Abstract
The Escherichia coli Rsd protein forms complexes with the RNA polymerase σ70 factor, but its biological role is not understood. Transcriptome analysis shows that overexpression of Rsd causes increased expression from some promoters whose expression depends on the alternative σ38 factor, and this was confirmed by experiments with lac fusions at selected promoters. The LP18 substitution in Rsd increases the Rsd-dependent stimulation of these promoter-lac fusions. Analysis with a bacterial two-hybrid system shows that the LP18 substitution in Rsd increases its interaction with σ70. Our experiments support a model in which the role of Rsd is primarily to sequester σ70, thereby increasing the levels of RNA polymerase containing the alternative σ38 factor.
The requirement of σ factors for the recognition of bacterial promoters by RNA polymerase (RNAP) is well known (9). Most bacteria contain a principal σ factor which assures the expression of most genes and minor σ factors which are needed for the expression of subsets of genes, often in response to specific stresses (10). Thus, in Escherichia coli K-12, σ70, encoded by rpoD, is the principal σ factor, while six alternative σ factors are present at lower levels. One of these alternatives, σ38, encoded by rpoS, accumulates when cell growth ceases and in response to certain stresses, and is regarded as the stationary-phase σ factor (11). Although the role of σ38 is understood, it is not clear how it captures sufficient RNAP in order to ensure expression of the σ38 regulon. This is because σ38 has a weaker affinity for RNAP than σ70, and its level is always less than that of σ70 (14, 22). Studies from several laboratories have identified different factors that might favor the formation of RNAP containing σ38, and its activity, in stationary phase (7, 8, 17). One such factor, discovered by Jishage and Ishihama (15), is the Rsd protein (regulator of sigma D) that was found to accumulate in stationary phase and to bind to free σ70. Jishage and Ishihama (16) showed that Rsd could increase expression from the σ38-dependent bolA promoter and reduce expression from certain σ70-dependent promoters and argued that by sequestering σ70, Rsd permits σ38, and possibly other alternative σ factors, to access RNAP. Recent studies showed that, as well as forming a 1:1 complex with σ70, Rsd can also interact with RNAP in the absence of σ, raising the possibility that Rsd might also affect gene expression in E. coli by direct interactions with RNAP (13, 27). Thus, in this work, we used transcriptomics to assess possible roles for Rsd and genetic analysis to investigate the mechanism of action of Rsd. We also describe the use of the bacterial adenylyl cyclase two-hybrid system (BACTH; reference 18) to investigate Rsd-σ70 and Rsd-Rsd interactions.
MATERIALS AND METHODS
Bacterial strains.
The starting E. coli K-12 strains used in this work were MG1655 (2), the Δlac strain MC4100 (24), and the cya strain BTH101 (19). The method of Datsenko and Wanner (5) was used to delete the rsd gene of MG1655 and insert a kanamycin resistance cassette. To do this, the kan cassette in plasmid pKD4 was amplified by PCR with primers RSD P1 and RSD P2 (see Table 2) and the product was electroporated into MG1655. The Δrsd::kan insertion was confirmed by PCR with the flanking primers RSD P1 screen and RSD P2 screen. The Δrsd::kan allele was transferred into MC4100 by P1 transduction. The Datsenko and Wanner (5) method was also used to remove the kan insertion, and the rpoS-359 allele (rpoS::kan [4]) was introduced by P1 transduction to generate an MC4100 Δrsd rpoS::kan derivative.
TABLE 2.
