L. xyli subsp. cynodontis. Precise fusion of the truncated forms of the toxin-antitoxin and IS1237 sequences at the upstream site-specific deletion site of IS1237 suggested that this is a hot site for DNA rearrangement, and it is reasonable to suspect that the direct repeat sequences play a role in such a DNA rearrangement.
No IS1237 transposition activity was detected in E. coli.
Multiple copies of IS1237 have been detected in the L. xyli subsp. cynodontis genome, suggesting that this IS element could be active in transposition. Sequence analysis revealed that a putative transposase is encoded by IS1237 if a +1 frameshift occurs between ORF1 and ORF2 during translation (23). It has been reported that IS1 and members of the IS3 family are capable of producing an active transposase by a −1 frameshift between two adjacent ORFs (30). As mentioned previously, IS1237 belongs to the IS5 family, and it has been noted that most members of the IS427 subgroup of the IS5 family have two partially overlapping ORFs in a potential −1 frameshift window (30). Therefore, it would be interesting to determine whether IS1237 transposase is produced by a +1 frameshift rather than by a −1 frameshift.
Because convenient plasmid systems are not available for working with L. xyli subsp. cynodontis, the hypothesis that there is an active IS1237 was tested using E. coli. If IS1237 transposes through a cut-and-paste mechanism, the transposition intermediates, including a linear IS1237 element or a circular IS1237 element with abutted terminal repeats that are joined, should be detected. As a result of the transposition, the donor plasmid loses the IS and the restriction site hosting the IS is regenerated. In order to detect the transposition event, three plasmids containing the wild-type and two mutant forms of the putative transposase genes of IS1237 in pET28a were constructed (Fig. 1). Both mutants (pETIS2 and pETIS3) allowed production of the full-length putative transposase without a requirement for frameshifting. These plasmids were cotransformed with plasmid pBAD33 containing the complete IS1237 sequence. If an active transposase was expressed upon IPTG induction, the circular transposition intermediate could be detected by PCR amplification using primers d15 and d18 (Fig. 1). Protein gel analysis showed that both the wild-type and mutant forms of the transposase proteins were expressed from the transformed plasmids (data not shown). However, no circular intermediate was detected, which indicates that the transposase encoded by IS1237 is inactive, at least in E. coli.
Simple breaking and opening of the cells after transposase induction followed by examination of the transposition intermediates has been successfully used to detect active transposition of IS150 and IS911 (9, 16). However, this method failed to detect any intermediates after IPTG induction of the IS1237 transposases (data not shown).
Because the intermediates were not readily detected by the methods described above, another system was used to detect the regeneration of the closure of the donor DNA backbone after IS1237 was removed by transposition. This system allowed us to determine whether IS transposition did not occur through mechanisms involving a circular or linear intermediate (16). An IS1237::Amp cassette was created in which an active ampicillin resistance gene was inserted into two BamHI sites of IS1237 and replaced about 539 bp of the IS element. The IS1237::Amp cassette was inserted into an SmaI site of the kanamycin resistance gene of pETIS1, pETIS2, and pETIS3 to obtain plasmids pETIS1-IR, pETIS2-IR, and pETIS3-IR. These new plasmids did not exhibit the kanamycin resistance phenotype but did exhibit ampicillin resistance, and transposition of IS1237 by the cut-and-past mechanism should have yielded a kanamycin resistance clone. The pETIS1-IR, pETIS2-IR, and pETIS3-IR plasmids were individually transformed into E. coli strain BL21. After IPTG induction of the expression of the different forms of the IS1237 transposase, no kanamycin-resistant clone was detected, supporting the conclusion that the IS1237 element from the L. xyli subsp. cynodotis genome does not translate to an active transposase in E. coli. However, we cannot exclude the possibility that this IS element encodes the functional transposase in its native host. It is also possible that some factor encoded by the native host is required for IS1237 transposition. Another possible explanation is that the ancient IS1237 element encoded an active transposase and the present coding form is a degenerate form.
Analysis of the upstream and downstream flanking sequences reveals that IS1237 frequently inserts in other ISs.
The DNA fragments upstream and downstream of IS1237 were amplified by TAIL-PCR and sequenced. Thirteen and ten distinct flanking sequences were found upstream and downstream of IS1237, respectively (Fig. 4). The upstream sequences consisted of 98 bp of the 5′-terminal IS1237 and 89 to 733 bp of the Leifsonia xyli subsp. xyli genome sequence. The downstream sequences consisted of 67 bp of the 3′-terminal IS1237 and 113 to 740 bp of the L. xyli subsp. xyli genome sequence.
