Abstract
Biogenesis of the large ribosomal subunit requires the coordinate assembly of two rRNAs and 33 ribosomal proteins. In vivo, additional ribosome assembly factors, such as helicases, GTPases, pseudouridine synthetases, and methyltransferases, are also critical for ribosome assembly. To identify novel ribosome-associated proteins, we used a proteomic approach (isotope tagging for relative and absolute quantitation) that allows for semiquantitation of proteins from complex protein mixtures. Ribosomal subunits were separated by sucrose density centrifugation, and the relevant fractions were pooled and analyzed. The utility and reproducibility of the technique were validated via a double duplex labeling method. Next, we examined proteins from 30S, 50S, and translating ribosomes isolated at both 16°C and 37°C. We show that the use of isobaric tags to quantify proteins from these particles is an excellent predictor of the particles with which the proteins associate. Moreover, in addition to bona fide ribosomal proteins, additional proteins that comigrated with different ribosomal particles were detected, including both known ribosomal assembly factors and unknown proteins. The ribosome association of several of these proteins, as well as others predicted to be associated with ribosomes, was verified by immunoblotting. Curiously, deletion mutants for the majority of these ribosome-associated proteins had little effect on cell growth or on the polyribosome profiles.
Assembly of the bacterial ribosome requires the coordinated synthesis of three rRNAs (5S, 16S, and 23S) and 55 ribosomal proteins, processing and modification of the rRNAs and proteins, and assembly into functional units. Both the small and large ribosomal subunits can be assembled in vitro using purified ribosomal proteins and rRNAs, but the conditions required for assembly in vitro are much more stringent than the in vivo physiological environment (42, 43, 56, 61). It is likely that, as in eukaryotes, in vivo ribosome biogenesis in prokaryotes depends on additional assembly proteins to promote ribosome maturation.
Recent studies that combined biochemical affinity purification methods with proteomic techniques have revealed more than 170 nonribosomal proteins that transiently associate with different preribosomal particles in Saccharomyces cerevisiae (for reviews, see references 21, 22, 34, and 52). In contrast, however, only a few ribosome assembly factors have been identified in Escherichia coli. In general, these proteins were discovered through conventional genetic approaches or were predicted on the basis of their similarity to other ribosome-associated proteins. These proteins include GTPases, methyltransferases, pseudouridine synthases, RNA helicases, chaperones, and proteins with unknown function (1, 6, 9, 12, 13, 19, 27, 28, 31, 51, 54). Additional ribosome-associated proteins can be predicted from a relatively recent high-throughput protein complex interaction study in E. coli that yielded a list of proteins that copurify either with ribosomal proteins or with known ribosome assembly factors (10).
Our long-term goal is to delineate the cellular events that lead to the generation of the bacterial 50S ribosomal subunit. Toward this end, we describe the identification of a number of novel ribosome-associated proteins using the semiquantitative proteomic approach, iTRAQ (isotope tagging for relative and absolute quantitation), which allows for multiplex analysis of complex protein mixtures (50, 66). This proteomic technique combines standard multidimensional protein identification technology (MudPIT) (two-dimensional [2D] separation of tryptic peptides from complex mixtures, followed by identification of parent ions by tandem mass spectrometry [MS-MS]), with the use of isobaric tags added to the amine groups of tryptic peptides. Due to the different isobaric masses of the iTRAQ reporter groups, the relative amounts of proteins from different samples, expressed as ratios, are obtained (50, 66). We show that this is a robust approach for the study of bacterial ribosomes and that it is an efficient method by which to identify and assign proteins to distinct ribosomal particles. Furthermore, we measured the subunit association of ribosome-associated proteins and potential ribosome assembly factors. The subunit association of uncharacterized proteins, as well as additional proteins we predicted to associate with ribosomes, was verified by immunoblotting. Polysome profiling revealed that several mutants displayed ribosome assembly defects, consistent with a role in ribosome assembly.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and growth measurements.
E. coli strains used are listed in Table 1. E. coli cells were grown at the indicated temperatures in Luria-Bertani (LB) broth (10 g of tryptone, 5 g of yeast extract, 10 g of NaCl per liter) or on LB agar plates (LB plus 1.5% agar) containing antibiotics as required. Culture growth was monitored by measuring the absorption at 600 nm. Antibiotics were used at the following concentrations: 100 μg/ml ampicillin, 30 μg/ml kanamycin, and 20 μg/ml chloramphenicol.
TABLE 1.
E. coli strains used in this study
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| BW25113 | lacIqrrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH3 ΔrhaBADLD78 | 17 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal lon dcm (DE3 [lacI lacUV5-T7 gene 1 ind-1 sam-7 nin-5]) | Novagen |
| W3110 | (λ− F−) | 3 |
| JM4688 | ΔnudH-1 | 2 |
| JM4682 | ΔrluB | 2 |
| JM4889 | ΔrluC | 2 |
| JM4678 | ΔybcJ | 2 |
| JM4679 | ΔybeB | 2 |
| JM4681 | ΔycbY | 2 |
| JM4687 | ΔyfiF | 2 |
| JM4690 | ΔyhbY | 2 |
| JM4692 | ΔyibL | 2 |
| JM4508 | yibL::Kan | 33 |
| JM4195 | nudH with SPA tag | 10 |
| JM4197 | rluB with SPA tag | 10 |
| JM4196 | rluC with SPA tag | 10 |
| JM4198 | ybcJ with SPA tag | 10 |
| JM4310 | BL21(DE3), pJM21 ybeB with M2 tag | This work |
| JM4200 | ycbY with SPA tag | 10 |
| JM4202 | yfiF with SPA tag | 10 |
| JM4199 | yhbY with SPA tag | 10 |
| JM4991 | yibL with SPA tag | 10 |
PCR and cloning.
For ribosomal localization, ybeB was amplified by colony PCR using E. coli MG1655 cells as the source of template DNA and primers ybeBNSac1 (5′-ATGAGCTCGCCGCAACGACCATT-3′) and ybeBCBamHI (5′-GCGGATCCACTCCAGAGTTTTTCCAGT-3′) and Triplemaster DNA polymerase (Eppendorf) on an PTC-100 programmable thermal controller (MJ Research, Inc.). The ybeB gene was cloned into pJM21 (35) to form an in-frame fusion to a C-terminal M2 tag sequence. Thus, ybeB was under the control of its native promoter and expressed in E. coli BL21(DE3).
Cold sensitivity analysis of deletion strains.
Two 2-ml cultures of each strain were grown overnight at 37°C in LB, diluted to an optical density at 600 nm (OD600) of 0.1 in 5 ml LB, and grown to an OD600 of 0.4 to 0.5 at 37°C. Cultures were diluted to an OD600 of 0.15, four serial dilutions of 1:10 were made, and 5 μl of each dilution were dropped onto duplicate LB agar plates. Plates were grown at 37°C overnight or at 18°C for 3 to 5 days.
Preparation of cell lysates for ribosome profiles and immunoblotting.
All E. coli deletion strains were grown in LB at 18°C. The remaining strains were grown in LB at 37°C. Chloramphenicol (Fisher Biotech) was added to a final concentration of 200 μg/ml 30 seconds before harvest. Cells were harvested at an OD600 of 0.4 to 0.8 by centrifugation at 10,000 × g and 4°C for 10 min in an SLA-1500 rotor (Sorvall). The cell pellet was resuspended in 1 ml lysis buffer (10 mM Tris-Cl, pH 7.5, 10 mM MgCl2, 30 mM NH4Cl, 100 μg/ml chloramphenicol) per 100 ml of culture. The cell lysate was mixed with an equal volume of glass beads (300 μm; Sigma) and vortexed for 5 min at 4°C. The lysate was clarified by a 10-min centrifugation at 32,000 × g and 4°C in an SA-600 rotor (Sorvall). The supernatant was carefully collected and quantified by UV absorbance at 260 nm.