Primers used in this work
| Primer use and name | Sequence |
|---|---|
| Creation and screening of rsd::kan allele | |
| RSD P1 | 5′-TGAGCAGTTTTTGAATACAAACTTGCGGAGTCAATCGTGTAGGCTGGAGCTGCTTC-3′ |
| RSD P2 | 5′-GCATTGAATGTAAATTACGCGTTAACAGCGCAGAACCATATGAATATCCTCCTTAG-3′ |
| RSD P1 screen | 5′-GATCCATAGCTCTTGCACTACC-3′ |
| RSD P2 screen | 5′-GATTAACTCTTGTTCCCTTCGC-3′ |
| Constructs for BACTH analysis | |
| RSD NT Xba | 5′-GACTCTAGAGATGCTTAACCAGCTCG-3′ |
| RSD CT Kpn | 5′-CTCGGTACCCGAGCAGGATGTTTGACGC-3′ |
| RSD CT Kpn-Stop | 5′-CTCGGTACCCGTCAAGCAGGATGTTTGACGC-3′ |
| RpoD NT Xba | 5′-GACTCTAGAGATGGAGCAAAACCCGCAG-3′ |
| RpoD CT Kpn-Stop | 5′-TTAGGTACCCGTTAATCGTCCAGGAAGCT-3′ |
| RIPF | 5′-GCATCTGCTCGTGGCT-3′ |
| RIPB | 5′-AAAACCTGTTGAAACTC-3′ |
| Cloning of promoters | |
| pgadAF | 5′-GCAGAATTCAGCAATGTTTGGGCGATTTTTATTAC-3′ |
| pgadAR | 5′-GCAAGCTTTATTTGAAGGCAATAAAAAAGTAGG-3′ |
| pgadBF | 5′-GCAGAATTCGATAATTCAGGAGACACAGAATGC-3′ |
| pgadBR | 5′-GCAAGCTTATGATTGGATCGCATTAAAAAAGTAGG-3′ |
| pybaSF | 5′-CGCAGAATTCAGGGTCAGGTCGATAGTTTGTG-3′ |
| pybaSR | 5′-GCACAAGCTTCCACTGCCTGCTGTAATTTG-3′ |
| Construction of pACYC RsdB and mutagenesis | |
| D49244 | 5′-CGATAAGCTTGCTGTTGTACTAACCAAACAGGTTC-3′ |
| D49245 | 5′-CGATCGGATCCAGATCTGCTCAGTGAGAAATGTAAAAACCATG-3′ |
| D49382 | 5′-GAGCAGATCTTGAATACAAACTTGCGGAGTCAATCATGC-3′ |
| D49383 | 5′-TGGTGCATGCGTTAACAGCGCAGAACTCAAG-3′ |
BACTH assays.
The E. coli cya strain BTH101 was transformed with derivatives of plasmids pU-T18 and pK-T25 encoding the T18 and T25 fragments of Bordetella pertussis adenylyl cyclase (Table 1). Transformants were plated on MacConkey maltose plates containing 100 μg/ml ampicillin and 50 μg/ml kanamycin. For β-galactosidase assays, transformants were grown aerobically at 30°C in LB medium containing 100 μg/ml ampicillin and 50 μg/ml kanamycin. Overnight cells were harvested and lysed with toluene, and activities were measured as described by Miller (23).
TABLE 1.
Plasmids used in this work
| Bacterial plasmid | Description | Reference |
|---|---|---|
| pRW50 | Broad-host-range lacZ fusion vector for cloning promoters on EcoRI-HindIII fragments, contains RK2 origin of replication, encodes Tetr | 21 |
| pACYC184 | Cloning vector; Cmr Tetr | 3 |
| pACYC184ΔHN | Derivative of pACYC184 with HindIII-NruI fragment removed; Cmr Tets | This work |
| pACYC-Rsd | E. coli rsd gene cloned between BamHI and SphI sites of pACYC184 | 16 |
| pACYC-RsdB | Derivative of pACYC-Rsd with shorter rsd insert cloned between HindIII and SphI sites of pACYC184, unique BglII site immediately upstream of rsd start codon | This work |
| pU-T18 | Contains B. pertussis cya gene T18 fragment, encodes Ampr | 18 |
| pK-T25 | Contains B. pertussis cya gene T25 fragment, encodes Kanr | 19 |
pU-T18 derivatives encoding Rsd-T18 fusions were made by cloning PCR-amplified DNA fragments encoding rsd into vector pU-T18 that had been digested with XbaI and KpnI. Wild-type and mutant rsd inserts were amplified with primers RSD NT Xba and RSD CT Kpn (Table 2), and the products were cleaved with XbaI and KpnI and purified prior to cloning. pK-T25 derivatives encoding T25-Rsd fusion derivatives were made by a similar procedure with the RSD NT Xba and RSD CT Kpn-Stop primers. The DNA sequences of recombinants were checked with two rsd internal primers with opposite orientations, RIPF and RIPB. Primers RpoD NT Xba and RpoD CT Kpn-Stop were used to amplify the full-length MG1655 rpoD gene by PCR. The product was cleaved with XbaI and KpnI and cloning into pK-T25 to create a derivative encoding a T25-σ70 fusion protein.
