Abstract
Grixazone (GX), which is a diffusible yellow pigment containing a phenoxazinone chromophore, is one of the secondary metabolites under the control of A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) in Streptomyces griseus. GX production is also induced by phosphate starvation. The whole biosynthesis gene cluster for GX was cloned and characterized. The gene cluster consisting of 13 genes contained six transcriptional units, griT, griSR, griR, griAB, griCDEFG, and griJIH. During cultivation in a phosphate-depleted medium, the six promoters were activated in the order (i) griR, (ii) griC and griJ, and (iii) griT, griS, and griA. Disruption of griR, which encodes a SARP family transcriptional regulator, abolished the transcriptional activation of all other genes in the cluster. In addition, ectopic expression of griR from a constitutively active promoter resulted in GX overproduction even in the absence of AdpA, a key transcriptional activator in the A-factor regulatory cascade, and in the presence of phosphate at a high concentration. GriR monomers bound direct repeat sequences in the griC and griJ promoters in a cooperative manner. Therefore, the early active genes (griCDEFG and griJIH), all of which, except for griG (which encodes a transporter-like protein), encode the GX biosynthesis enzymes, were directly activated by GriR. The transcription of griR was greatly reduced in the presence of phosphate at a high concentration and was hardly detected in the absence of AdpA. These findings showed that both A-factor and phosphate depletion signals were required for griR transcription and both signals were transmitted to the GX biosynthesis genes solely via the griR promoter.
A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone; see Fig. 7 for its structure) is a chemical signaling molecule, or a microbial hormone, that triggers secondary metabolism and cell differentiation in Streptomyces griseus (10, 25). A-factor is gradually accumulated in a growth-dependent manner by the activity of AfsA, which is the key enzyme for A-factor biosynthesis. We have recently established the whole A-factor biosynthesis pathway, including the function of AfsA, which catalyzes β-ketoacyl transfer between dihydroxyacetone phosphate and 8-methyl-3-oxononanoyl-acyl carrier protein (12). When the concentration of A-factor reaches a critical level at or near the middle of the exponential growth phase, it binds the A-factor receptor protein (ArpA), which has bound and repressed the promoter of adpA, and dissociates ArpA from the promoter, thus inducing transcription of adpA (26). AdpA then activates a number of genes required for secondary metabolism and morphological differentiation, forming an AdpA regulon (13, 27). Members of the AdpA regulon that are involved in secondary metabolism include strR, the pathway-specific transcriptional activator for streptomycin biosynthesis (38), and an open reading frame (ORF) that encodes a probable pathway-specific regulator for a polyketide compound (39).
FIG. 7.
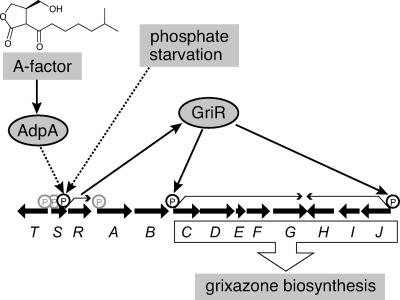
Model of the regulation of GX biosynthesis by A-factor and phosphate starvation. Both A-factor and phosphate depletion signals are required for the transcription of griR. GriR binds and activates the griC and griJ promoters, causing the transcription of all of the GX biosynthesis enzymes and production of GX. Because AdpA does not bind the upstream region of griR, AdpA is assumed to activate the griR promoter indirectly. The phosphate depletion signal is also transferred to the griR promoter, although its signaling pathway remains to be elucidated.
Grixazone (GX) is a diffusible yellow pigment containing a phenoxazinone chromophore produced by S. griseus (24). This yellow pigment actually consists of two structurally related compounds named GX-A and GX-B (see Fig. 1B for their structures). Because GX is one of the secondary metabolites under the control of A-factor, we were interested in the regulation of GX biosynthesis. In addition, GX production is also induced by phosphate starvation; GX is hardly produced in medium containing phosphate at concentrations of more than 2.5 mM (24). The negative control exerted by inorganic phosphate on the biosynthesis of many different types of antibiotics and other secondary metabolites has been observed for many years. However, the molecular mechanism of the regulation of secondary metabolism by phosphate remained obscure. Therefore, GX biosynthesis by S. griseus makes a good target to study the regulation of secondary metabolite formation not only by A-factor but also by phosphate.
FIG. 1.
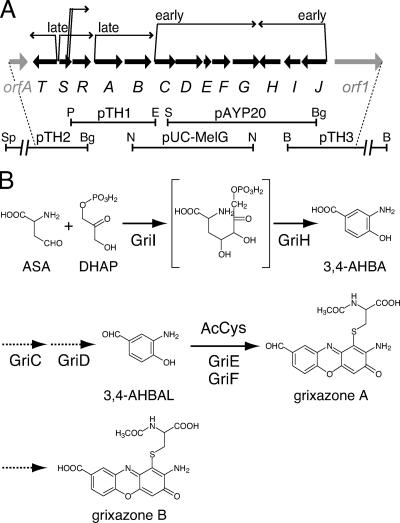
Organization of the GX biosynthesis gene cluster (A) and a proposed GX biosynthesis pathway (B). (A) Bold arrows indicate the positions and directions of the GX biosynthesis genes (griT, -S, -R, and -A to -J) and two neighboring genes (orfA and orf1). The extents of the DNA fragments cloned on vectors are shown below the arrows. Six transcriptional units are also shown above the arrows. Restriction enzyme abbreviations: B, BamHI; Bg, BglII; E, EcoRI; N, NcoI; P, PstI; S, Sau3AI; Sp, SphI. (B) The catalytic functions of GriI, GriH, GriE, and GriF have been elucidated by in vitro analysis with purified enzymes, and their steps in GX biosynthesis are indicated by solid arrows. The functions of GriC and GriD have been elucidated by in vivo analysis, and their steps are indicated by dotted-line arrows. A key intermediate in GX biosynthesis, 3,4-AHBA, is synthesized from two primary metabolites, aspartate-4-semialdehyde (ASA) and dihydroxyacetone phosphate (DHAP), by the action of GriI and GriH. 3,4-AHBA is reduced to 3,4-AHBAL by the action of GriC and GriD, probably in an ATP-dependent manner. GX-A is synthesized from two molecules of 3,4-AHBAL and N-acetylcysteine (AcCys) by the action of GriE and GriF. Conversion of GX-A to GX-B is presumably catalyzed by an oxidase that is not specific to GX-A and is encoded out of the GX biosynthesis gene cluster.