Polysome fractionation and quantification.
Sucrose gradients were formed in buffer I (10 mM Tris-Cl, pH 7.5, 10 mM MgCl2, 100 mM NH4Cl) using a gradient maker (SG15 or SG50; Hoeffer). Approximately 13 OD260 units of the cell lysates were loaded onto the top of the 10-ml 7 to 47% sucrose gradients, and the gradients were centrifuged in a Beckman SW41 Ti rotor for 3 h at 41,000 rpm (210,000 × g). The resulting gradients were fractionated manually using a Brandel gradient fractionator (model BR-186) connected to a syringe pump (model SYR-101). The syringe was filled with 60% sucrose, and the pump flow rate was set at 0.75 ml/min for the 10-ml gradients or at 1.5 ml/min for the 35-ml gradients. The UV absorbance (254 nm) of the samples was monitored and recorded by an ISCO UA-5 detector. To quantify the ribosomal subunit ratios, the lowest point on each polysome profile (between the 70S monosome and the first polysome) was used as the baseline, and the peak height for each subunit was determined as the distance between the highest point of each peak and the baseline. At least three independent polysome profiles were used to measure the peak heights and calculate the ratios between different peaks.
Immunoblot analysis.
All fractions were precipitated with 15% trichloroacetic acid and 0.03% deoxycholic acid, resuspended in sodium dodecyl sulfate (SDS) loading buffer, separated by SDS-polyacrylamide gel electrophoresis, and subjected to immunoblot analysis. Proteins separated by SDS-polyacrylamide gel electrophoresis were transferred to polyvinylidene difluoride membranes (NEN Life Science Products) with a Hoeffer TE77 semidry transfer apparatus (GE Healthcare). The immunoblot analyses were carried out as previously described (38). The following antibodies and concentrations were used: anti-Flag (Eastman Kodak), 1:2,000; goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Pierce), 1:20,000; rabbit anti-mouse immunoglobulin G conjugated to horseradish peroxidase (Sigma), 1:5,000.
iTRAQ sample preparation.
Cell lysates were obtained as previously described (62). Overnight saturated E. coli MG1655 cells were diluted 1:100 and were grown in LB either at 16°C (double duplex labeling and 16°C experiments) or 37°C (37°C experiment) until the OD600 reached 0.4 to 0.6. Cell lysates were sedimented either through 5 to 20% sucrose gradients in a Beckman SW28 rotor for 7 h at 28,000 rpm (double duplex labeling and 16°C experiments; see Fig. 1A and Fig. 3A) or through 15 to 45% gradients for 15 h at 24,000 rpm (37°C experiment; see Fig. 2A) and fractionated as previously described (31, 62). Fractions representing the desired ribosome particles from multiple gradients were pooled and precipitated with trichloroacetic acid as previously described (31, 62). The pellet was washed twice with acetone and resuspended in double-distilled H2O, and the amount of protein was estimated by Bradford assay (Bio-Rad). Protein samples were lyophilized by a speed vacuum prior to tryptic digestion and iTRAQ labeling (31). For the double duplex labeling experiment, iTRAQ reagents 114 and 116 were used to label tryptic peptides from the 30S particles, and reagents 115 and 117 were used to label tryptic peptides from the 70S particles. For particles obtained at 16°C, iTRAQ reagents 114, 116, and 117 were used to label 30S, 50S, and 70S, respectively. For particles obtained at 37°C, iTRAQ reagents 114, 115, 116, and 117 were used to label polysomes, 30S, 50S, and 70S, respectively. Peptides were separated by two-dimensional liquid chromatography (2DLC), and parent ions were identified on a Sciex/ABI 4700 matrix-assisted laser desorption ionization-tandem time of flight mass spectrometer as described previously (31). A number of proteins identified multiple times but by only a single parent ion were verified by de novo sequencing.
FIG. 1.
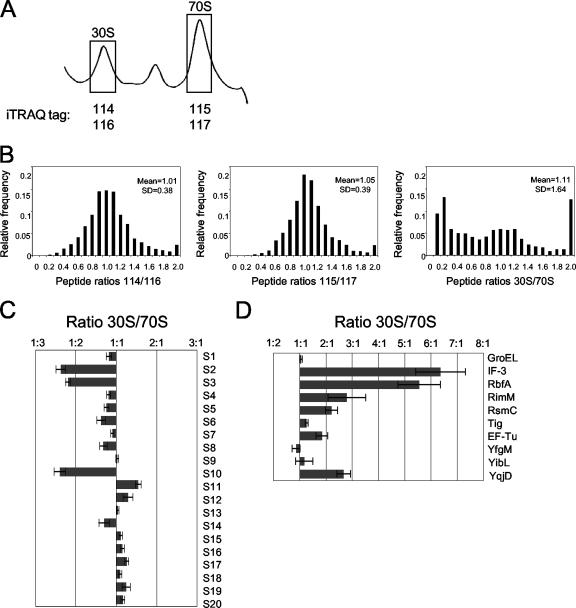
Relative ratios obtained using isobaric tags are reproducible in a double duplex labeling experiment. (A) Representative ribosomal profile trace of E. coli MG1655 grown at 16°C. Fractions representing 30S and 70S ribosomal particles (shown in boxes) were pooled and labeled with isobaric tags 114/116 and 115/117, respectively. (B) Relative distribution of peptides quantified with isobaric tags. Shown are the distributions of the peptide ratios for isobaric tags 114/116 (duplicate 30S), 115/117 (duplicate 70S), and (114 + 116)/(115 + 117) (30S/70S). For each plot, the mean and standard deviation (SD) are given. (C and D) The relative levels of small ribosomal proteins (C) and key associated proteins (D) are shown, expressed as the average ratio of proteins in 30S/70S particles plus standard error of the mean (error bars).
FIG. 3.
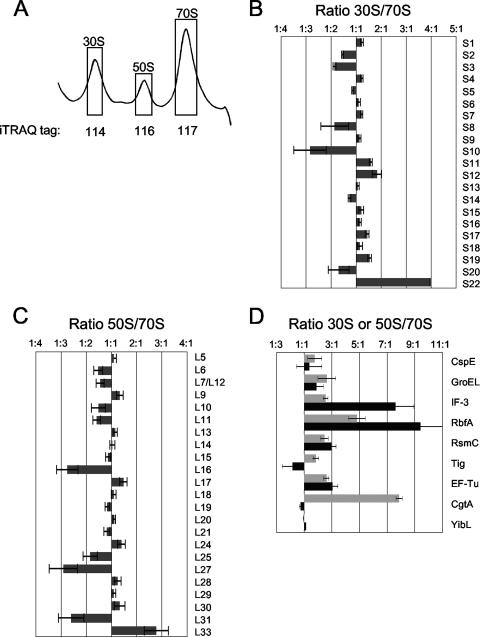
iTRAQ analysis of ribosomal particles isolated from E. coli MG1655 cells grown at 16°C. (A) Representative ribosomal profile trace of E. coli MG1655 grown at 16°C. Fractions representing 30S, 50S, and 70S ribosomal particles (shown in boxes) were pooled and labeled with isobaric tags 114, 116, and 117, respectively. Tag 115 was left blank. (B and C) Ratios of small ribosomal proteins (30S/70S) (B) and large ribosomal proteins (50S/70S) (C) are shown. (D) The ratios (30S/70S [black] or 50S/70S [dark gray]) of certain proteins are given. Error bars show standard errors of the means.