Transcriptomics.
To prepare RNA from bacteria in stationary phase, a 0.2-ml aliquot of an overnight culture grown aerobically at 37°C in LB medium (pH 7.4) supplemented with antibiotics as appropriate was inoculated into 50 ml of fresh medium in a 300-ml flask and grown to stationary phase while shaking at 180 rpm at 37°C for 8 h. The culture was mixed with 2 volumes of RNAprotect (QIAGEN) and incubated for 5 min, and cells were collected by centrifugation and stored at −80°C. Total RNA was purified with a QIAGEN RNeasy mini-kit according to the manufacturer's instructions. For each experiment, 20 μg total RNA was used to produce Cy3- and Cy5-labeled cDNA with the Amersham Cyscribe Postlabeling kit. To analyze the RNA, an E. coli oligonucleotide library (Sigma-Genosys) was printed by a Lucidea microarray spotter on Corning UltraGAPS slides. Full details of the preparation of the arrays, prehybridization, hybridization, and washing procedures were described by Kershaw et al. (20). In our experiments, plus and minus Rsd samples were labeled with Cy5 and Cy3 dyes, respectively. After hybridization and washing, microarray slides were scanned with a Fuji FLA8000 scanner and data for 4,289 genes were collated in scatter plots. Probes for each gene were spotted in duplicate, and all experiments were performed twice; hence, four datum points were measured for each gene. Data were analyzed and are presented as MA plots (described in reference 12) that display the effects of Rsd as a function of signal strength for each transcript, in order to identify genes where the expression changes are most significant. Our cutoff criterion for Rsd-dependent up-regulation was an intensity value of >1 and a relative log ratio (with and without Rsd) of >0.4 for at least three datum points.
Promoter cloning and activity measurements.
The E. coli gadA, gadB, and ybaS promoters were amplified by PCR from MG1655 genomic DNA with the primer pairs pgadAF and pgadAR, pgadBF and pgadBR, and pybaSF and pybasR. The PCR products were cleaved with EcoRI and HindIII, and the resulting fragments were purified and cloned between the EcoRI and HindIII sites of the broad-host-range lac expression vector pRW50 (Table 1) to create promoter-lac fusions. The control pmelR EcoRI-HindIII fragment was as described by Webster et al. (26). To assay expression from the different promoters, host strains were transformed with pRW50 derivatives and β-galactosidase activities were measured with the Miller protocol (23). Cells were grown aerobically in LB medium containing 35 μg/ml tetracycline (to maintain pRW50 derivatives) and 35 μg/ml chloramphenicol (to maintain pACYC-Rsd and derivatives). β-Galactosidase activities are reported as nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of dry cell mass. Each experiment was performed independently three times.
Multicopy plasmids encoding wild-type and mutant Rsd.
Vector plasmids pACYC184 and pACYC-Rsd were a gift from Akira Ishihama (Table 1). For use as a control, a Cmr Tets derivative of pACYC184, pACYC184ΔHN, was constructed by removing DNA between the HindIII and NruI sites. pACYC-RsdB is a derivative of pACYC-Rsd carrying a shorter insert carrying rsd and its regulatory region, with a unique BglII site immediately upstream of the rsd translation initiation codon. To construct pACYC-RsdB, the rsd regulatory region was amplified by PCR with primers D49244 and D49245 (Table 2) and the product was digested with HindIII and BglII. Similarly, the rsd open reading frame (ORF) was amplified with primers D49382 and D49383 and the product was cleaved with BglII and SphI. The HindIII-BglII and BglII-SphI fragments covering the rsd regulatory region and the rsd ORF were purified and cloned between the HindIII and SphI sites of pACYC184 to give pACYC-RsdB. To make libraries of random mutations in rsd, error-prone PCR by the protocol described by Barne et al. (1) was used, with primers D49382 and D49383 and Taq DNA polymerase (Bioline) to amplify BglII-SphI fragments covering the rsd ORF. These fragments were then recloned into pACYC-RsdB, and the resulting plasmid libraries were screened for mutations that altered Rsd function. Western blotting, with rabbit anti-Rsd sera kindly provided by Akira Ishihama, was used to check levels of Rsd expressed from mutant pACYC-RsdB derivatives. The Western blot assay was calibrated with purified Rsd prepared according to Westblade et al. (27).