To investigate the regulation of GX biosynthesis by A-factor and phosphate, we cloned the GX biosynthesis gene cluster. As described previously (36), we generated a GX-deficient mutant strain, named M31, from S. griseus IFO13350 by UV mutagenesis and used it as a host for cloning the GX production genes. We obtained a 7.5-kb DNA fragment that caused mutant M31 to produce a GX-like yellow pigment by shotgun cloning with a chromosomal DNA library of the wild-type S. griseus strain (36). Characterization of several genes on this fragment revealed that this fragment was part of the GX biosynthesis gene cluster (35, 36). We then cloned and sequenced several DNA fragments including upstream and downstream regions of the fragment to cover the whole GX biosynthesis gene cluster. In this paper, we describe (i) identification of the whole GX biosynthesis gene cluster composed of 13 genes; (ii) transcriptional analyses of the GX biosynthesis genes; (iii) disruption of griR, which encodes a SARP (Streptomyces antibiotic regulation protein) family transcriptional regulator; (iv) binding of recombinant GriR to the griC and griJ promoters in a cooperative manner; and (v) production of GX by ectopic expression of griR in an adpA disruptant and in the presence of phosphate at a high concentration. On the basis of these results, we concluded that GriR served as the pathway-specific transcriptional activator for GX biosynthesis and both A-factor and phosphate depletion signals are transmitted to the GX biosynthesis genes solely via the promoter of griR. A vivid contrast with the A-factor control on streptomycin biosynthesis (38) was that AdpA indirectly activated the griR promoter, perhaps via an activator under the control of AdpA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. griseus IFO13350 was obtained from the Institute of Fermentation, Osaka, Japan (IFO). S. griseus mutants ΔadpA (26) and M31 (36) were described previously. S. griseus strains were grown at 30°C in YMPD medium (yeast extract [Difco], 0.2%; meat extract [Kyokuto], 0.2%; Bacto peptone [Difco], 0.4%; NaCl, 0.5%; MgSO4·7H2O, 0.2%; glucose, 1%; pH 7.2) and SMM medium [glucose, 0.9%; l-asparagine, 0.9%; (NH4)2SO4, 0.2%; Trizma base, 0.24%; NaCl, 0.1%; K2SO4, 0.05%; MgSO4·7H2O, 0.02%; CaCl2, 0.01%; trace element solution (9), 1%; pH 7.2] containing 0.25 or 2.5 mM KH2PO4. The precultured cells in YMPD were washed twice with SMM and inoculated into fresh SMM at 2% (vol/vol). For solid medium, 2.2% agar was added. Escherichia coli JM109 and vector pUC19 for DNA manipulation were purchased from Takara Biomedicals. E. coli JM110 containing dam and dcm mutations was used for preparing nonmethylated Streptomyces DNA used for gene disruption. Thiostrepton (25 μg/ml) and neomycin (10 μg/ml) were added when necessary. Media and growth conditions for E. coli were described by Maniatis et al. (17). Ampicillin (50 μg/ml) and kanamycin (50 μg/ml) were used for E. coli when necessary.
General recombinant DNA studies.
Restriction enzymes, T4 DNA ligase, and other DNA-modifying enzymes were purchased from Takara Biomedicals. [α-32P]dCTP (110 TBq/mmol) for DNA labeling with the Takara BcaBest DNA labeling system and [γ-32P]ATP (220 TBq/mmol) for end labeling at 5′ ends with T4 polynucleotide kinase were purchased from Amersham Biosciences. DNA was manipulated in Streptomyces (9, 15) and in E. coli (4, 17) as described earlier. Nucleotide sequences were determined by the dideoxy-chain termination method with the Thermo Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham) or the CEQ DTCS Quick Start Kit (Beckman Coulter) on an automated DNA sequencer.
Cloning of the DNA fragments carrying the GX biosynthesis genes.
The isolation of pAYP20, which had a 7.5-kb Sau3AI fragment containing six complete (griD to -I) and two truncated (griC and griJ) ORFs, was described previously (36). pUC-MelG, which had a 6.1-kb NcoI fragment containing four complete (griC-F) and two truncated (griB and griG) ORFs, was obtained from K. Ueda (7). We cloned three DNA fragments containing the flanking regions by a standard method including Southern hybridization and colony hybridization (see Fig. 1A). With the 1.2-kb NcoI-EcoRI fragment on pUC-MelG as a 32P-labeled probe, a 4.3-kb PstI-EcoRI fragment was cloned in pUC19, resulting in pTH1. With the 0.8-kb PstI-BglII fragment on pTH1 as a probe, a 6.3-kb SphI-BglII fragment was cloned into pUC19, resulting in pTH2. Furthermore, with the 1.0-kb PmaCI-BglII fragment on pAYP20 as a probe, an 8.7-kb BamHI fragment was cloned into pUC19, resulting in pTH3. The nucleotide sequences of these fragments were determined.
S1 nuclease mapping.
Total RNA was isolated with ISOGEN (Nippon Gene) from cells grown in SMM. The method of S1 nuclease mapping was described previously (14). Hybridization probes were prepared by PCR with a pair of 32P-labeled and nonlabeled primers. Table 1 lists the forward (F) and reverse (R) primer sequences. The PCR primers used for low-resolution S1 mapping were glkA-F and glkA-R* for glkA, griT-F and griT-R* for griT, griS-F and griS-R* for griS, griR-F and griR-R* for griR, griA-F and griA-R* for griA, griB-F and griB-R* for griB, griC-F and griC-R* for griC, griE-F and griE-R* for griE, griG-F and griG-R* for griG, griH-F and griH-R* for griH, griI-F and griI-R* for griI, griJ-F and griJ-R* for griJ, orf1-F and orf1-R* for orf1, and orfA-F and orfA-R* for orfA. Primers with asterisks were labeled at the 5′ end with [γ-32P]ATP by using T4 polynucleotide kinase before PCR. glkA, which encodes a glucokinase (1; our unpublished data), was used to check the purity and amount of RNA used. Primers used for high-resolution S1 mapping were griT-H-F and griT-H-R* for griT, griS-H-F and griS-H-R* for griS, griR-H-F and griR-H-R* for griR, griA-H-F and griA-H-R* for griA, griC-H-F and griC-H-R* for griC, and griJ-F and griJ-H-R* for griJ. Protected fragments were analyzed on 6% polyacrylamide DNA sequencing gels by the method of Maxam and Gilbert (19).
TABLE 1.