FIG. 2.
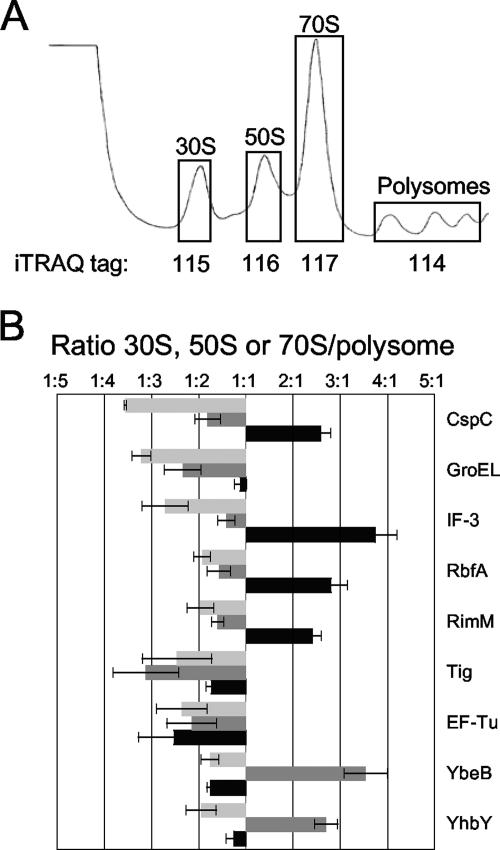
iTRAQ analysis of ribosomal particles isolated from E. coli MG1655 cells grown at 37°C. (A) Representative polysome trace of E. coli MG1655 grown at 37°C. Fractions representing polysomes and 30S, 50S, and 70S ribosomal particles (shown in boxes) were pooled and labeled with isobaric tags 114, 115, 116, and 117, respectively. (B) The relative ratios (30S/polysome, [black], 50S/polysome [dark gray], and 70S/polysome [light gray]) of select proteins are shown. Error bars show standard errors of the means.
iTRAQ quantification.
Data with >95% confidence of identification were used for peptide and protein identification calculations. Prior to iTRAQ quantification, any data set that contained a zero or null ratio was eliminated. For the double duplex labeling experiment, the ratios of (iTRAQ reagents 114 + 116)/(iTRAQ reagents 115 + 117) (30S/70S) were calculated for each parent ion. Data were normalized using the 30S/70S ratios of the average of all values for peptides derived from 30S subunit proteins. Ratios of iTRAQ reagent 114/116 and 115/117 were also calculated for each parent ion. These were normalized using the ratios of the average of all values for peptides derived from 30S proteins and total ribosomal proteins, respectively. For the 37°C experiment, the ratios of 30S/polysome, 50S/polysome, and 70S/polysome were calculated for each parent ion. Data were normalized using the subunit/polysome ratio of the average of all values for peptides derived from ribosomal proteins found in that subunit. Thus, data were normalized such that the 70S: polysome ratios were normalized against total ribosomal (large and small) proteins. Similarly, 30S/polysome ratios were normalized using only small ribosomal proteins, while 50S/polysome ratios were normalized against total large ribosomal proteins. For the 16°C experiment, the ratios of 30S/70S and 50S/70S were calculated for each parent ion. Data were normalized using the subunit/monosome ratio of the average of all values for peptides derived from ribosomal proteins found in that subunit. For all experiments, normalized ratios for all peptides of each protein were then averaged, and the standard deviation was calculated. Outliers were removed by application of the Grubbs' test with an alpha value of 0.05 (http://secure.graphpad.com/quickcalcs/Grubbs1.cfm). The data are displayed as ratios between select pairwise samples. Other relative ratios can be obtained by calculating the desired ratio from the data in our figures.
Proteomic data accession number.
The ProteomeCommons.org Tranche network (http://tranche.proteomecommons.org/) is used to provide fast downloads of verifiable, exact copies of the data described in the manuscript. The Tranche accession number for this data set is hgjMH/f+T7xmofYeZ15GdQRB/Pgyp9AUgrmXN8KprZuv2HG7MzD51LC7izKkGzeXxa+hE/hoUhChBuS6Lwvrtt/oY5g AAAAAAOQU1A==.
RESULTS
The iTRAQ approach is a powerful technique to study large complexes, such as ribosomes.
To identify novel ribosome-associated proteins, we combined a multiplexed protein semiquantitation method, iTRAQ (50), with ribosomal particle isolation via traditional sucrose density centrifugation. The power of this new technique is the ability to simultaneously identify proteins and compare the relative protein levels from up to four different protein complex mixtures (50). Using this method, we expected not only to identify potential ribosome-associated proteins but also to assign the proteins to individual ribosomal particles. In the iTRAQ approach, isobaric tags modify the N terminus and lysyl side chains of every peptide in a complex mixture. Isobaric tags have the same mass but fragment in a tandem mass spectrometer to provide differential mass tags. There are four different tags, each with a charged reporter (114 to 117 Da) and a neutral loss balancer (mass = 145 Da − reporter) coupled to a peptide reactive group (50), allowing up to four different protein samples to be labeled with a unique tag. In the mass spectrometer, identical, intact peptides labeled with different tags will have the same masses. After collision-induced dissociation in a tandem mass spectrometer, however, the resulting reporter ions differ by 1 to 4 Da and are easily detected as peaks at a mass-to-charge ratio (m/z) of 114, 115, 116, and 117. Semiquantitative information is obtained from the relative intensities of the reporter ions, while protein identification is based on the fragmentation pattern of the peptide that is available from the same MS-MS spectrum. Thus, semiquantitative information is obtained for every parent ion used for protein identification. Although iTRAQ is highly accurate (<6% error when testing on protein mixtures of known proportions), variations of up to 25% compared to the known values occur, as has been noted previously (30, 50).
We first performed a double duplex isobaric labeling experiment to validate the reproducibility of data obtained using the iTRAQ approach from this multiprotein particle. Ribosomal particles of E. coli cells grown at 16°C were isolated, and tryptic fragments of proteins obtained from 30S ribosomal fractions were labeled with 114 or 116 isobaric tag, while those from 70S fractions were labeled with 115 or 117 isobaric tag (Fig. 1A). Samples were mixed and separated by 2DLC, and parent ions were identified by MS-MS analysis and quantified by examining the ratios of released reporter ions for each peptide. A total of 1,922 peptides (>95% confidence) were identified, of which 462 were unique. Ninety-six proteins were identified with two or more independent peptide measurements or by a single measurement confirmed by de novo sequencing (see Table S1 in the supplemental material). Proteins identified by only a single measurement of a single peptide are listed for reference purposes (see Table S2 in the supplemental material) but were not included in subsequent analyses. As expected, comparison of the peptide ratios for all peptides identified shows a tight distribution between the duplicate 30S (114/116) and 70S (115/117) samples (Fig. 1B). Both ratios showed a mean around the expected value of 1.0 and a relatively small variation (standard deviations being 0.38 and 0.39 for 30S and 70S samples, respectively) (Fig. 1B). In contrast to the tight correlations observed between the duplicate 30S and 70S samples, however, but as expected when assessing dissimilar particles, such as the 30S and 70S ribosomes, the ratios observed comparing 30S to 70S subunits varied widely (mean of 1.11; standard deviation of 1.64; Fig. 1B). Taken together, these observations support the reproducibility of the iTRAQ method in the context of ribosomal particles.