Nucleotide sequence accession number.
Raw data from the transcriptome experiments described here (see Fig. 2) were deposited in the NCBI GEO database under accession number GSE5981.
FIG. 2.
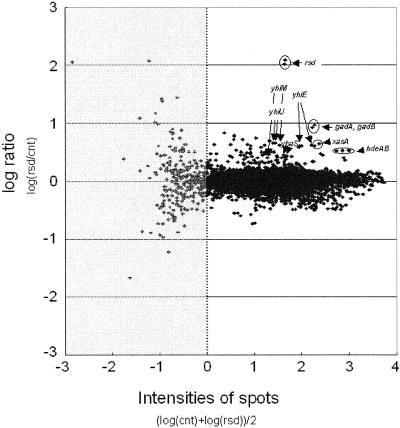
Transcript analysis of the effects of Rsd. Shown is a graphic representation of an experiment done to compare the abundance of RNA corresponding to 4,289 probes in MC4100 Δrsd::kan cells grown to stationary phase and carrying either pACYC-Rsd or the empty vector. Data are plotted vertically as the log of the relative ratio of the signal for each RNA in the presence (rsd) or absence (cnt) of overexpressed Rsd. The data are plotted horizontally as a function of the overall signal intensity (as in reference 12), thus permitting the identification of transcripts for which changes are significant. The subset of data where the signal intensity is insufficient for reliable analysis is shaded light gray. Fifteen genes were judged to be significantly up-regulated by Rsd (see text). Of these 15, 9 are known to be dependent on σ38, and the corresponding datum points are identified and annotated.
RESULTS AND DISCUSSION
A two-hybrid system can be used to detect Rsd-σ70 and Rsd-Rsd interactions.
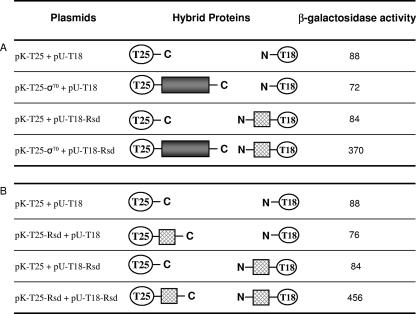
Biochemical studies with purified proteins have shown that Rsd can form a 1:1 complex with σ70 or a 1:1 complex with itself to form a dimer (27, 28). To measure Rsd-σ70 and Rsd-Rsd interactions in vivo, we used the BACTH assay system, which relies on the fact that activity of B. pertussis adenylyl cyclase requires that two independently folding domains, T18 and T25, be brought together (18). Compatible plasmids pU-T18 and pK-T25 encode T18 and T25, respectively, and contain restriction sites to permit the cloning of DNA encoding Rsd or σ70 to give protein fusions to T18 and T25. Figure 1 illustrates the different fusions that were constructed. Combinations of pU-T18 and pK-T25 and their derivatives were introduced into E. coli cya lac+ strain BTH101, and functional adenylyl cyclase activity, reconstituted from the T18 and T25 fragments, was measured by the induction of the lac operon (which is dependent upon cyclic AMP). Figure 1A lists the β-galactosidase activities measured in an experiment designed to monitor Rsd-σ70 interactions; activities above the background level were found only in cells carrying both an Rsd-T18 and a T25-σ70 fusion. Similarly, Fig. 1B lists the β-galactosidase activities measured in an experiment designed to monitor Rsd-Rsd interactions; activities above the background level were found only in cells carrying both an Rsd-T18 and a T25-Rsd fusion.
FIG. 1.
BACTH experiments to investigate Rsd-σ70 and Rsd-Rsd interactions. Each line illustrates an experiment in which β-galactosidase expression was measured in BTH101 cya cells containing pK-T25 and pU-T18 and derivatives. The first column lists the plasmids present in each experiment, and the extreme right-hand column lists the measured β-galactosidase activity. The central part illustrates the different proteins encoded by the plasmids. The B. pertussis adenylyl cyclase T18 and T25 domains are illustrated as ovals. Rsd fused to T18 or T25 is illustrated by hatched squares with N and C indicating the N- and C-terminal ends of fusion proteins. Similarly, σ70 fused to T18 or T25 is illustrated by a dark-shaded rectangle. Panels A and B illustrate experiments designed to monitor Rsd-σ70 and Rsd-Rsd interactions, respectively. Data shown are the average of six independent experiments.