Primers used in this study
| Gene and primer | Positionsa | Sequenceb (5′ to 3′) |
|---|---|---|
| glkA | ||
| glkA-F | −144 to −125 | CACCGAACGCATCGACCTGG |
| glkA-R | +130 to +111 | TCGCGTCGACGATGCCTTCG |
| griT | ||
| griT-F | −233 to −214 | CGGTCCGCCAGCAGCACCGC |
| griT-R | +227 to +208 | CAGTAGACGGCGCAGCCGCC |
| griT-H-F | −182 to −163 | GCAAGCCCCAACATTCCCGC |
| griT-H-R | +57 to +38 | CCGGGAGAGCAGTTCCGCCC |
| griS | ||
| griS-F | −277 to −258 | CCGCGTCGTACGGCATGAGC |
| griS-R | +104 to +85 | CGTCCGATGCTCCGGTCCGC |
| griS-H-F | −121 to −102 | GTCGTCCCGCTCCAACCCCC |
| griS-H-R | +92 to +73 | CGGTCCGCCAGCAGCACCGC |
| griR | ||
| griR-F | −639 to −620 | ATGTTGGGGCTTGCGGTCGC |
| griR-R | −193 to −212 | CTCCACGACGGCCACCTTGG |
| griR-H-F | −475 to −456 | GATCGTGGTCTTCGCGATCG |
| griR-H-R | −304 to −323 | CTTGACCCACCGCACCTCCG |
| griA | ||
| griA-F | −312 to −293 | AGCGGGAGATCGCGGACCTG |
| griA-R | +146 to +126 | TGCCGCCCGATCCGGTTGAAC |
| griA-H-F | −233 to −214 | CTGACCGGCGTCCGCCATCG |
| griA-H-R | +13 to −7 | CAAGGCGCGGCGGCGATGAG |
| griB | ||
| griB-F | −255 to −236 | CTGAACTTCCTCGCGGGCGC |
| griB-R | +119 to +100 | GCGGTGACGCCGAAGACGAC |
| griB-RT | +1342 to +1323 | GCCGCCCCAGGGTGAAGGCG |
| griC | ||
| griC-F | −279 to −260 | TGGCGAAGTCGATGTCGCTC |
| griC-R | +131 to +112 | TCGGTGACGGGATCGAAACC |
| griC-H-F | −118 to −99 | GTTCCCCGACCCGCGGACAC |
| griC-H-R | +67 to +48 | CACCGAACTGCCAGGCCATC |
| griC-PCR-F | +12 to +34 | CGATGATCCCGAATTCGACAGTC |
| C1-F | −134 to −115 | CCGTGCGGGCGGAAGCGTTC |
| C1-R | −35 to −72 | GCGGTACGGCACCCATCGTG |
| C2-R | −106 to −125 | CGGGTCGGGGAACGCTTCCG |
| C3-F | −62 to −38 | CCCGCCCGCACGATGGGTGCCGTAC |
| griE | ||
| griE-F | −395 to −376 | GGAGACCGTCGTGGAGCTGC |
| griE-R | +43 to +24 | CCCCGGTGGTGGCCATGACC |
| griF | ||
| griF-RT | +439 to +420 | TCAGGGCCGGATCCTCGGGG |
| griG | ||
| griG-F | −261 to −242 | GACGTGCCCCACTACCTGCC |
| griG-R | +154 to +135 | CGAAGGTGGTCCAGCTGCCG |
| griG-RT | +956 to +937 | GTGGCCAGCAGGGCCAGCCC |
| griH | ||
| griH-F | −143 to −124 | TCATCCACAACAGCGGCACC |
| griH-R | +260 to +241 | TCCTCCGGGATCTGCTTCCC |
| griH-RT | +908 to +889 | CCCGTCTTCAGTTCGCTCAG |
| griI | ||
| griI-F | −294 to −274 | CGCCAAGGTCTCGATCGTCGG |
| griI-R | +107 to +87 | CCGTCGGTGATCGAGTGGTCG |
| griJ | ||
| griJ-F | −133 to −114 | ACCGCGTCCGCACTGTCCCG |
| griJ-R | +163 to +144 | TGTCGCCCTGTGCGGAGACC |
| griJ-H-R | +39 to +20 | CACCACGTCGCCCGTGGGAC |
| J1-R | −34 to −52 | CCCGAGCACTGCGACGAGC |
| J2-F | −395 to −376 | CAGCGGACCCGGGCATTCGG |
| J2-R | −107 to −129 | TGCGTATCGGGACAGTGCGGACG |
| J3-F | −62 to −40 | CCTCCTGGAAGCTCGTCGCAGTG |
| J1-m-F | −133 to −90 | ACCGCGTCCGCACTGTCCCGATACGCACGTGGCGACCCGCCGGT |
| orfA | ||
| orfA-F | −68 to −49 | CAACTCTTGCGGAGGCCGAC |
| orfA-R | +338 to +319 | GCTCCGGTCGTCTTCAGTCC |
| orf1 | ||
| orf1-F | −334 to −310 | CCGTCTCTCCGTCGCACCGGCGGGC |
| orf1-R | +132 to +108 | GGGGTCCTCGGGCGGCAGCGCGAAG |
Nucleotide positions are relative to the first nucleotide of the start codon, which is +1.
The mutated nucleotides are underlined.
Reverse transcription (RT)-PCR.
Total RNA was isolated with the RNAqueous phenol-free total RNA isolation kit (Ambion) from cells grown in SMM containing 0.25 mM KH2PO4. cDNA was synthesized with Super Script II reverse transcriptase (Gibco BRL) with the purified RNA and the reverse primers, according to the manual of the supplier. Table 1 lists the primer sequences. RNA was removed by digestion with RNase H. cDNAs were then used to synthesize double-stranded DNAs by PCR with EX-Taq polymerase. For detection of transcripts for griAB, primer griB-RT was used for RT, and primers griB-F and griB-R were used for amplification of the cDNA. For griCDEF, griF-RT was used for RT and griC-PCR-F and griF-RT were used for amplification of the cDNA. For griFG, griG-RT was used for RT and griG-F and griG-R were used for amplification of the cDNA. For griJIH, griH-RT was used for RT and griI-F and griH-R were used for amplification of the cDNA.
Gene disruption.
For disruption of griR, a 5.6-kb SphI-BamHI fragment was excised from pTH2 and placed between the SphI and BglII sites of pTH1, resulting in pUC-ΔgriR. pUC-ΔgriR contained a deleted griR sequence that encodes GriRΔLeu32-Glu253 with the 5.6-kb upstream and 3.3-kb downstream regions from griR. A 1.1-kb HindIII fragment carrying the kanamycin and neomycin resistance gene cassette from Tn5 (5) was placed at the HindIII site in the multicloning sequence of pUC-ΔgriR, resulting in pUC-ΔgriR-Km. This plasmid was denatured by alkali (23) and introduced by protoplast transformation into S. griseus IFO13350, and neomycin (5 μg/ml)-resistant colonies were isolated. After one of the neomycin-resistant transformants had been cultured in the absence of neomycin, neomycin-sensitive colonies were isolated as candidates for true griR disruptants (ΔgriR). Correct disruption was checked by Southern hybridization with appropriate regions as 32P-labeled probes (data not shown).