Subunit association of known and novel ribosome-associated proteins.
From the double duplex labeling experiment, both ribosomal proteins and ribosome-associated proteins were identified. The identification of ribosomal proteins was excellent (39% of the identified proteins were ribosomal proteins) and included 20 of 22 small-subunit proteins and 24 of 33 large-subunit proteins (see Table S1 in the supplemental material). An additional three large ribosomal proteins were identified by only one peptide once (see Table S2 in the supplemental material). In general, the distribution of the individual small ribosomal proteins was similar in the 30S and 70S fractions with some key exceptions (Fig. 1C). The small ribosomal proteins S2, S3, and S10 were underrepresented in the 30S fraction (Fig. 1C). As all three of these proteins are assembled late in the in vitro 30S reconstitution system (15, 16), one possibility is that under the conditions used in this study (low temperatures), the in vivo incorporation of these three small ribosomal proteins into the 30S particles occurs late or is not fully achieved until the 70S particle is formed.
In addition to proteins from large complexes (such as dehydrogenases) that comigrate with ribosomal particles in sucrose gradients, a number of proteins involved in either ribosome assembly or translation, including GroEL, InfC (initiation factor 3 [IF-3]), RbfA, RimM, RsmC, Tig (Trigger factor), and TufA (EF-Tu), were identified. In the studies reported here, we focused on proteins that had multiple independent measurements and for which we had other data to suggest a role in ribosome function. Additional proteins identified in our studies are listed in Table S1 in the supplemental material. The assignment of these proteins to the ribosomal particles by iTRAQ is consistent with their function. For example, IF-3 is known to bind to 30S ribosomes and ensure the efficiency and fidelity of codon-specific formation of 30S initiation complexes (44). The formation of the 70S ribosome causes IF-3 to be released from the 30S particle (8, 44), agreeing with the relative enrichment of IF-3 in the 30S fractions seen by iTRAQ (Fig. 1D). Both RbfA and RimM have been implicated in 16S rRNA processing and 30S subunit biogenesis (11, 28, 29, 39, 64) and were accordingly found enriched in the 30S fractions (Fig. 1D). RsmC is a methyltransferase that specifically methylates m2G1207 of the 16S rRNA (57) and is also more abundant in the 30S fractions (Fig. 1D).
A number of uncharacterized proteins were also identified and may represent novel ribosome-associated proteins. Of these proteins, YibL could be expected to associate with the ribosome on the basis of its copurification with ribosomal proteins and known ribosome assembly factors (10) (Table 2). Iterative PSI-BLAST searches (not shown) indicate that YfgM shares sequence similarity with DnaJ (a cochaperone of DnaK, which is involved in assembly of both ribosomal subunits at high temperatures [20]). YqjD is uncharacterized but appears to associate primarily with the 30S fractions (Fig. 1D).
TABLE 2.
Summary of potential ribosome-associated proteins/ribosome assembly factors analyzed in this study
| Protein | iTRAQ location | Polysome locationa | Copurifying proteinsb | Mutant phenotype(s)c | Comment |
|---|---|---|---|---|---|
| NudH (YgdP) | Not found | Slightly larger than 30S | CsdA, SrmB, YcbY, RluB, RluC, and YfiF | Increased 30S and decreased 70S | (Di)nucleoside polyphosphate hydrolase |
| RluB (YciL) | Not found | ∼40S | CspA, CspC, CspD, CspE, CsdA, NudH, RluC, SrmB, YcbY, YfiF, YhbY, and YibL | Cold sensitive, increased 30S and 50S, decreased 70S | Pseudouridine synthase |
| RluC (YceC) | Not found | ∼40S | CspA, CspC, CspD, CsdA, NudH, RluB, SrmB, YbcJ, YcbY, YfiF, YhbY, and YibL | Cold sensitive, mild polysome defect | Pseudouridine synthase |
| YbcJ | Not found | Small portion with the 50S | CsdA, SrmB, YcbY, RluB, RluC, YfiF, and YibL | None | S4-like domain |
| YbeB | 50S | 50S | Large ribosomal proteins | None | |
| YcbY | Not found | ∼30S | CspA, CspC, CspD, CspE, CsdA, NudH RluB, RluC, SrmB, YbcJ, and YfiF | None | Putative methyltransferase |
| YfiF | Not found | ∼30S | CspC, CspD, CsdA, NudH, RluB, RluC, YcbY, YhbY, and YibL | None | Putative tRNA/rRNA methyltransferase |
| YhbY | 50S | 50S | CspC, RluB, RluC, YfiF, and YibL | Cold sensitive, increased 30S and 50S, decreased 70S | RNA binding protein |
| YibL | Unclear | Slightly larger than 30S | RluB, RluC, YbcJ, YfiF, and YhbY | None |
Additional iTRAQ studies reveal more candidates for ribosome-associated proteins.
We performed two additional iTRAQ experiments to identify additional proteins associated with the 30S, 50S, or translating (70S and polysomes) ribosomes. First, 30S, 50S, 70S, and polysome samples obtained from cells grown at 37°C were each individually labeled with a unique isobaric tag (Fig. 2A). Two independent 2DLC MS-MS experiments resulted in the identification of 2,960 total peptides (>95% confidence) with 349 unique peptides and 77 proteins. Seventy-two proteins had at least two measurements or were confirmed by de novo sequencing. Of these, 51 (71%) were ribosomal proteins. All small-subunit proteins except S22 were identified, as well as all large ribosomal subunit proteins with the exception of the small L34, L35, and L36 proteins (see Table S1 in the supplemental material), although L34 was detected once (see Table S2 in the supplemental material). As expected, all small ribosomal proteins are underrepresented in the 50S fractions compared to the polysomes (with an average relative 116/114 [50S/polysome] ratio of 0.496), as are large ribosomal proteins in the 30S fraction (with an average relative 115/114 [30S/polysome ratio] of 0.281). These ratios indicate that the fractions collected from the sucrose gradients are only modestly contaminated with adjoining particles (Fig. 2A).
A number of proteins, such as EF-Tu, GroEL, IF-3, RbfA, RimM, and Tig, were identified both in this study and in the double duplex labeling study (Fig. 1D; see Table S1 in the supplemental material). Additional proteins that were absent from the double duplex labeling analysis, such as CspC, GroES, YbeB, and YhbY, were found in this study (Fig. 2B; see Table S1 in the supplemental material). Several of these proteins associated with a specific ribosomal subunit, and the 30S association of IF-3, RbfA, and RimM were confirmed by this method (Fig. 2B). CspC also appears to be a 30S binding protein (Fig. 2B).
Of particular interest to us are YbeB and YhbY, both of which appear to associate with the 50S subunit (Fig. 2B). YbeB is a highly conserved 69-amino-acid protein, listed as one of the most common unknown proteins (23). It is similar to a plant protein, Iojap, which is involved in the stability of chloroplast ribosomes (60); therefore, its association with the 50S subunit is not unexpected. YhbY is a 97-amino-acid protein with a crystal structure that reveals an RNA binding fold similar to that of the 30S binding protein IF-3 (45). YhbY has recently been shown to associate with a pre-50S particle (4), consistent with our assignment to the 50S fractions.