Attempts to study rsd function by transcriptomics.
As a first step to studying the function of Rsd in vivo, a derivative of E. coli strain MG1655 in which the rsd gene was deleted and replaced with a kanamycin resistance cassette was made. Bacteriophage P1 transduction was then used to transfer the Δrsd::kan allele into E. coli strain MC4100. The profiles of transcripts in MC4100 and the Δrsd::kan mutant were then compared in cells growing exponentially and in stationary phase in LB medium. To do this, RNA was extracted from wild-type and mutant cells, differentially labeled, and analyzed with microarrays (see Materials and Methods). This analysis revealed no significant differences due to the rsd disruption (data not shown). The experiment was therefore repeated with the MC4100 Δrsd::kan strain carrying pACYC-Rsd, a multicopy plasmid carrying the rsd gene and regulatory region, or a control empty plasmid. The results of this experiment, illustrated in Fig. 2, show that the expression of a small number of transcripts in stationary phase is increased 3- to 10-fold by the overexpression of Rsd. Fifteen genes satisfied our criteria for increased expression (see Materials and Methods), i.e., ybaS, rmf, b0964, xasA, gadA, gadB, sufS, yhiM, hdeB, hdeA, yhiE, yhiU, yiiE, mopB, and pyrL. These include nine genes whose expression is known to be controlled by σ38, i.e., ybaS, xasA, gadA, gadB, yhiM, hdeB, hdeA, yhiE, and yhiU. Many of the products of these genes play key roles in ensuring the survival of E. coli in low-pH media (6, 25). These effects were observed in stationary phase but not in exponentially growing cells (data not shown).
Effects of Rsd measured by lac fusions.
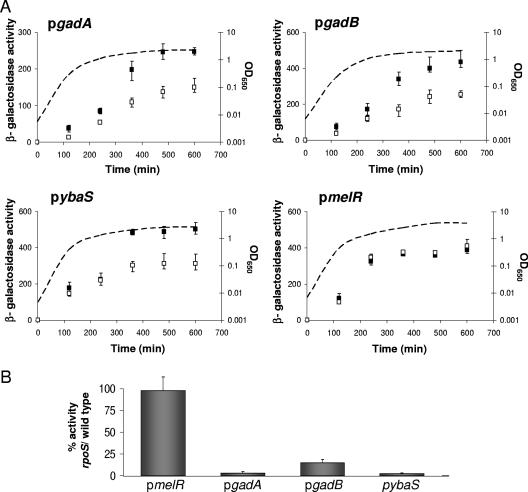
To confirm some of the effects of Rsd found by transcriptome analysis, PCR was used to amplify DNA fragments carrying the gadA, gadB, and ybaS promoters and the different fragments were cloned into pRW50, a broad-host-range promoter probe vector, to create promoter-lac fusions (Table 1). The resulting recombinant plasmids were transformed into the MC4100 Δrsd::kan strain carrying pACYC-Rsd or pACYC184ΔHN, and β-galactosidase activities were measured throughout the growth of each strain in LB medium. The results, illustrated in Fig. 3A, show that Rsd has a clear stimulatory effect on the gadA, gadB, and ybaS promoters. As a control, the experiment was repeated with the melR promoter, and as expected, Rsd had little or no effect on expression of the pmelR::lacZ fusion. Data from complementary experiments, illustrated in Fig. 3B, show that expression from the gadA, gadB, and ybaS promoters is substantially reduced by mutation of the rpoS gene, while expression from the melR promoter is unaffected.
FIG. 3.
Effects of Rsd on expression from different promoters. The four parts of panel A illustrate measurements of the expression of gadA::lac, gadB::lac, ybaS::lac, and melR::lac fusions during the growth of MC4100 Δrsd::kan cells carrying either pACYC-Rsd (with Rsd; filled squares) or the empty vector pACYC184ΔHN (without Rsd; open squares). Activities were measured at different times (x axes), and the corresponding A650 values of cultures are indicated by the dashed lines. Panel B shows the effects of rpoS disruption on the activity of the melR::lac, gadA::lac, gadB::lac, and ybaS::lac fusions after overnight growth in MC4100 Δrsd::kan cells carrying pACYC-Rsd. The histogram illustrates activities measured in the rpoS-359 mutant strain as a percentage of the activities in cells carrying wild-type rpoS. OD650, optical density at 650 nm.