The other gri genes and the flanking genes, orfA and orf1, were also disrupted by deletion, by frameshift mutation, or by insertion of the neomycin and kanamycin resistance gene cassette on Tn5. An in-frame mutation of the region corresponding to Pro-108 to Asp-444 was introduced into griA. griB was disrupted by replacement of the griB sequence (Ser-8 to Ala-419) by the neomycin and kanamycin resistance gene cassette. An in-frame mutation of the region corresponding to Asp-10 to Gly-350 was introduced into griC. The chromosomal griD sequence corresponding to Pro-6 to Ala-454 was replaced by a short linker, resulting in production of only the first five amino acids of GriD (Met-Ser-Thr-Val-Ala), followed by a Leu. The chromosomal griG sequence was separated into two fragments that overlapped partially, +1 to +1030 and +857 to +1360, relative to the first nucleotide of the ATG start codon at +1, by insertion of the neomycin and kanamycin resistance gene cassette. The chromosomal griJ sequence corresponding to Tyr-181 to Gln-387 was replaced by a short linker, resulting in the production of a truncated GriJ protein (Met-1 to Ile-180 plus Leu) lacking the COOH-terminal 247 amino acid residues. A frameshift mutation (insertion of TAGA between +44 and +45, relative to the first nucleotide of the ATG start codon at +1) was introduced into griS. griT was disrupted by replacement of the griT sequence (Gly-3 to Arg-396) by the neomycin and kanamycin resistance gene cassette. orfA was disrupted by replacement of the orfA sequence with its upstream region, from nucleotide position −133 to the sequence corresponding to Gly-287 (relative to the first nucleotide of the ATG start codon at +1), by the neomycin and kanamycin resistance gene cassette. orf1 was disrupted by replacement of the orf1 sequence (Gly-17 to Arg-449) by the neomycin and kanamycin resistance gene cassette.
Production and purification of GriR.
The griR sequence was amplified by PCR with primers 5′-tggaagcttcatATGTCTACGCACCATCAATTCTCCG-3′ (griR-NdeI-F; with the HindIII site underlined, the NdeI site in the italics, and the start codon of griR in boldface) and 5′-tggtctagaccctcgagGCCGGAGCGCCCTT-3′ (with the XbaI site underlined, the XhoI site in italics, and the C-terminal Gly-307 codon of griR in boldface). The amplified DNA fragment was cloned between the HindIII and XbaI sites of pUC19, resulting in pUC19-griR-X. The griR sequence was excised as an NdeI-XhoI fragment from pUC19-griR-X and inserted between the NdeI and XhoI sites of pET26b (Novagen), resulting in pET26-griR.
E. coli BL21 (DE3)/pLysS harboring pET26-griR was cultured in Luria-Bertani medium at 37°C for 12 h. A portion (1 ml) of the culture was inoculated into 100 ml Luria-Bertani medium in a 500-ml Sakaguchi flask and cultured at 30°C with reciprocal shaking. When the optical density at 595 nm of the culture reached about 0.6, isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 1 mM. After an additional 4 h of cultivation, the E. coli cells were harvested by centrifugation. GriR with a His tag at its C terminus (GriR-H) was purified from the soluble fraction with an Ni-nitrilotriacetic acid spin column (QIAGEN) according to the manual of the manufacturer. GriR-H was eluted from the column with buffer A (50 mM NaHPO4, 300 mM NaCl, 10% glycerol, pH 8.0) containing 250 mM imidazole. Protein concentrations were measured with a Bio-Rad protein assay kit with bovine serum albumin as the standard. A Sephadex G-200 gel filtration column (Sigma) equipped in a fast protein liquid chromatography system (Pharmacia) was used to determine the molecular mass (flow rate, 0.2 ml/min; running buffer, 50 mM KH2PO4-150 mM NaCl, pH 8.0).
Gel mobility shift assay.
The DNA fragments used for 32P-labeled probes were amplified by PCR and 32P labeled with T4 polynucleotide kinase. Various regions upstream of the coding sequences of griC and griJ were used as 32P-labeled probes. The regions upstream of the coding sequences of griA and griR and the intervening region between griT and griS were also used as 32P-labeled probes. Table 1 lists the primer sequences for preparing these probes. Three probes, C1 to C3 (see Fig. 6A), for griC were prepared, i.e., C1-F and C1-R for probe C1, griC-F and C2-R for probe C2, and C3-F and griC-R for probe C3. Three probes for griJ (see Fig. 6A) were prepared, i.e., griJ-F and J1-R for probe J1, J2-F and J2-R for probe J2, and J3-F and griJ-R for probe J3. For griA, griA-H-F and griA-H-R were prepared for probe A. For griR, griR-H-F and griR-H-R were prepared for probe R. For griT and griS, griT-H-F and griT-H-R were prepared for probe TS. In addition to these probes, a mutant griJ probe was amplified by J1-m-F and J1-R. The gel mobility shift assay method used was described previously (39). Purified GriR-H (0.2 to 3.0 μg) was incubated with a 32P-labeled probe in buffer A containing 1 μg poly(dI-dC)·poly(dI-dC) and 0.01% bovine serum albumin.
FIG. 6.
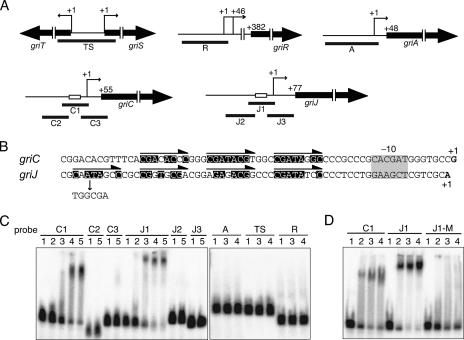
Binding of GriR-H to the griC and griJ promoters. (A) Schematic representation of the positions of probes used in panel C. The open boxes in the griC and griJ promoters represent the direct repeats. The transcriptional start point for each gene (see Fig. S1 in the supplemental material) is shown as +1. (B) Nucleotide sequences of the griC and griJ promoters. The direct repeats composed of an 8-bp sequence are indicated by arrows above the sequences. The nucleotides matching the consensus sequence, 5′-CGATACGC-3′, are highlighted. The transcriptional start points are shown in the boldface letters and numbered +1. The −10 element is indicated by a gray box. The mutation of one of the repeated sequences in the griJ promoter is also shown. (C) Gel mobility shift assay to determine the binding of GriR-H to the regions upstream of griC and griJ. GriR-H bound only to the C1 and J1 probes. GriR-H did not bind the regions upstream from griA (probe A) or griR (probe R) or the intervening region between griT and griS (probe TS). The amounts of GriR-H used were 0 μg (lane 1), 0.2 μg (lane 2), 0.4 μg (lane3), 0.8 μg (lane 4), and 1.5 μg (lane 5). (D) Gel mobility shift assay to determine the binding of GriR-H to the mutated sequence J1-M. GriR-H did not bind probe J1-M, whereas it bound control probes C1 and J1. The amounts of GriR-H used were 0 μg (lane 1), 0.3 μg (lane 2), 1.0 μg (lane3), and 3.0 μg (lane 4).