Several ribosome assembly factors have been shown to be cold shock proteins whose abundance greatly increases upon lowering the growth temperature or proteins necessary for ribosome assembly only at cold temperatures. These include large ribosomal subunit assembly factors SrmB and CsdA and the small ribosomal subunit assembly factor RbfA (12, 13, 28, 64). Consistently, we obtained more RbfA peptides from cells grown at 16°C than at 37°C (22 and 8 proteins, respectively; see Table S1 in the supplemental material). Moreover, we failed to detect RsmC, YfgM, YibL, and YqjD from the 37°C samples. Therefore, we performed a further iTRAQ experiment from cells grown at a lower temperature, this time collecting the 30S, 50S, and 70S peaks (Fig. 3A). We identified 1,769 parent ions (508 unique) and obtained 101 protein identifications. Of these 101, 88 proteins had at least two independent measurements or a single measurement confirmed by de novo sequencing and were examined further. All of the small ribosomal proteins, except S21, and 25 of the large ribosomal proteins were identified (see Table S1 in the supplemental material), accounting for 52% of identified proteins. Of these, S22 is a newly identified component of the 30S ribosome, whose level is dramatically increased during stationary phase (59). Our results indicate that this protein is more abundant in the 30S fractions than in the 70S fractions (Fig. 3B), implying that S22 is not a core component of translating ribosomes and may be better described as a ribosome-associated protein. This is also consistent with S22 being observed at only 0.1 to 0.2 copy per 30S ribosome during exponential phase (59). As seen in the double duplex labeling experiment (Fig. 1C), S2, S3, and S10 were enriched in the 70S. In addition, the levels of three large ribosomal proteins that assemble late onto the 50S particle in vitro (26), L16, L27, and L31, were also reduced in the 50S fractions (Fig. 3C), consistent with their late assembly onto the maturing 50S particle in vivo. L33 appears to be enriched on the 50S particle (Fig. 3C), although we currently do not have an explanation for this. Equally mysterious is that although the large ribosomal proteins L1, L2, L3, L4, L22, and L23 were robustly identified from 50S particles from cells grown at 37°C, they were not identified in either study from cells grown at 16°C (see Table S1 in the supplemental material). These results may indicate differences between 50S particles formed in the cold compared to normal growth conditions. Consistent with this hypothesis, we consistently observe lower levels of free 50S particles than 30S particles for cells grown in the cold (Fig. 1A and 3A), an observation previously reported by others (12, 13).
A few additional ribosome-associated proteins were found in this study. Not unexpectedly, several cold shock or cold shock-like proteins, such as CspA, CspC, and CspE, were observed (see Table S1 in the supplemental material). This family of proteins share a cold shock domain that is involved in RNA binding (53). Ribosome association of proteins from this family has also been reported (41). CspA is a major cold shock protein that may facilitate translation by destabilizing secondary structure of mRNA at cold temperatures (31). CspC and CspE, however, are present constitutively even at 37°C (CspC was also identified in our 37°C iTRAQ analysis; see Table S1 in the supplemental material) and are proposed to be involved in the regulation of RpoS and UspA, two stress response proteins (46). Both proteins have RNA binding/melting activity and may also perform essential functions in the cold shock responses (47, 48, 63). Therefore, the ribosome association of Csp family proteins is not unexpected.
Verification of predicted ribosome-associated proteins via immunoblotting.
We predicted that several of the uncharacterized proteins that we identified by iTRAQ analysis (YbeB, YhbY, and YibL) were bona fide ribosome-associated proteins, as these proteins copurified with either ribosomal proteins, ribosome-associated proteins, or each other in a high-throughput affinity purification scheme (10). Moreover, several additional uncharacterized proteins, including NudH (YgdP), YbcJ, YcbY, and YfiF, also showed similar affinity purification partners (10) (Table 2), and we predicted that these proteins may also represent heretofore unknown ribosome-associated proteins. Therefore, to provide an independent assay to support their ribosome association, we examined the association of these proteins with ribosomal subunits by immunoblotting. For this purpose, we either episomally affinity tagged (C-terminal M2 tag) the protein or obtained the sequential peptide affinity (SPA)-tagged strains (10) and examined the locations of these tagged proteins using sucrose gradients followed by immunoblot analyses (Fig. 4). These studies, in general, confirmed our predictions for ribosome association. YhbY has recently been shown to be associated with a pre-50S particle (4). In our hands, YhbY appears to peak coincident with the 50S peak, although a trail of protein is seen in lighter fractions. YbeB is clearly associated with a 50S particle, although the majority of the protein is found at the top of the gradient and likely represents free protein. Curiously, YibL was associated with an early pre-50S particle, although the iTRAQ data suggested that it associated with 30S, 50S, and 70S particles equivalently (Fig. 3D). A possible explanation for this discrepancy is that in the iTRAQ experiment, we did not collect the fractions that contain the majority of YibL. If this is true, we may have been measuring the lower levels of protein that migrated in other fractions.
FIG. 4.
Ribosome association of select E. coli proteins. Extracts from cells expressing the indicated tagged proteins were separated by sucrose density centrifugation, and fractions were probed with relevant antibodies. The proteins were tagged with the SPA or M2 tag. The positions of the 30S, 50S, 70S, and polysome fractions, as determined by absorbance at 254 nm, are shown. The leftmost lane is a loading control (1/100 of the extract loaded).
Two pseudouridine synthases that modify the 23S rRNA (14, 18), RluB and RluC, were exclusively associated with particles migrating slightly slower than 50S particles (Fig. 4). This association pattern resembles that of the 40S ribosome-associated proteins CsdA and SrmB (12, 13), suggesting that the modification function of these two proteins occurs during 50S particle maturation. NudH (YgdP) was found to comigrate with a particle slightly bigger than the 30S particle (Fig. 4). NudH is involved in hydrolyzing diadenosine pentaphosphate, an alarmone signaling molecule related to stress response (5). YcbY comigrates with a 30S particle (Fig. 4). YcbY (renamed RimL) has recently been shown to methylate m2G2445 on the 23S rRNA (37), and therefore, the 30S particle is likely a very early pre-50S intermediate. YfiF (also encoding a methyltransferase) also appears to have a peak at the 30S peak, although the majority of the protein is in lighter fractions (Fig. 4). YbcJ, which has an S4-like RNA binding domain (58), displays weak association with the 50S particle (Fig. 4). Taken together, these data confirm the ribosome association of these proteins.
Deletion mutants of certain E. coli ribosome-associated proteins are cold sensitive.
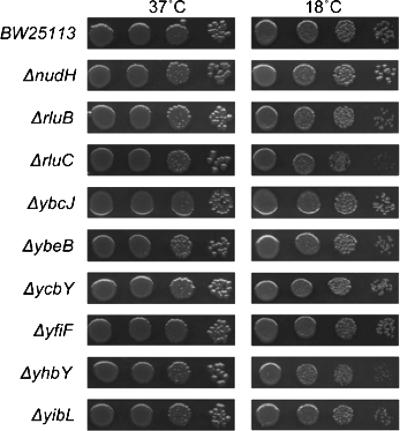
As many ribosome assembly mutants have shown cold sensitivity (9, 12, 13, 24, 25), deletion mutants with the nonessential potential ribosome-associated proteins identified in our study deleted were examined for their growth at 18°C by serial dilutions of exponentially growing cells (Fig. 5). Most of these mutants, including the ΔycbY mutant, which was reported to have a very slight growth defect in liquid (37), grew similarly to the isogenic wild-type control strains at all temperatures examined. Growth of ΔyhbY, ΔrluB, and ΔrluC mutants was impaired in the cold (Fig. 5). Interestingly, nudH mutants grew slightly better at low temperatures than the wild-type control did (Fig. 5).