FIG. 4.
Diagram of 14 distinct upstream flanking sequences and 10 distinct downstream flanking sequences of IS1237 in the L. xyli subsp. cynodontis strain #3 genome. The recognition sequences TAA and TTA are indicated by bold type. Translational start codons are underlined. The shaded arrows with GenBank accession numbers represent contiguous flanking ORFs. The shaded boxes indicate the truncated genes which were interrupted by IS1237. Brief descriptions of flanking ORFs, where GenBank numbers are indicated, are given to the left or right of the flanking ORFs. The spacers between IS1237 elements and start or stop codons of the flanking genes are represented by bidirectional arrows, and their lengths are indicated. ID, identity; aa, amino acids.
It has been reported previously that IS1237 contains a duplicated TTA target sequence (23). All 13 upstream sequences were adjacent to the 5′ intact IS1237, and the target site of IS1237 insertion was TAA for seven genes and TTA for five genes. However, such a target site was not found in the fifth upstream sequence that encodes a putative transposase. All downstream sequences were adjacent to the intact 3′ IS1237, and the target site of IS1237 insertion was TAA for seven sequences and TTA for 3′ sequences. The presence of TAA and TTA target insertion sites both upstream and downstream of IS1237 indicates that IS1237 is present in multiple locations in the genome of L. xyli subsp. cynodotis because of transposition.
BLASTX searching of the GenBank database with the upstream sequences showed that nine of the upstream sequences encoding putative proteins exhibited significant levels of homology (45 to 97%) with known or putative protein-encoding genes of L. xyli subsp. xyli sequenced strain CTCB07. Homologous genes were identified for eight of the downstream sequences.
It is interesting that many sequences flanking IS1237 exhibit higher levels of homology with genes from gram-positive bacteria other than L. xyli subsp. xyli (Fig. 4), suggesting that there is a certain amount of genome diversity between L. xyli subsp. cynodontis and L. xyli subsp. xyli.
We noticed that the fourth, fifth, and sixth upstream sequences were very similar. However, the differences in the translational start codons of ORFs and the internal spacers between the ORFs and IS1237 demonstrated that these sequences were from three distinct loci of the L. xyli subsp. cynodontis genome. A 10-bp spacer and a TTA target sequence are present between the fourth upstream sequence and IS1237. A 56-bp spacer is present between the fourth upstream sequence and IS1237, and the TTA target sequence is missing. Although the internal spacer of the sixth sequence is the same as the internal spacer of the fourth sequence, the sixth sequence utilizes the ATG start codon, while the fourth sequence uses GTG. Nevertheless, these three sequences have an ORF that is significantly homologous to the sequence encoding the transposase of the known transposasble elements ISLxx4 and Tn5564. Tn5564 was identified in R-plasmid pTP10 of Corynebacterium striatum M82B. It has 22-bp inverted repeats flanked by 6-bp target site duplications, and the central region encodes the chloramphenicol resistance gene and a putative transposase (39).
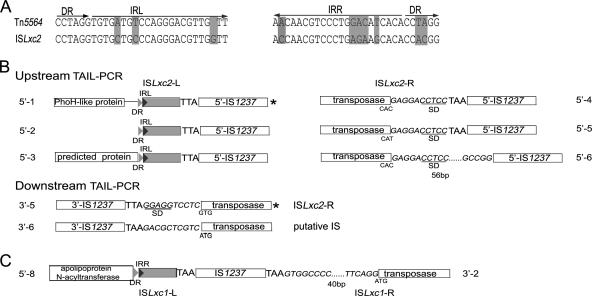
TAIL-PCR was performed twice to amplify the complete sequence of the L. xyli subsp. cynodontis transposase with specific primers Tn5564-p1, Tn5564-p2, Tn5564-p3, Tn5564-p4, Tn5564-p5, and Tn5564-p6 and arbitrary degenerate primers. In a comparison with the sequence of Tn5564, sequence analysis of the DNA fragments obtained revealed that both the full-length transposase and the downstream IR sequences essential for an active transposon were present in all three loci, indicating that a novel IS element of L. xyli subsp. cynodontis, designated ISLxc2 (GenBank accession no. EF176596), exists. However, the upstream IR sequence of ISLxc2 was not found between the spacer of its transposase and the adjacent IS1237, suggesting that IS1237 might insert inside ISLxc2. The downstream IR sequence of ISLxc2 was highly homologous (86%) to the sequence of Tn5564 (Fig. 5).