Construction of pHR20 for expression of griR.
A 0.4-kb DNA fragment containing the constitutively active promoter of hrdB, which encodes a principal sigma factor of RNA polymerase (31), was amplified by PCR with the S. griseus chromosomal DNA as a template and primers 5′-tggaagcttCGCGCCGCGCGAGCACTGAC-3′ (positions −234 to −215 with respect to the transcriptional start point of hrdB with the HindIII site underlined) and 5′-tgggaattccatatgCAACCTCTCGGAACGATGGAAAC-3′ (positions +119 to +97 with the EcoRI site underlined, the NdeI site in italics, and the start codon of hrdB in boldface). The amplified DNA fragment was cloned between the HindIII and EcoRI sites of pUC19ΔNdeI, resulting in pUC19-PhrdB. pUC19ΔNdeI was a pUC19 derivative in which an NdeI site (CATATG) was changed to CATATATG. The griR sequence was amplified by PCR with primers griR-NdeI-F and 5′-tgggaattcTCAGCCGGAGCGCCCTT-3′ (with the EcoRI site underlined and the termination codon of griR in boldface). The amplified DNA fragment was cloned between the HindIII and EcoRI sites of pUC19, resulting in pUC19-griR-E. The griR sequence excised from pUC19-griR-E with NdeI and EcoRI was placed between the NdeI and EcoRI sites of pUC19-PhrdB, resulting in pUC19-PhrdB-griR. Finally, the griR sequence with the hrdB promoter, excised from pUC19-PhrdB-griR with HindIII and EcoRI, was cloned into pKUM20, resulting in pHR20. pKUM20 is a low-copy-number shuttle vector between E. coli and Streptomyces with a multicloning HindIII-PmaCI-BamHI-PstI-EcoRI-XhoI sequence at the original PstI site of pKU209 (11) in the orientation opposite to that of the thiostrepton resistance gene.
Nucleotide sequence accession number.
We have deposited the 19-kb sequence including 13 gri genes and 2 neighboring genes (see Fig. 1A) in the DDBJ, EMBL, and GenBank DNA databases under accession no. AB259663.
RESULTS
Cloning of the GX biosynthesis gene cluster.
As described previously (36), we cloned part of the GX biosynthesis gene cluster (a 7.5-kb Sau3AI fragment on pAYP20; Fig. 1A) by shotgun cloning with GX-deficient mutant strain M31 as the host. In addition, Ueda and colleagues (7) had cloned a DNA fragment (a 6.1-kb NcoI fragment on pUC-MelG; Fig. 1A) containing melC1 and melC2 homologues (griE and griF) in the course of their study of melanogenesis in S. griseus. These two DNA fragments overlapped. We further cloned three DNA fragments containing their flanking regions (a 4.3-kb PstI-EcoRI fragment of pTH1, a 6.3-kb SphI-BglII fragment of pTH2, and an 8.7-kb BamHI fragment of pTH3; Fig. 1A) by a standard gene walking method including Southern hybridization and colony hybridization.
We determined the nucleotide sequences of these fragments and found 18 complete ORFs in the 25-kb region. Of these ORFs, we identified 13 genes, named griT, griS, griR, griA, griB, griC, griD, griE, griF, griG, griH, griI, and griJ, as GX biosynthesis genes. These 13 genes were all activated by A-factor and phosphate depletion signals, as described below. In addition, disruption of griT, griS, griR, griB, griC, griD, griEF, or griJ resulted in no GX production, although disruption of griA or griG had almost no effect on GX production (data not shown). The two neighboring genes (orfA and orf1) turned out to have no effect on GX biosynthesis; disruption of orfA and orf1 had no effect on GX production (data not shown).
Function of each gri gene product.
We so far have characterized the function of several gri gene products and revealed the GX biosynthesis pathway (Fig. 1B). The functions of GriI/GriH (36), GriC/GriD (our unpublished results), and GriE/GriF (35) have been biochemically elucidated. GriI and GriH are responsible for the biosynthesis of 3-amino-4-hydroxybenzoic acid (3,4-AHBA) from C3 and C4 primary metabolites (36). GriI, belonging to the class I aldolase family, catalyzes aldol condensation between l-aspartate-4-semialdehyde and dihydroxyacetone phosphate to form a C7 product, probably 2-amino-4,5-dihydroxy-6-one-heptanoic acid-7-phosphate. GriH, which is similar to the UPF0245 family members, converts the C7 compound to 3,4-AHBA. GriJ, an aspartokinase homologue, may catalyze conversion of aspartate to β-aspartyl phosphate, which is subsequently converted to l-aspartate-4-semialdehyde by the aspartate semialdehyde dehydrogenase for primary metabolism. 3,4-AHBA is reduced by the action of GriC and GriD, probably in an ATP-dependent manner, to form 3-amino-4-hydroxybenzaldehyde (3,4-AHBAL) (unpublished data). GriE and GriF are responsible for GX-A formation from two molecules of 3,4-AHBAL and N-acetylcysteine (35). GriE activates GriF by transferring Cu ions to GriF, as is observed for a Streptomyces melanogenesis system in which the MelC1 copper chaperon transfers Cu ions to MelC2 tyrosinase. GriF oxidizes 3,4-AHBAL to form an o-quinone imine, two molecules of which are then nonenzymatically coupled to form a phenoxazinone. In the presence of N-acetylcysteine, GX-A is formed from 3,4-AHBAL by the GriF reaction. GriF is thus responsible for the formation of the phenoxazinone chromophore and the introduction of an N-acetylcysteine molecule into the chromophore in the GX biosynthesis pathway. GX-A is oxidized by an oxidase to GX-B independently of the gene products in the GX biosynthesis gene cluster (our unpublished data). The enzymes, GriI/GriH, GriC/GriD, and GriE/GriF, that are required for GX-A synthesis from the primary metabolites are all encoded in the GX biosynthesis gene cluster. As described below, all of these genes were classified into the early active genes and transcribed by two mRNA species (Fig. 1A).