Substitutions that affect the activity of Rsd at σ38-dependent promoters.
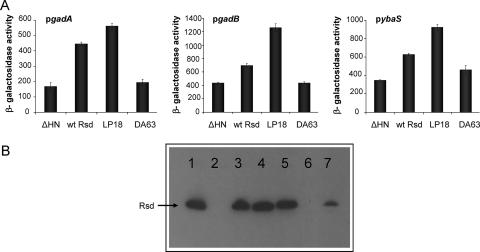
Two different mechanisms might account for the Rsd-dependent stimulation of expression from σ38-dependent promoters. One possibility is that the sequestration of σ70 by Rsd permits σ38 to capture more RNAP. The second possibility is that the direct interaction of Rsd with RNAP favors the recognition of certain σ38-dependent promoters. To distinguish between these possibilities, we searched for an Rsd mutant that was better able to stimulate expression from σ38-dependent promoters. Such a mutant would show improved binding to σ70 if effects are due to σ70 sequestration. Alternatively, if Rsd functions via interaction with RNAP, it would be unlikely that the mutant has improved affinity for σ70. To search for such mutations, we constructed a derivative of pACYC-Rsd carrying a BglII site immediately upstream of the rsd ORF and an SphI site immediately downstream. With this derivative, pACYC-RsdB, as a template, error-prone PCR was used to amplify the BglII-SphI fragment carrying the rsd gene. This fragment was then cloned back into pACYC-RsdB to give a library of plasmids carrying random base changes in the rsd gene. Three independent libraries were constructed, and these were transformed into the MC4100 Δrsd::kan strain carrying pRW50 with the pybaS::lacZ fusion and screened on MacConkey lactose indicator plates. Transformants carrying pACYC-RsdB and the control pACYC184ΔHN appear pink and white, respectively. After screening 1,000 colonies from each of the libraries, we identified 1 colony that exhibited a deeper pink Lac+ phenotype. Further investigation showed that this phenotype was due to a mutation that created the LP18 substitution in Rsd encoded by pACYC-RsdB. After purification, the pACYC-RsdB derivative encoding LP18 was transformed into MC4100 Δrsd::kan carrying the pybaS::lac, pgadA::lac, or pgadB::lac fusion cloned in pRW50. Figure 4A illustrates the results of an experiment designed to quantify the effect of the LP18 substitution at the ybaS, gadA, and gadB promoters. These results show that the Rsd-dependent stimulation of expression from each promoter is enhanced by the LP18 substitution. Western blotting was used to check the levels of wild-type and LP18 mutant Rsd in this experiment. Data in Fig. 4B show that the level of Rsd is unaffected by the LP18 substitution.
FIG. 4.
Effects of Rsd substitutions on expression from different promoters. The bar charts in panel A illustrate measurements of expression of gadA::lac, gadB::lac, and ybaS::lac fusions during the growth of MC4100 Δrsd::kan cells carrying either the empty vector pACYC184ΔHN; pACYC-RsdB, which encodes Rsd; or a derivative carrying the LP18 or DA63 substitution. β-Galactosidase activities were measured after overnight growth. Panel B illustrates a Western blot assay in which a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel was probed with anti-Rsd antibodies. Purified Rsd (27) was loaded in lane 1. The other lanes contain extracts from stationary-phase cultures of MC4100 Δrsd::kan pACYC184ΔHN (lane 2), MC4100 Δrsd::kan pACYC-RsdB (lane 3), MC4100 Δrsd::kan pACYC-RsdB LP18 (lane 4), MC4100 Δrsd::kan pACYC-RsdB DA63 (lane 5), MC4100 Δrsd::kan (lane 6), and wild-type MC4100 (lane 7).
To use the BACTH system to assess the effect of the LP18 substitution, a derivative of pU-T18 that encodes an LP18 Rsd-T18 fusion was constructed. Table 3 shows the results of a BACTH experiment designed to investigate the effects of the LP18 change on Rsd-σ70 and Rsd-Rsd interactions. The data show that the LP18 substitution increases the signal due to the Rsd-σ70 interaction while reducing the signal due to Rsd-Rsd contacts.
TABLE 3.