FIG. 5.
(A) Comparison of IR and direct repeat (DR) sequences of ISLxc2 and Tn5564. The residues of ISLxc2 that are different from Tn5564 residues are shaded. (B) Loci of IS1237 inserted into ISLxc2. TAIL-PCR analysis of the upstream and downstream sequences of IS1237 identified eight unique flanking sequences related to ISLxc2, and two of them (indicated by asterisks) were from the same locus. The shaded boxes indicate the DR and left IR (IRL) sequences of ISLxc2 and the adjacent ISLxc2 sequence before the 5′ transposition site of IS1237. The genes upstream of ISLxc2 are indicated by open boxes, and the putative proteins are indicated in the boxes. The direct repeat sequences of IS1237, TAA and TTA, are indicated by bold type. The spacer sequence and the lengths of the IS1237 and ISLxc2 transposase start codons (ATG and GTG) are shown. The underlined sequences are putative ribosome binding sites (SD). ISLxc2-L, left terminal sequence of ISLxc2; ISLxc2-R, transposase ORF and right terminal sequence of ISLxc2. (C) Locus of IS1237 inserted into ISLxc1. See above for an explanation. IRR, right IR sequence. ISLxc1-L, left terminal sequence of ISLxc1; ISLxc1-R, transposase ORFs and right terminal sequence of ISLxc1.
Interestingly, we found that the second upstream sequence (103 bp) was almost identical to a significant fraction of the first and third sequences, as well as to the upstream sequence of Tn5564, including the upstream IR sequence (Fig. 5). This observation indicated that IS1237 is inserted at a position opposite ISLxc2 in the loci containing the first, second, and third sequences. Obviously, the coding region of the third sequence was interrupted by an ISLxc2::IS1237 cassette. Full-length ISLxc2 with IS1237 and full-length ISLxc2 without IS1237 were both amplified from the L. xyli subsp. cynodotis genome, and the sequences were the same. This finding revealed a new IS element in L. xyli subsp. cynodotis that is highly homologous to Tn5564, and the newly identified ISLxc2 may transpose independent of or together with IS1237.
Strikingly, three downstream sequences, the second, fifth, and sixth sequences, all have the potential to encode transposases. The putative transposase encoded by the fifth sequence is identical to that encoded by the newly identified ISLxc2 element, supporting the hypothesis that there is a high frequency of transposition of IS1237 into ISLxc2. The sixth putative transposase gene exhibits 59% amino acid identity with the fifth putative transposase gene, indicating that the sixth sequence represents an ISLxc2-like IS element in L. xyli subsp. cynodontis. The putative transposase encoded by the second sequence does not exhibit significant homology with the transposase of ISLxc2, but it exhibits a high level of homology with a putative transposase encoded by the Rhodococcus erythropolis genome, indicating that the sequence is a novel transposon of L. xyli subsp. cynodontis, namely ISLxc1 (GenBank accession no. DQ191803). The transposase-encoding sequences and IR sequences of this novel putative transposon have been identified and will be published elsewhere.
Ten of the 13 upstream and 10 downstream sequences at loci that IS1237 occupies by transposition (as judged by the presence of TAA and TTA target sites) contained the sequences of three other IS elements and assembled to form IS1237::ISLxc(x) fusion cassettes; seven of these sequences are IS1237::ISLxc2 and two are IS1237::ISLxc1 (Fig. 5), suggesting that IS1237 has a high tendency to transpose into other IS elements.
IS1237 is a high-copy-number transposon in the chromosome of L. xyli subsp. cynodontis.