griG, which encodes a benzoate transporter-like protein, was shown to be involved in the uptake of exogenous 3,4-AHBA (unpublished data). Although a griG disruptant produced as much GX as the wild-type strain, it is possible that GriG serves as an importer of 3,4-AHBA that might be leaked from the cells. In fact, 3,4-AHBA was leaked or excreted without the function of GriG when griI and griH were simultaneously overexpressed and 3,4-AHBA was overproduced (36). GriB, similar to membrane efflux proteins, may serve as an exporter of GX into the medium, although a griB disruptant produced no GX in the cell or in the medium (data not shown). GriT contains a C-terminal helix-turn-helix DNA-binding domain that is similar to that of the LuxR family members. In agreement with the idea that GriT is an activator, disruption of griT resulted in abolishment of the transcription of all of the gri genes (data not shown). We therefore speculate that GriT acts as a regulator of GX synthesis in an as-yet-unknown way.
On the other hand, the functions of the other gri genes are hardly predicted by their homology with known proteins registered in the databases. A homology search predicted that GriA is a putative flavin adenine dinucleotide-dependent oxygenase. Since a griA disruptant produced as much GX as the wild-type strain (data not shown), the function of GriA is not clear. GriS shows weak sequence similarity to oxidoreductases. However, a frameshift mutation at +45, relative to the first nucleotide of the ATG start codon of griS at +1, abolished the transcription of all of the other gri genes. Because the transcriptional start points of griR (+286 and +331) were more than 240 bp downstream from the mutation point (+45) (see below), this frameshift mutation in griS appeared not to have a direct influence on griR transcription. We have no plausible explanation for the effect of the griS mutation on the other gri genes.
The two neighboring genes, orfA and orf1, were disrupted individually. The mutant strains still produced a yellow pigment. These findings suggest no involvement of these genes in GX biosynthesis, although we cannot exclude the possibility that the gene products are concerned with GX biosynthesis, for example, in minor modifications of GXs.
Six transcriptional units covering the whole GX biosynthesis gene cluster.
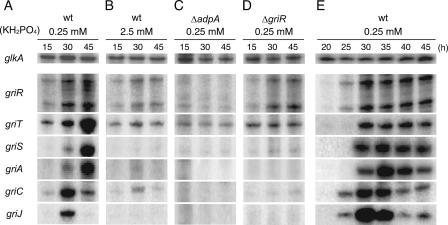
Because the termination codon of griC overlapped the start codon of griD, we assumed that griC and griD were cotranscribed and cotranslated. The functional relationship between griE and griF was evident, suggesting that griE and griF were also cotranscribed. We therefore examined transcription from the putative promoters of griT, griS, griR, griA, griB, griC, griE, griG, griH, griI, and griJ by low-resolution S1 mapping. RNA was prepared from wild-type strain IFO13350 grown at 30°C for 15, 30, and 45 h in SMM containing 0.25 mM KH2PO4. In this culture, the strain entered the stationary phase at about 30 h and began to produce GX. glkA, which encodes glucokinase, was used as an internal control.
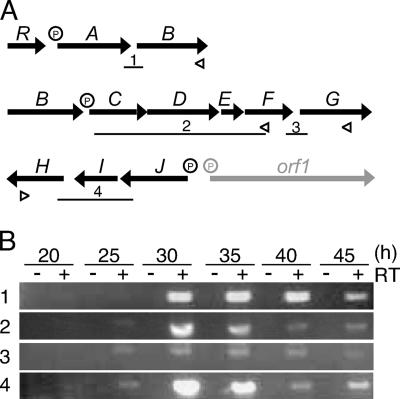
We detected transcripts from the promoters in front of griT, griS, griR, griA, griC, and griJ, all of which were activated at 30 h and thereafter (Fig. 2A). The time courses of their transcription will be described in detail below. When the probes for griB, griE, griG, griH, and griI were used, the full-length protection of the probes was observed, indicating that these genes were transcribed by read-through from their preceding genes (data not shown). To confirm this, we performed RT-PCR analysis (Fig. 3). With the cDNA that had been synthesized by reverse transcriptase from a primer annealing to the griB mRNA as the template, an intervening region between griA and griB was amplified by PCR (Fig. 3B, part 1). The amplification of the expected 374-bp fragment at 30 h and thereafter indicated that griA and griB were cotranscribed from the griA promoter. Similarly, with the cDNA synthesized with a primer annealing to the griH mRNA, a region covering a 3′ portion of griJ, griI, and a 5′ portion of griH was amplified (Fig. 3B, part 4). Therefore, griJ, griI, and griH were cotranscribed from the griJ promoter. For analysis of the long transcript covering griCDEFG, we divided it into two parts. With the cDNA synthesized from a primer annealing to the griF mRNA, a region covering a 3′ portion of griC, griD, griE, and a 5′ portion of griF was amplified (Fig. 3B, part 2). In addition, with the cDNA synthesized from a primer annealing to the griG mRNA, an intervening region between griF and griG was amplified (Fig. 3B, part 3). These results indicated that griC, griD, griE, griF, and griG were cotranscribed from the griC promoter. The weak amplification of the intervening region between griF and griG (Fig. 3B, part 3) may be due to weak transcriptional termination caused by a large inverted repeat sequence (ACGCGGCGGCGCCGGaCAGCGGat*caCCGCTGcCCGGCGCCGCCGCGT; * indicates a dyad axis; lowercase letters indicate mismatched nucleotides) that is present just downstream of griF. This sequence may act as a transcriptional terminator. griR was transcribed by two mRNA species, one from the griS promoter and the other from its own promoter (see below). We thus revealed six transcriptional units (griT, griSR, griR, griAB, griCDEFG, and griJIH) covering the whole GX biosynthesis gene cluster, as shown in Fig. 1A.
FIG. 2.
Time courses of griR, griT, griS, griA, griC, and griJ transcription as determined by low-resolution S1 nuclease mapping. RNA was prepared from the wild-type (wt) strain grown in SMM containing 0.25 mM (A and E) or 2.5 mM (B) KH2PO4. Transcription of the gri genes in SMM containing 0.25 mM phosphate in mutants ΔadpA (C) and ΔgriR (D) was also determined. Cultivation times (hours) are shown above the panels.
FIG. 3.