FIG. 5.
Deletion of potential ribosome-associated proteins results in cold sensitivity in E. coli. The isogenic wild-type BW25113 and deletion strains of nonessential candidate proteins were grown to saturation, diluted into fresh medium to an OD600 of ∼0.1, and grown for 2 or 3 generations. Equivalent serial dilutions for each strain were plated onto LB and grown overnight at 37°C or for 3 days at 18°C.
Polysome profiling of deletion mutants discovers potential ribosome assembly factors.
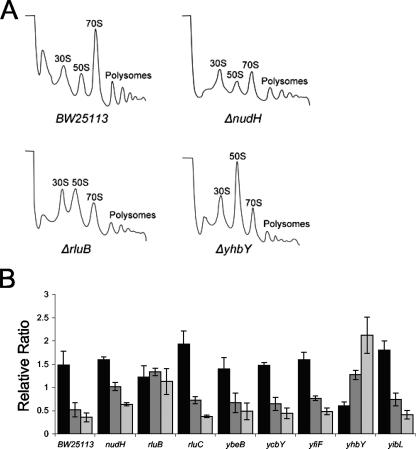
To further explore the function of these ribosome-associated proteins, we examined whether deletion of these proteins resulted in polysome defects as assayed by sucrose density centrifugation. An observed defect would be a strong indication that a particular protein is a ribosome assembly factor. Among the ribosome-associated proteins examined, three mutants showed dramatically altered polysome profiles. Deletion of NudH causes an evident increase in the 30S particles (Fig. 6A). Given that NudH is seen cosedimenting with a particle slightly bigger than the 30S particle (Fig. 5) and copurifies with large ribosome-associated proteins (Table 2), the accumulated particle may represent combined 30S and pre-50S particles. Deletion of the pseudouridine synthase RluB also results in the accumulation of free 30S and 50S particles, with a relative decrease in 70S particles and polysomes (Fig. 6A). Whether it is the modification of the 23S rRNA or the protein RluB itself that is important for the correct ribosome assembly remains to be determined. The ΔyhbY mutant had increased levels of 30S and 50S particles, with decreases in the levels of 70S particles and polysomes (Fig. 6A), consistent with its cold-sensitive phenotype (Fig. 5). This polysome defect is very similar to the defects observed in an ΔrrmJ mutant (9) and a cgtA mutant (31) and has recently been reported by others (4). Interestingly, yhbY is transcribed divergently from rrmJ, and overexpression of CgtA suppresses the polysome defect of the ΔrrmJ mutant (54), suggesting a close functional relationship among these three proteins. Deletion of the other genes had either no effect or modest effects on the polysome profiles (Fig. 6B).
FIG. 6.
Polysome profiles of select potential ribosome assembly factor mutants. Extracts from isogenic wild-type BW25113 and select mutants were separated by sucrose density centrifugation, and the positions of the 30S, 50S, 70S, and polysome fractions were monitored by UV absorbance (254 nm). (A) Polysome profiles of the wild type (BW25113) and ΔnudH, ΔrluB, and ΔyhbY mutants. (B) The average relative peak height ratios of 30S/50S, 30S/70S, and 50S/70S (black, dark gray, and light gray, respectively) from the indicated strains are shown. Error bars show standard errors of the means.
DISCUSSION
We show that the iTRAQ technology is a robust methodology to study the distribution of proteins in a large complex, such as the bacterial ribosome. This multiplex method allows for comparison of levels of multiple proteins between different samples simultaneously. Moreover, protein ratios are often the result of independent multiple measurements of different tryptic peptides, lending high confidence to the results obtained. The yield of ribosomal proteins identified here was excellent, and their assignments to the proper subunit were unambiguous. Moreover, a few ribosomal proteins (S2, S3, S10, L16, L27, and L31) that associate with the assembling ribosomal particle late (15, 26) were found to be underrepresented in the subunit fractions compared to the 70S monosomes, consistent with the subunit fractions containing late assembly intermediates as well as mature particles.
We show that the presence of nonribosomal proteins in our crude particle preparations is not problematic when using the iTRAQ approach to study ribosomal complexes isolated from sucrose gradients. In each of the studies reported here, approximately 15% of the identified peptides were from large protein complexes, such as dehydrogenases (see Table S1 in the supplemental material). One of the strengths of the iTRAQ approach is that the data are obtained from MS-MS fragmentation of specific peptide ions and, therefore, are independent of each other. Once the data are normalized to account for differences in protein levels in different samples, the presence of irrelevant proteins in the fractions examined do not affect the data for proteins of interest (in this case, ribosomal proteins and ribosome-associated proteins). Thus, we were able to accurately assign association to the proper subunit for all ribosomal proteins as well as known and discovered ribosome-associated proteins. Clearly, an advantage of working with sucrose gradient fractions is the ability to detect ribosome-associated proteins that may dissociate from the ribosome under more-stringent purification conditions.
In addition to ribosomal proteins, several known ribosome-associated proteins were identified, and their subunit association was assigned on the basis of the resulting isobaric tag ratios (Fig. 1D, 2B, and 3D). As with other proteomic studies, it appears that we detected only the most abundant ribosome-associated proteins, as a number of known helicases, pseudouridylases, and GTPases were not identified in this study. A likely possibility is that since the majority of the ribosomal particles examined in this study were mature particles, proteins that associate transiently or only with assembling ribosomal subunits would not be very abundant in these samples and were, therefore, missed using this approach. Regardless, we did identify several potential new ribosome-associated proteins, including YbeB, YfgM, YibL, YqjD, and YhbY. An improvement on this approach would be to examine intermediates that accumulate in select assembly mutant backgrounds.
Relatively recently, a high-throughput protein complex interaction study was performed in E. coli (10). Chromosomal copies of ∼850 proteins were tagged (with either the yeast-based TAP [tandem affinity purification] tag or the newly developed SPA tag), ∼650 protein complexes were affinity purified, and the interacting proteins were identified by MS (10). Included in this study are poorly characterized proteins that have been found to copurify either with ribosomal proteins or with known ribosome assembly factors. These results initiated our search for potential novel ribosome-associated proteins and potential ribosome assembly factors (Table 2). Toward this end, we verified the ribosome association of NudH, RluB, RluC, YbcJ, YcbY, and YfiF (Fig. 4). Because all of these proteins copurify with ribosomal proteins and with one or more of each other, we predict that they are all pre-50S particle-associated proteins.
The pre-50S particle association of several of these proteins is not unexpected. RluB and RluC are both pseudouridylases, and YcbY is a methyltransferase. These three proteins modify the 23S rRNA (14, 18, 37). Their association with particles smaller than 50S particles suggests that they function during 50S particle assembly with YcbY, on the basis of its association with a smaller particle, functioning prior to the pseudouridylases. YfiF is also predicted to be a methyltransferase, and we also predict that it functions very early in 50S particle synthesis. Two other proteins have predicted RNA binding motifs. YbcJ has an S4-like domain (58), whereas YhbY resembles the C terminus of IF-3 (45) and has a small motif shared with KH RNA binding domains (4). Very recently, YhbY has been shown by others to be associated with pre-50S subunits and required for ribosome assembly (4).