In addition to the TA-3′IS1237 fusion cassette that we found in the chromosome and plasmid of L. xyli subsp. xyli, 13 unique upstream and 10 unique downstream sequences of IS1237 were identified. The small difference in the number of sequences recovered upstream and downstream of IS1237 using the same experimental procedure might have been due to the fact that two shorter variants of IS1237 were covered for upstream sequence identification but not for downstream sequence identification because some specific primers for the downstream TAIL-PCR were in the deleted region of IS1237d1 and IS1237d2. In order to determine how many unique loci of IS1237 are present in L. xyli subsp. cynodontis, high-fidelity PCR amplification using primers pairing the upstream and downstream sequences was performed to determine how many of the unique sequences were from the same loci. All the upstream and downstream sequences containing the same IS1237 target site (TAA or TTA) were grouped together, and each upstream primer was combined with all downstream primers in the same group to perform PCR amplification. Only two pairs of primers specifically yielded PCR products that were the predicted length, and sequence analysis confirmed that the corresponding sequences were from the same loci. The upstream first and downstream fifth sequences constitute an IS1237::ISLxc2 locus, and the upstream eighth and downstream second sequences constitute an IS1237::ISLxc1 locus.
The identified 13 upstream and 10 downstream sequences of IS1237 were randomly amplified from the genome by TAIL-PCR, and two pairs of sequences happened to be from the same loci. Assuming that the possibilities of random PCR amplification from all of the IS1237 loci were the same, the maximum possible number of IS1237 loci was estimated to be 65.
IS1237 activates transcription of flanking genes by outwardly directed promoters at the terminus.
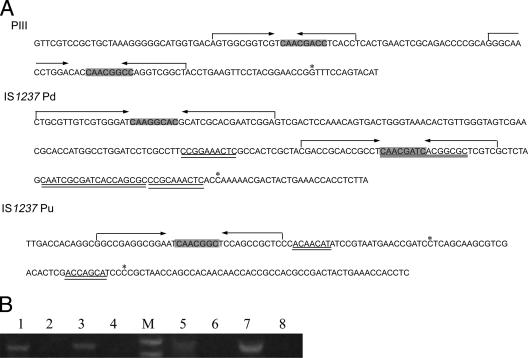
Many transposons and IS elements carry promoters that can activate transcription of flanking genes (3, 4, 6, 18, 19, 30, 36). Three strong promoters in the L. xyli subsp. cynodontis genome have been isolated by Haapalainen et al. (14). Direct repeat and reverse repeat sequences are rich in these promoters. The repeats preceding the transcription start site might be recognized and bound by the transcription regulatory proteins. Direct and reverse repeat sequences were found at the two termini of IS1237, and they may serve as promoters (Fig. 6A); the downstream promoter (Pd) contains two reverse repeat sequences resembling the L. xyli subsp. cynodontis strong promoter PIII, while the upstream promoter (Pu) contains only one reverse repeat. The organizations of these putative promoters are interesting; they are oriented in opposite outward directions and drive transcription of the adjacent genes upstream and downstream of IS1237.
FIG. 6.
(A) Comparison of IS1237 downstream promoter (Pd) and IS1237 upstream promoter (Pu) with a strong L. xyli subsp. cynodontis promoter, PIII. The arrows indicate the IR sequence. The direct repeat sequences are double underlined. The shaded sequences are the conserved motifs between IR sequences. The potential transcription start sites are indicated by asterisks. (B) RT-PCR to detect transcription of the GUS gene. In the RT reaction we used primer GUS7. Lanes 1 and 3, RT-PCR results for Pd obtained using 5′ PCR primers Pd1 and Pd2, respectively; lanes 2 and 4, negative controls; lanes 5 and 7, RT-PCR results for Pu obtained using 5′ PCR primers Pu1 and Pu2, respectively; lanes 6 and 8, negative controls; lane M, marker. The results demonstrate that Pu and Pd can promote transcription of the GUS gene.