RT-PCR analysis of the polycistronic mRNA in the GX biosynthesis gene cluster. (A) Schematic representation of the positions of primers for RT reactions and DNA fragments amplified by PCR. The positions of primers used for synthesis of cDNA in RT reactions are shown by open triangles. The lengths and positions of the DNA fragments amplified by PCR in panel B are indicated by bars below the ORFs. P represents a promoter. (B) RNA was prepared from the wild-type strain grown in SMM containing 0.25 mM KH2PO4. Cultivation times in hours are shown above the panel. Control experiments with no RT (lanes −) confirmed that the RNA samples contained no chromosomal DNA. The amplified fragments were analyzed by agarose gel electrophoresis and stained with ethidium bromide.
We next determined transcriptional start points from the six promoters by high-resolution S1 nuclease mapping (see Fig. S1 in the supplemental material). The transcriptional start points of griT and griS were both determined to be the first nucleotides of their translational start codons. There were two transcriptional start points for griR, which were determined to be 381 and 336 nucleotides upstream of the translational start codon. Both transcriptional start points of griR were located in the coding sequence of griS. This means that there is no transcriptional terminator in the intervening region between griS and griR and the transcript starting from the griS promoter extends to griR. The transcriptional start points of griA (47 nucleotides upstream of the translational start codon), griC (43 nucleotides), and griJ (43 nucleotides) were also determined.
Expression profiles of the gri genes.
As mentioned above, the six promoters were all activated at 30 h and thereafter (Fig. 2A). To determine the detailed expression profiles of the six transcriptional units covering the GX biosynthesis gene cluster, we performed S1 nuclease mapping with RNA prepared from the wild-type strain grown for 20, 25, 30, 35, 40, or 45 h in SMM containing 0.25 mM KH2PO4 (Fig. 2E). Under the cultural conditions used, the cells entered the stationary phase at about 25 h and began to produce GX. From the transcription profiles shown in Fig. 2A and E, the six transcriptional units were classified into three groups. The first group included only griR. The transcription of griR, starting during the late exponential phase, reached a maximum in the early stationary phase and continued thereafter. The second group included griCDEFG and griJIH. The transcription from the griC and griJ promoters, starting just after the transcription of griR, reached a maximum in the early stationary phase and decreased rapidly. The third group included griT, griSR, and griAB. The griT, griS, and griA promoters were activated after the transcriptional activation of the second group genes, although griT was apparently transcribed at a basal level during the early growth stage. griCDEFG and griJIH can be called “early” active genes, and griT, griS, and griAB are “late” active genes in GX biosynthesis. On the other hand, griR is an “initially” active gene in the gene cluster, although griR is also cotranscribed with griS from the griS promoter in the late stage. These results are in good agreement with the fact that griR encodes the pathway-specific transcriptional activator for the gene cluster and that GriR directly activates the griC and griJ promoters, as described below.
Disruption of griR.
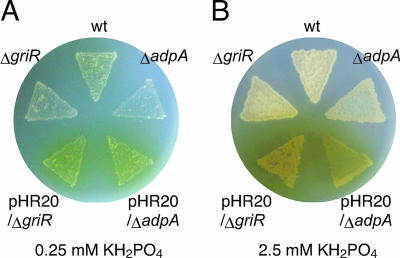
griR encodes a 307-amino-acid protein showing significant sequence similarity (30 to 40% identity) to SARP family transcriptional activators (40). To determine the possible role of GriR in the regulation of the GX biosynthesis genes, we constructed a strain containing in-frame griR deletion (ΔgriR), in which the region coding for amino acids Leu-32 to Glu-253 was deleted. Mutant ΔgriR did not produce GX (Fig. 4A). Introduction of pHR20 carrying griR under the control of the hrdB promoter restored GX production of mutant ΔgriR (Fig. 4A), confirming that the failure of ΔgriR to produce GX resulted from the inactivation of griR. Consistent with this observation, no transcriptional activation of griT, griS, griA, griC, or griJ occurred in mutant ΔgriR when determined with RNA prepared from the cells grown for 15, 30, or 45 h in SMM containing 0.25 mM KH2PO4 (Fig. 2D). griT was transcribed at a basal level. Transcription from the two griR promoters was not affected by the deletion of the griR-coding sequence, indicating the absence of self-regulation of griR (Fig. 2D). All of these findings showed that GriR was the pathway-specific transcriptional activator for GX biosynthesis.
FIG. 4.
GX production on SMM agar medium. SMM agar contained 0.25 mM (A) or 2.5 mM (B) KH2PO4. The wild-type (wt) strain, the strain with adpA disrupted (ΔadpA), ΔadpA harboring pHR20 (pHR20/ΔadpA), the strain with griR disrupted (ΔgriR), and ΔgriR harboring pHR20 (pHR20/ΔgriR) were cultured at 28°C for 4 days. The wild-type strain produced a detectable amount of the yellow pigment GX on medium containing 0.25 mM phosphate but not on medium containing 2.5 mM phosphate. Mutants ΔadpA and ΔgriR produced no yellow pigments on medium containing 0.25 or 2.5 mM phosphate. Plasmid pHR20 carrying griR under the control of the hrdB promoter conferred yellow pigment productivity on both mutants. The amounts of GX produced by both mutants harboring pHR20 on medium containing 2.5 mM phosphate are larger than those produced on medium containing 0.25 mM phosphate. The mycelium of the strains grown on medium containing 2.5 mM phosphate is more abundant than that of the strain grown on medium containing 0.25 mM phosphate.
Binding of GriR to the griC and griJ promoters.
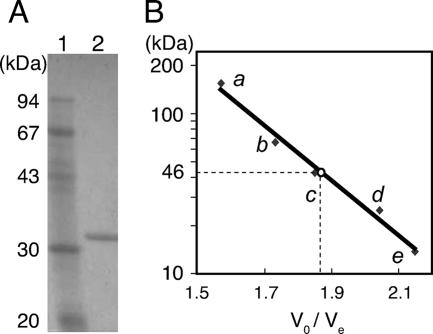
For purification of GriR, we constructed pET26-griR carrying the griR sequence under the control of the T7 promoter. The recombinant GriR (GriR-H), having a structure of GriR-Leu-Glu-(His)6, was produced in E. coli and purified with an Ni-nitrilotriacetic acid spin column (Fig. 5A). We used gel filtration chromatography to determine the subunit structure of GriR-H under nondenaturing conditions (Fig. 5B). A single peak representing GriR-H eluted at 46 kDa when determined on the basis of the value of V0/Ve (V0, exclusion volume of the column and Ve, elution volume). Because GriR-H had a calculated molecular mass of 34.6 kDa, we concluded that this protein was monomeric.