The roles of NudH and YibL, two other proteins shown to be associated with pre-50S particles, are unclear. We find the ribosome association of NudH particularly intriguing. NudH is a Nudix protein responsible for catalyzing the alarmone Ap5A (5). Alarmones are synthesized as by-products of tRNA charging aminoacyl tRNA synthetase reactions (65). In bacteria, Ap5A is synthesized to high levels under heat shock or oxidative stress (7, 36) and binds to heat shock and oxidative stress proteins (32, 55). We previously reported that SpoT, a hydrolase responsible for catalyzing another stress response alarmone, pppGpp, is associated with the ribosome-associated GTPase CgtA (62). One exciting possibility is that the state of the ribosome modulates the activities of these alarmone hydrolases, thereby playing a role in the bacterial stress response.
One striking finding is that many of the ribosome-associated proteins identified thus far are not required for ribosome assembly or cell growth. This includes many of the modifying enzymes (14, 18, 40, 49) and several conserved GTPases (S. M. Sullivan and J. R. Maddock, unpublished data). A likely scenario is that these proteins and/or their activities play more subtle roles in ribosome function, perhaps only under growth conditions not examined to date.
Taken together, the experiments described here demonstrate the power of the combination of traditional methods, such as sucrose gradients, with newer proteomics technologies, such as iTRAQ. The novel ribosome-associated proteins identified and confirmed here provide a starting point for protein-specific investigation of ribosome assembly processes. Furthermore, the advantage of the iTRAQ methodology is that it allows identification of proteins from entire fractions and/or entire particles that include integral controls, in the form of the known ribosomal proteins, to ensure that the separations of particles are effective and that the identifications of proteins are reliable. Moreover, the ability to obtain semiquantitative data describing the relative abundance of a given protein among the ribosomal particles provides information that extends beyond assignment of a protein to a specific subunit. Application of this combination of techniques to particles collected in a more specialized fashion, as for example, from assembly intermediates obtained from assembly mutants, will potentially not only identify more assembly factors but will also provide information as to the order of function of assembly factors, which is at the core of describing the process of in vivo ribosome assembly.
Supplementary Material
Acknowledgments
We warmly thank Nishi Patel and Matt Buszek for technical assistance and George Michailidis for helpful discussions.
P.C.A. is funded by NIH/NCRR National Resource for Proteomics and Pathways grant P41-18627.
Footnotes
Published ahead of print on 2 March 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alix, J. H., and M. F. Guerin. 1993. Mutant DnaK chaperones cause ribosome assembly defects in Escherichia coli. Proc. Natl. Acad. Sci. USA 90:9725-9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmann, B. J. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36:525-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barkan, A., L. Klipcan, O. Ostersetzer, T. Kawamura, Y. Asakura, and K. P. Watkins. 2007. The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein. RNA 13:55-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bessman, M. J., J. D. Walsh, C. A. Dunn, J. Swaminathan, J. E. Weldon, and J. Shen. 2001. The gene ygdP, associated with the invasiveness of Escherichia coli K1, designates a Nudix hydrolase, Orf176, active on adenosine (5′)-pentaphospho-(5′)-adenosine (Ap5A). J. Biol. Chem. 276:37834-37838. [DOI] [PubMed] [Google Scholar]
- 6.Bharat, A., M. Jiang, S. M. Sullivan, J. R. Maddock, and E. D. Brown. 2006. Cooperative and critical roles for both G domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA. J. Bacteriol. 188:7992-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bochner, B. R., P. C. Lee, S. W. Wilson, C. W. Cutler, and B. N. Ames. 1984. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell 37:225-232. [DOI] [PubMed] [Google Scholar]
- 8.Boelens, R., and C. O. Gualerzi. 2002. Structure and function of bacterial initiation factors. Curr. Protein Pept. Sci. 3:107-119. [DOI] [PubMed] [Google Scholar]
- 9.Bugl, H., E. B. Fauman, B. L. Staker, R. Zheng, S. R. Kushner, M. A. Saper, J. C. Bardwell, and U. Jakob. 2000. RNA methylation under heat shock control. Mol. Cell 6:349-360. [DOI] [PubMed] [Google Scholar]
- 10.Butland, G., J. M. Peregrin-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emil. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531-537. [DOI] [PubMed] [Google Scholar]
- 11.Bylund, G. O., L. C. Wipemo, L. A. Lundberg, and P. M. Wikstrom. 1998. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J. Bacteriol. 180:73-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charollais, J., M. Dreyfus, and I. Iost. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 32:2751-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charollais, J., D. Pflieger, J. Vinh, M. Dreyfus, and I. Iost. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 48:1253-1265. [DOI] [PubMed] [Google Scholar]
- 14.Conrad, J., D. Sun, N. Englund, and J. Ofengand. 1998. The rluC gene of Escherichia coli codes for a pseudouridine synthase that is solely responsible for synthesis of pseudouridine at positions 955, 2504, and 2580 in 23 S ribosomal RNA. J. Biol. Chem. 273:18562-18566. [DOI] [PubMed] [Google Scholar]
- 15.Culver, G. M. 2003. Assembly of the 30S ribosomal subunit. Biopolymers 68:234-249. [DOI] [PubMed] [Google Scholar]
- 16.Culver, G. M., and H. F. Noller. 2000. In vitro reconstitution of 30S ribosomal subunits using complete set of recombinant proteins. Methods Enzymol. 318:446-460. [DOI] [PubMed] [Google Scholar]
- 17.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Campo, M., Y. Kaya, and J. Ofengand. 2001. Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA 7:1603-1615. [PMC free article] [PubMed] [Google Scholar]
- 19.El Hage, A., and J. H. Alix. 2004. Authentic precursors to ribosomal subunits accumulate in Escherichia coli in the absence of functional DnaK chaperone. Mol. Microbiol. 51:189-201. [DOI] [PubMed] [Google Scholar]
- 20.El Hage, A., M. Sbai, and J. H. Alix. 2001. The chaperonin GroEL and other heat-shock proteins, besides DnaK, participate in ribosome biogenesis in Escherichia coli. Mol. Gen. Genet. 264:796-808. [DOI] [PubMed] [Google Scholar]
- 21.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 22.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 23.Galperin, M. Y., and E. V. Koonin. 2004. ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res. 32:5452-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustafsson, S., and B. C. Persson. 1998. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J. Bacteriol. 180:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutgsell, N. S., M. P. Deutscher, and J. Ofengand. 2005. The pseudouridine synthase RluD is required for normal ribosome assembly and function in Escherichia coli. RNA 11:1141-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herold, M., and K. H. Nierhaus. 1987. Incorporation of six additional proteins to complete the assembly map of the 50S subunit from Escherichia coli ribosomes. J. Biol. Chem. 262:8826-8833. [PubMed] [Google Scholar]
- 27.Himeno, H., K. Hanawa-Suetsugu, T. Kimura, K. Takagi, W. Sugiyama, S. Shirata, T. Mikami, F. Odagiri, Y. Osanai, D. Watanabe, S. Goto, L. Kalachnyuk, C. Ushida, and A. Muto. 2004. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res. 32:5303-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue, K., J. Alsina, J. Chen, and M. Inouye. 2003. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol. Microbiol. 48:1005-1016. [DOI] [PubMed] [Google Scholar]