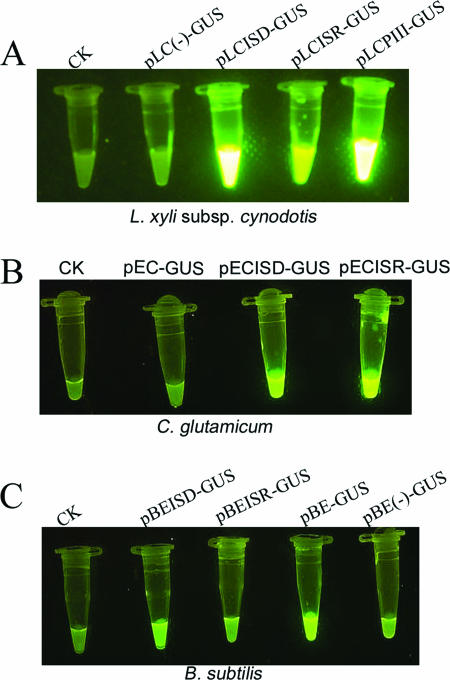
To examine if the putative IS1237 promoters at both ends could promote transcription bidirectionally, two IS1237-GUS elements in which IS1237 was placed in the same direction as the GUS gene or in the opposite direction were cloned into plasmids pLC011, pEC-K18mob2, and pBE2 in L. xyli subsp. cynodontis, C. glutamicum, and B. subtilis, respectively, for the promoter activity assay. The L. xyli subsp. cynodontis strong promoter PIII and an unrelated 850-bp sequence were used as the positive and negative controls, respectively, and Fig. 7 shows that IS1237 promoted the expression of the GUS gene regardless of the orientation in which it was inserted, but the GUS activity was much stronger when the downstream promoter was used than when the upstream promoter was used. These results suggested that the promoters at both ends of IS1237 are capable of driving GUS gene transcription in L. xyli subsp. cynodontis, while the downstream promoter is stronger than the right promoter, correlating with the finding that Pd has two reverse repeats while Pu has only one reverse repeat. In C. glutamicum, both IS1237 promoters activated transcription of the GUS gene; however, the extent of activation was less than that in L. xyli subsp. cynodontis (Fig. 7B). The activity of the IS1237 downstream promoter, but not the activity of the upstream promoter, was evident in B. subtilis. The activity of the downstream promoter activity was even lower than the activity of the SP6 promoter (Fig. 7C).
FIG. 7.
Promoter activity of IS1237 Pd and IS1237 Pu in L. xyli subsp. cynodontis, C. glutamicum, and B. subtilis. Strains without a plasmid were used as negative controls (CK). The plasmids harbored by the strains are indicated above the images.
The presence of predicted GUS mRNA in C. glutamicum was also confirmed by RT-PCR analysis (Fig. 6B). To determine the transcription start sites for Pu and Pd, 5′ RACE RT-PCR was performed to identify the 5′ end of the GUS mRNA. Interestingly, the sequencing results indicated that there is more than one unique 5′ end for each promoter (data not shown). The ends that occurred at a high frequency are likely to be the transcription start sites, which are shown in Fig. 6A. Interestingly, two transcription start sites are located 2 bp downstream of a direct repeat sequence. The repeated sequences might provide a binding site for the transcription regulatory protein to allow initiation of transcription immediately downstream.
We clearly demonstrated that both IS1237 terminal promoters are active in L. xyli subsp. cynodontis. The downstream terminal promoter Pd is as strong as the L. xyli subsp. cynodontis strong promoter PIII, while the upstream promoter Pu is less efficient. These IS1237 terminal promoters exhibit moderate activity in C. glutamicum and weak activity in B. subtilis, which could be due to differences in the homology of these gram-positive bacteria to L. xyli subsp. cynodontis. To the best of our knowledge, this is the first report of the presence of two active promoters at the termini of an IS in bacteria. Our results suggest that IS1237 may benefit its host organism, L. xyli subsp. cynodontis, by providing active promoters to transcribe the adjacent genes, which is consistent with our observation that most IS1237 copies are close to the coding region of the adjacent upstream and downstream genes. The strong IS1237 downstream promoter also provides a choice for efficient expression of desired genes in L. xyli subsp. cynodontis and C. glutamicum.
Acknowledgments
We thank Y. Zhou for his help in calculating the number of IS1237 loci. We acknowledge P. Yin and Wan-Ting Chen for their help.