Effects of LP18 and DA63 substitutions in Rsd measured by BACTHa
| Protein encoded by pU-T18 | β-Galactosidase activity (Miller units) |
|---|---|
| In BTH101 cells carrying pK-T25 derivative encoding T25-σ70 | |
| T18 | 71 |
| WT Rsd-T18 | 341 |
| LP18 Rsd-T18 | 764 |
| DA63 Rsd-T18 | 77 |
| In BTH101 cells carrying pK-T25 derivative encoding T25-Rsd | |
| T18 | 75 |
| WT Rsd-T18 | 370 |
| LP18 Rsd-T18 | 78 |
| DA63 Rsd-T18 | 319 |
β-Galactosidase activities were measured in BTH101 cya cells carrying pK-T25 derivatives encoding a T25-σ70 or a T25-Rsd fusion protein and pU-T18 derivatives encoding T18 Rsd-T18 fusions. Cells were grown aerobically overnight at 30°C in LB medium supplemented with kanamycin and ampicillin. Data listed are averages of five to nine independent measurements that differed by no more than 20%. WT, wild type.
As a control for this experiment, we needed an Rsd mutant that was defective in the activation of expression from σ38-dependent promoters. To find this, we screened for pACYC-RsdB derivatives that were unable to stimulate expression of the pybaS::lacZ fusion carried by pRW50 in MC4100 Δrsd::kan cells. This experiment yielded pACYC-RsdB derivatives carrying either the LP134 substitution or a nonsense triplet at codon 43. Western blotting showed that the LP134 substitution resulted in a 10-fold reduction in Rsd levels (data not shown), and hence neither mutant was studied further. Prompted by the high-resolution structure of the Rsd-σ70 domain 4 complex, which shows that D63 of Rsd makes a key contact with σ70 (G. A. Patikoglou, L. F. Westblade, E. A. Campbell, V. Lamour, W. J. Lane, and S. A. Darst, personal communication), a derivative of pACYC-RsdB encoding DA63 Rsd was constructed. Since Western blotting showed that the DA63 substitution does not alter Rsd levels (Fig. 4B), the effect of DA63 substitution on the ybaS, gadA, and gadB promoters was measured. Data in Fig. 4A show that Rsd carrying the DA63 substitution is defective in activation of expression from these promoters. Complementary BACTH assays (Table 3) confirmed that the DA63 substitution destroys the Rsd-σ70 interaction while hardly affecting Rsd-Rsd contacts.
Conclusions.
At first sight, the function of the E. coli Rsd protein as an anti-σ70 factor appears simple. However, its low level relative to σ70, the absence of a phenotype in rsd mutants, and its interaction with RNAP lacking σ70 suggest that it might play a more subtle role (13, 16). In our study, we were able to measure biological effects of Rsd only when it was overproduced. Under these conditions, the expression of some σ38-dependent promoters was increased. Our observation that these effects were enhanced by a substitution in Rsd that also improves its interaction with σ70 argues that, here, Rsd is functioning by biasing the competition between σ70 and σ38 for available RNAP by sequestering σ70. The high-resolution structure of the Rsd-σ70 domain 4 complex shows that Rsd residue L18 plays no direct role in the Rsd-σ70 domain 4 interface (Patikoglou et al., personal communication). Thus, the effects of LP18 may be due to its destabilization of the Rsd dimer, thereby generating more monomer species to interact with σ70, or due to an improvement with another domain of σ70. Our results suggest that the role of Rsd may be to favor the expression of σ38-dependent genes that are needed for E. coli to respond to lower-pH conditions (6, 25). However, it is unclear why Rsd-dependent activation is restricted to a subset of σ38-dependent genes. We suggest that there may be some conditions under which the balance of levels of Rsd, σ70, and σ38 is set such that the presence of Rsd makes an important difference.
Acknowledgments
This work was funded by the Wellcome Trust and by the United Kingdom BBSRC via the Exploiting Genomics initiative and a Japan partnering award.
We are especially grateful to Akira Ishihama for generously supplying anti-Rsd sera and plasmids encoding Rsd, Seth Darst for communicating results prior to publication, and Naotake Ogasawara and Ryosuke Ito for help with transcriptome analysis and handling of data.
Footnotes
Published ahead of print on 9 March 2007.