- 29.Inoue, K., J. Chen, Q. Tan, and M. Inouye. 2006. Era and RbfA have overlapping function in ribosome biogenesis in Escherichia coli. J. Mol. Microbiol. Biotechnol. 11:41-52. [DOI] [PubMed] [Google Scholar]
- 30.Jagtap, P., G. Michailidis, R. Zielke, A. K. Walker, N. Patel, J. R. Strahler, A. Driks, P. C. Andrews, and J. R. Maddock. 2006. Early events of Bacillus anthracis germination identified by time-course quantitative proteomics. Proteomics 6:5199-5211. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, M., K. Datta, A. Walker, J. Strahler, P. Bagamasbad, P. C. Andrews, and J. R. Maddock. 2006. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 188:6757-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnstone, D. B., and S. B. Farr. 1991. AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J. 10:3897-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang, Y., T. Durfee, J. D. Glasner, Y. Qiu, D. Frisch, K. M. Winterberg, and F. R. Blattner. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186:4921-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kressler, D., P. Linder, and J. de la Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai, E.-M., U. Nair, N. D. Phadke, and J. R. Maddock. 2004. Proteomic screening and identification of the differentially distributed membrane proteins in Escherichia coli. Mol. Microbiol. 52:1029-1044. [DOI] [PubMed] [Google Scholar]
- 36.Lee, P. C., B. R. Bochner, and B. N. Ames. 1983. AppppA, heat-shock stress, and cell oxidation. Proc. Natl. Acad. Sci. USA 80:7496-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lesnyak, D. V., P. V. Sergiev, A. A. Bogdanov, and O. A. Dontsova. 2006. Identification of Escherichia coli m2G methyltransferases. I. The ycbY gene encodes a methyltransferase specific for G2445 of the 23 S rRNA. J. Mol. Biol. 364:20-25. [DOI] [PubMed] [Google Scholar]
- 38.Lin, B., D. A. Thayer, and J. R. Maddock. 2004. The Caulobacter crescentus CgtAC protein cosediments with the free 50S ribosomal subunit. J. Bacteriol. 186:481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lovgren, J. M., G. O. Bylund, M. K. Srivastava, L. A. Lundberg, O. P. Persson, G. Wingsle, and P. M. Wikstrom. 2004. The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits. RNA 10:1798-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lövgren, J. M., and P. M. Wikström. 2001. The rlmB gene is essential for formation of Gm2251 in 23S rRNA but not for ribosome maturation in Escherichia coli. J. Bacteriol. 183:6957-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikulik, K., Q. Khanh-Hoang, P. Halada, S. Bezouskova, O. Benada, and V. Behal. 1999. Expression of the Csp protein family upon cold shock and production of tetracycline in Streptomyces aureofaciens. Biochem. Biophys. Res. Commun. 265:305-310. [DOI] [PubMed] [Google Scholar]
- 42.Nierhaus, K. H. 1991. The assembly of prokaryotic ribosomes. Biochimie 73:739-755. [DOI] [PubMed] [Google Scholar]
- 43.Nierhaus, K. H., and F. Dohme. 1974. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl. Acad. Sci. USA 71:4713-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Connor, M., S. T. Gregory, U. L. Rajbhandary, and A. E. Dahlberg. 2001. Altered discrimination of start codons and initiator tRNAs by mutant initiation factor 3. RNA 7:969-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostheimer, G. J., A. Barkan, and B. W. Matthews. 2002. Crystal structure of E. coli YhbY: a representative of a novel class of RNA binding proteins. Structure 10:1593-1601. [DOI] [PubMed] [Google Scholar]
- 46.Phadtare, S., and M. Inouye. 2001. Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J. Bacteriol. 183:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phadtare, S., and M. Inouye. 1999. Sequence-selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol. Microbiol. 33:1004-1014. [DOI] [PubMed] [Google Scholar]
- 48.Phadtare, S., M. Inouye, and K. Severinov. 2002. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 277:7239-7245. [DOI] [PubMed] [Google Scholar]
- 49.Raychaudhuri, S., L. Niu, J. Conrad, B. G. Lane, and J. Ofengand. 1999. Functional effect of deletion and mutation of the Escherichia coli ribosomal RNA and tRNA pseudouridine synthase RluA. J. Biol. Chem. 274:18880-18886. [DOI] [PubMed] [Google Scholar]
- 50.Ross, P. L., Y. N. Huang, J. N. Marchese, B. Williamson, K. Parker, S. Hattan, N. Khainovski, S. Pillai, S. Dey, S. Daniels, S. Purkayastha, P. Juhasz, S. Martin, M. Bartlet-Jones, F. He, A. Jacobson, and D. J. Pappin. 2004. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3:1154-1169. [DOI] [PubMed] [Google Scholar]
- 51.Sato, A., G. Kobayashi, H. Hayashi, H. Yoshida, A. Wada, M. Maeda, S. Hiraga, K. Takeyasu, and C. Wada. 2005. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells 10:393-408. [DOI] [PubMed] [Google Scholar]
- 52.Saveanu, C., A. Namane, P.-E. Gleizes, A. Lebreton, J.-C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sommerville, J. 1999. Activities of cold-shock domain proteins in translation control. Bioessays 21:319-325. [DOI] [PubMed] [Google Scholar]
- 54.Tan, J., U. Jakob, and J. C. Bardwell. 2002. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 184:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanner, J. A., M. Wright, E. M. Christie, M. K. Preuss, and A. D. Miller. 2006. Investigation into the interactions between diadenosine 5′,5‴-P1,P4-tetraphosphate and two proteins: molecular chaperone GroEL and cAMP receptor protein. Biochemistry 45:3095-3106. [DOI] [PubMed] [Google Scholar]
- 56.Traub, P., and M. Nomura. 1968. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl. Acad. Sci. USA 59:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tscherne, J. S., K. Nurse, P. Popienick, H. Michel, M. Sochacki, and J. Ofengand. 1999. Purification, cloning, and characterization of the 16S RNA m5C967 methyltransferase from Escherichia coli. Biochemistry 38:1884-1892. [DOI] [PubMed] [Google Scholar]
- 58.Volpon, L., C. Lievre, M. J. Osborne, S. Gandhi, P. Inannuzzi, R. Larocque, M. Cygler, K. Gehring, and I. Ekiel. 2003. The solution structure of YbcJ from Escherichia coli reveals a recently discovered αL motif involved in RNA binding. J. Bacteriol. 185:4204-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wada, A. 1998. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells 3:203-208. [DOI] [PubMed] [Google Scholar]
- 60.Walbot, V., and E. H. Coe. 1979. Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc. Natl. Acad. Sci. USA 76:2760-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wireman, J. W., and P. S. Sypherd. 1974. In vitro assembly of 30S ribosomal particles from precursor 16S RNA of Escherichia coli. Nature 247:552-554. [DOI] [PubMed] [Google Scholar]
- 62.Wout, P., K. Pu, S. M. Sullivan, V. Reese, S. Zhou, B. Lin, and J. R. Maddock. 2004. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 186:5249-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia, B., H. Ke, and M. Inouye. 2001. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 40:179-188. [DOI] [PubMed] [Google Scholar]
- 64.Xia, B., H. Ke, U. Shinde, and M. Inouye. 2003. The role of RbfA in 16S rRNA processing and cell growth at low temperature in Escherichia coli. J. Mol. Biol. 332:575-584. [DOI] [PubMed] [Google Scholar]
- 65.Zamecnik, P. C., M. L. Stephenson, C. M. Janeway, and K. Randerath. 1966. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 24:91-97. [DOI] [PubMed] [Google Scholar]
- 66.Zieske, L. R. 2006. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J. Exp. Bot. 57:1501-1508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.