This work was supported by the National Natural Science Foundation of China through grants 30270749 and 30470942 to T.-Y. Li and grant 30330170 to Y. Zhang.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search program. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blazey, D., and R. O. Burns. 1982. Transcriptional activity of the transposable elements Tn10 in the Salmonella typhimurum ilvGEDA operon. Proc. Natl. Acad. Sci. USA 79:5011-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlier, D., J. Piette, and N. Glansdorff. 1982. IS3 can function as a mobile promoter in E. coli. Nucleic Acids Res. 10:5935-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, L., T. Y. Li, and Y. Zhang. 2004. Rapid preparation of total nucleic acids from E. coli for multi-purpose applications. J. Biochem. Mol. Biol. 37:351-355. [DOI] [PubMed] [Google Scholar]
- 6.Ciampi, M. S., M. B. Schmid, and J. R. Roth. 1982. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc. Natl. Acad. Sci. USA 79:5016-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, M. J., and B. J. Augustin. 1984. Occurrence in Florida of the bacterium that cause bermudagrass stunting disease. Plant Dis. 68:1095-1097. [Google Scholar]
- 8.Davis, M. J., A. G. Gillaspie, Jr., A. K. Vidaver, and R. W. Harris. 1984. Clavibacter: a new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratton stunting disease of sugarcane and bermudagrass stunting disease. Int. J. Syst. Bacteriol. 34:107-117. [Google Scholar]
- 9.Duval-Valentin, G., C. Normand, V. Khemici, B. Marty, and M. Chandler. 2001. Transient promoter formation: a new feedback mechanism for regulation of IS911 transposition. EMBO J. 20:5802-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evtushenko, L. I., L. V. Dorofeeva, S. A. Subbotin, J. R. Cole, and J. M. Tiedje. 2000. Leifsonia poae gen. nov., sp. nov., isolated from nematode galls on Poa annua, and reclassification of “Corynebacterium aquaticum” Leifson 1962 as Leifsonia aquatica (ex Leifson 1962) gen. nov., nom. rev., comb. nov. and Clavibacter xyli Davis et al., 1984 with two subspecies as Leifsonia xyli (Davis et al., 1984) gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 50:371-380. [DOI] [PubMed] [Google Scholar]
- 11.Farabaugh, P. J., U. Schmeissner, M. Hofer, and J. H. Miller. 1978. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J. Mol. Biol. 126:847-863. [DOI] [PubMed] [Google Scholar]
- 12.Guo, X., H. Z. Xiong, M. Zhou, S. F. Jia, and Y. Xu. 1991. The construction of multifunctional shuttle vectors of Bacillus subtilis-Escherichia coli. Chin. J. Biotechnol. 7:224-229. [Google Scholar]
- 13.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haapalainen, M., M. Karp, and M. C. Metzler. 1996. Isolation of strong promoters from Clavibacter xyli subsp. cynodontis using a promoter probe plasmid. Biochim. Biophys. Acta 1305:130-134. [DOI] [PubMed] [Google Scholar]
- 15.Haapalainen, M. L., N. Kobet, E. Piruzian, and M. C. Metzler. 1998. Integrative vector for stable transformation and expression of a β-1,3-glucanase gene in Clavibacter xily subsp. cynodontis. FEMS Microbiol. Lett. 162:1-7. [DOI] [PubMed] [Google Scholar]
- 16.Haas, M., and B. Rak. 2002. Escherichia coli insertion sequence IS150: transposition via circular and linear intermediates. J. Bacteriol. 184:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallet, B., and D. J. sherratt. 1997. Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangement. FEMS Microbiol. Rev. 21:157-178. [DOI] [PubMed] [Google Scholar]
- 18.Hübner, A., and W. Hendrickson. 1997. A fusion promoter created by a new insertion sequence, IS1490, activates transcription of 2,4,5-trichlorophenoxyacetic acid catabolic genes in Burkholderia cepacia AC1100. J. Bacteriol. 179:2717-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kallastu, A., R. Horak, and M. Kivisaar. 1998. Identification and characterization of IS1411, a new insertion sequence which causes transcriptional activation of the phenol degradation genes in Pseudomonas putida. J. Bacteriol. 180:5306-5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchner, O., and A. Tauch. 2003. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 104:187-199. [DOI] [PubMed] [Google Scholar]
- 21.Kirillov, M. Y., Y. L. Shumakov, E. V. Nechaeva, S. Butcher, L. Sinjashina, K. Runeberg, M. Romantschuk, and G. I. Karataev. 1995. Repeated sequences isolated from Bordetella pertussis induce DNA rearrangements and deletions at high frequency. Gene 166:111-116. [DOI] [PubMed] [Google Scholar]