REFERENCES
- 1.Barne, K. A., J. A. Bown, S. J. W. Busby, and S. D. Minchin. 1997. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the “extended −10” motif at promoters. EMBO J. 16:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conter, A., C. Menchon, and C. Guttierrez. 1997. Role of DNA Supercoiling and RpoS sigma factor in the osmotic and growth phase-dependent induction of the gene osmE of Escherichia coli K12. J. Mol. Biol. 273:75-83. [DOI] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster, J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2:898-907. [DOI] [PubMed] [Google Scholar]
- 7.Gaal, T., M. J. Mandel, T. J Silhavy, and R. L. Gourse. 2006. Crl is a pro-sigma factor that stimulates RNA polymerase holoenzyme formation. J. Bacteriol. 188:7966-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gourse, R. L., W. Ross, and S. T. Rutherford. 2006. General pathway for turning on promoters transcribed by RNA polymerases containing alternative σ factors. J. Bacteriol. 188:4589-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross, C. A., C. Chan, A. Dombroski, T. Gruber, M. Sharp, J. Tupy, and B. Young. 1998. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harbor Symp. Quant. Biol. 63:141-155. [DOI] [PubMed] [Google Scholar]
- 10.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57:441-466. [DOI] [PubMed] [Google Scholar]
- 11.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σs (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirai, M. Y., M. Klein, Y. Fujikawa, M. Yano, D. B. Goodenowe, Y. Yamazaki, S. Kanaya, Y. Nakamura, M. Kitayama, H. Suzuki, N. Sakurai, D. Shibata, J. Tokuhisa, M. Reichelt, J. Gershenzon, J. Pappenbrock, and K. Saito. 2005. Elucidation of gene-to-gene and metabolite-to-gene networks on Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 280:25590-25595. [DOI] [PubMed] [Google Scholar]
- 13.Ilag, L. L., L. F. Westblade, C. Deshayes, A. Kolb, S. J. W. Busby, and C. V. Robinson. 2004. Mass spectrometry of Escherichia coli RNA polymerase: interactions of the core enzyme with σ70 and Rsd protein. Structure 12:269-275. [DOI] [PubMed] [Google Scholar]
- 14.Jishage, M., and A. Ishihama. 1995. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of σ70 and σ38. J. Bacteriol. 177:6832-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jishage, M., and A. Ishihama. 1998. A stationary phase protein in Escherichia coli with binding activity to the major σ subunit of RNA polymerase. Proc. Natl. Acad. Sci. USA 95:4953-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jishage, M., and A. Ishihama. 1999. Transcriptional organization and in vivo role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase sigma D. J. Bacteriol. 181:3768-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jishage, M., K. Kvint, V. Shingler, and T. Nyström. 2002. Regulation of σ factor competition by the alarmone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 95:5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karimova, G., A. Ullmann, and D. Ladant. 2001. Protein-protein interaction between Bacillus stearothermophilus tyrosyl tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 3:73-82. [PubMed] [Google Scholar]
- 20.Kershaw, C. J., N. L. Brown, C. Constantinidou, M. D. Patel, and J. L. Hobman. 2005. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology 151:1187-1198. [DOI] [PubMed] [Google Scholar]
- 21.Lodge, J., J. Fear, S. Busby, P. Gunasekaran, and N. R. Kamini. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 95:271-276. [DOI] [PubMed] [Google Scholar]
- 22.Maeda, H., N. Fujita, and A. Ishihama. 2000. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28:3497-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 24.Peters, J. E., T. E. Thate, and N. L. Craig. 2003. Definition of the Escherichia coli MC4100 genome by use of a DNA array. J. Bacteriol. 185:2017-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σs-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webster, C., K. Gaston, and S. Busby. 1988. Transcription from the Escherichia coli melR promoter is dependent on the cyclic AMP receptor protein. Gene 68:297-305. [DOI] [PubMed] [Google Scholar]
- 27.Westblade, L. F., L. L. Ilag, A. K. Powell, A. Kolb, C. V. Robinson, and S. J. W. Busby. 2004. Studies of the Escherichia coli Rsd-σ70 complex. J. Mol. Biol. 335:685-692. [DOI] [PubMed] [Google Scholar]
- 28.Westblade, L. F. 2004. Studies on Escherichia coli Rsd. Ph.D. thesis. University of Birmingham, Birmingham, United Kingdom.