- 22.Kiss, J., M. Szabó, and F. Olasz. 2003. Site-specific recombination by the DDE family member mobile element IS30 transposase. Proc. Nat. Am. Sci. USA 100:15000-15005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laine, M. J., Y.-P. Zhang, and M. C. Metzler. 1994. IS1237, a repetitive chromosomal element from Clavibacter xyli subsp. cynodontis, is related to insertion sequences from gram-negative and gram-positive bacteria. Plasmid 32:270-279. [DOI] [PubMed] [Google Scholar]
- 24.Lampel, J. S., G. L. Canter, M. B. Dimoch, J. L. Kelly, J. J. Anderson, B. B. Uratani, J. S. Foulke, Jr., and J. T. Turner. 1994. Integrative cloning, expression, and stability of the CrylA(c) gene from Bacillus thuringiensis subsp. kurstaki in a recombinant strain of Clavibacter xyli subsp. cynodontis. Appl. Environ. Microbiol. 60:501-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, T. Y., P. Yin, Y. Zhou, Y. Zhang, Y. P. Zhang, and T. A. Chen. 2004. Characterization of the replicon of a 51-kb native plasmid from the gram-positive bacterium Leifsonia xyli subsp. cynodontis. FEMS Microbiol. Lett. 236:33-39. [DOI] [PubMed] [Google Scholar]
- 26.Li, T. Y., H. L. Zeng, Y. Ping, H. Lin, X. L. Fan, Z. G. Guo, and C. F. Zhang. 2007. Construction of a stable expression vector for Leifsonia xyli subsp. cynodontis and its application in studying the effect of the bacterium as an endophytic bacterium in rice. FEMS Microbiol. Lett. 267:176-183. [DOI] [PubMed] [Google Scholar]
- 27.Liao, C. H., and T. A. Chen. 1981. Isolation, culture and pathogenicity to Sudan Grass of a corynebacterium associated with ratoon stunting of sugarcane and with Bermuda grass. Phytopathology 71:1303-1306. [Google Scholar]
- 28.Liu, Y. G., and N. Huang. 1998. Efficient amplification of insert end sequences from bacterial artificial chromosome clones by thermal asymmetric interlaced PCR. Plant Mol. Biol. Rep. 16:175-181. [Google Scholar]
- 29.Lorincz, M. C., M. K. Parente, M. Roederer, G. P. Nolan, Z. Diwu, D. I. Martin, L. A. Herzenberg, and J. H. Wolfe. 1999. Single cell analysis and selection of living retrovirus vector corrected mucopolysaccharidosis VII cells using a fluorescence activated cell sorting-based assay for mammalian β-glucuronidase enzymatic activity. J. Biol. Chem. 274:657-665. [DOI] [PubMed] [Google Scholar]
- 30.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metzler, M. C., Y. P. Zhang, and T. A. Chen. 1992. Transformation of the gram-positive bacterium Clavibacter xyli subsp. cynodontis by electroporation with plasmids from the IncP incompatibility group. J. Bacteriol. 174:4500-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagy, Z., and M. Chandle. 2004. Regulation of transposition in bacteria. Res. Microbiol. 155:387-398. [DOI] [PubMed] [Google Scholar]
- 33.Ni, X. Q., Z. Y. Chen, M. Q. He, and B. X. Cai. 1999. Detection of antibiotics sensitivity and plasmid of beneficial bacillus. Chin. J. Microecol. 11:4. [Google Scholar]
- 34.Ohtsubo, E., and Y. Sekine. 1996. Bacterial insertion sequence. Curr. Top. Microbiol. Immunol. 204:1-26. [DOI] [PubMed] [Google Scholar]
- 35.Pearson, W. R., and D. J. Lipman. 1998. Improved tools for biological sequence comparison. Proc. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds, A. E., J. Felton, and A. Wright. 1981. Insertion of DNA activates the cryptic bgl operon in E. coli K-12. Nature 293:625-629. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Schneider, D., and R. E. Lenski. 2004. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res. Microbiol. 155:319-327. [DOI] [PubMed] [Google Scholar]
- 39.Tauch, A., T. Z. Zheng, A. Puhler, and J. Kalinowski. 1998. Corynebacterium striatum chloramphenicol resistance transposon Tn5564: genetic organization and transposition in Corynebacterium glutamicum. Plasmid 40:126-139. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, J., R. S. Stearman, and B. B. Uratani. 1993. Development of a native plasmid as a cloning vector in Clavibacter xyli subsp. cynodontis. Plasmid 29:241-244. [DOI] [PubMed] [Google Scholar]
- 41.Yu, B., Q. W. Shen, and G. J. Zhu. 2005. An improved method for integrative electrotransformation of Corynebacterium glutamicum with xenogenetic DNA. Chin. Biotechnol. 25:78-81. [Google Scholar]