FIG. 5.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of purified GriR-H (A) and estimation of its molecular weight by gel filtration (B). (A) GriR-H used in this study was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Phosphorylase b (94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), and soybean trypsin inhibitor (20 kDa) were used as molecular mass standards. (B) The column used was a Sephadex G-200 column (Sigma) in a fast protein liquid chromatography system. Aldolase (158 kDa) (a), bovine serum albumin (67 kDa) (b), ovalbumin (43 kDa) (c), chymotrypsinogen A (25 kDa) (d), and RNase A (14 kDa) (e) were used for generation of a calibration line.
We examined binding of GriR-H to DNA fragments containing the promoter regions of griT, griS, griR, griA, griC, and griJ (Fig. 6A) by gel mobility shift assay. As shown in Fig. 6C, GriR-H bound the griC and griJ promoters (see below) but did not bind the griA, griR, griS, or griT promoter (no retardation with probe A, TS, or R). Three and four direct repeats composed of an 8-bp sequence with intervals of 3 bp were present in the griC and griJ promoters, respectively (Fig. 6B). The consensus sequence of the 8-bp repeat was CGATACGC. The presence of a direct repeat in which repeated sequences appear every 11 bp and the one most proximal to the transcriptional start point is located 8 bp apart from the −10 element is typical for the SARP-binding site (40). No such repeats were present in the griA, griR, griS, or griT promoter.
To confirm the binding of GriR-H to the direct repeats in the griC and griJ promoters, we prepared six probes, C1, C2, C3, J1, J2, and J3, of which C1 and J1 contained the direct repeats (Fig. 6A). GriR-H bound to only C1 and J1 (Fig. 6C). Interestingly, despite the same length of probes C1 and J1, the (GriR-H)-C1 complex ran faster than the (GriR-H)-J1 complex. We assumed that one molecule of GriR-H bound one repeat and therefore three and four molecules of GriR-H bound C1 containing three repeats and J1 containing four repeats, respectively, as is found for DnrI binding to its target site (30). We then introduced a mutation into one of the repeated sequences in J1; CAATAGCC was replaced with TGGCGACC (Fig. 6B). The mutated J1 (J1-M) probe that still contained the other three intact repeats, however, was not bound by GriR-H at all (Fig. 6D). This finding indicated that the repeat that was mutated was essential for GriR-H to bind J1 in a cooperative manner, as is also found for DnrI, the pathway-specific transcriptional activator for daunorubicin biosynthesis in Streptomyces peucetius (30). Although further experiments are necessary for elucidating the mechanism of GriR binding to C1 and J1, the present data clearly show that GriR binds the direct repeats in the griC and griJ promoters.
Dependence of the transcription of gri genes on A-factor and phosphate depletion.
Because GX production is under the control of A-factor, we determined the transcription of gri genes in an adpA-disrupted strain (ΔadpA) by low-resolution S1 nuclease mapping. RNA was prepared from S. griseus ΔadpA grown for 15, 30, or 45 h in SMM containing 0.25 mM KH2PO4. As we expected, griR was not transcribed in mutant ΔadpA and no transcriptional activation of any other gri genes occurred, although the transcription of griT at a basal level was still detected throughout growth (Fig. 2C).
GX is produced under phosphate depletion, and GX production is hardly detected in the presence of 2.5 mM phosphate. We determined transcription of gri genes in the presence of phosphate at a high concentration. RNA was prepared from the wild-type strain grown for 15, 30, or 45 h in SMM containing 2.5 mM KH2PO4. The amounts of the griR transcripts at 30 and 45 h were greatly reduced in comparison with those under the low-phosphate (0.25 mM) conditions (Fig. 2B). Furthermore, transcriptional activation of other gri genes was hardly detected, although only very weak transcription from the griC promoter, except for griT transcription at the basal level, was detected. We assumed that the reduced griR transcription in the presence of phosphate at a high concentration resulted in an insufficient supply of GriR to induce the other gri genes.
We could not detect the transcription of orfA, which encodes a necrosis-inducing protein (NPP1)-like protein, even under GX-producing conditions (data not shown). On the other hand, orf1, which encodes a large hypothetical protein containing tetratricopeptide repeats, was constitutively transcribed even in mutant ΔadpA and in the presence of 2.5 mM phosphate (data not shown). These results also supported the idea that orfA and orf1 have no relationship to GX biosynthesis.
Production of GX by ectopic griR expression.
Since all of the gri genes were found to be under the control of GriR, we expected that constitutive expression of griR would lead to GX production in the absence of A-factor and in the presence of a high concentration of phosphate. We constructed pHR20 that carried griR under the control of the hrdB promoter on low-copy-number plasmid pKUM20 and introduced it into mutant ΔgriR. Mutant ΔgriR harboring pHR20 produced a larger amount of a yellow pigment, which was confirmed to be GX by HPLC analysis, than the wild-type strain on SMM agar containing 0.25 mM KH2PO4 (Fig. 4A), probably because of the strong promoter activity of hrdB. Furthermore, on SMM agar containing 2.5 mM KH2PO4, the strain also produced a much larger amount of GX, whereas the wild-type strain never produced GX under the conditions tested (Fig. 4B). The greater amount of GX on SMM agar containing 2.5 mM phosphate than on 0.25 mM phosphate was due to more rapid and better growth in the former medium. Therefore, the phosphate-depleted signal was transmitted to the GX biosynthesis genes solely via the promoter of griR (Fig. 7).
S. griseus mutant ΔadpA harboring pHR20 produced an amount of GX similar to that produced by S. griseus mutant ΔgriR harboring pHR20 on SMM agar containing 0.25 or 2.5 mM KH2PO4 (Fig. 4A and B). These results clearly showed that GX was produced once griR was expressed even in the absence of AdpA, indicating that the A-factor signal was also transmitted to the GX biosynthesis genes solely via the griR promoter (Fig. 7). We observed no AdpA binding to regions upstream or downstream of the griR promoter (data not shown), suggesting indirect regulation of griR by AdpA.
DISCUSSION
The biosynthesis genes for a certain secondary metabolite usually consist of a gene cluster that includes a regulatory gene that encodes a pathway-specific transcriptional activator. Many pathway-specific transcriptional activators encoded in the biosynthetic gene cluster for a certain secondary metabolite in streptomycetes belong to the SARP family. SARPs, which are found only in actinobacteria, are characterized by their DNA-binding domains resembling that of OmpR, which consists of three α helices packed against two antiparallel β sheets, forming a winged helix-turn-helix (18). ActII-ORF4 for actinorhodin production (2) and RedD for undecylprodigiosin production (22) in Streptomyces coelicolor A3(2), DnrI for daunorubicin production in S. peucetius (30), MtmR for mithramycin production in Streptomyces argillaceus (16), and CcaR for cephamycin and clavulanic acid production in Streptomyces clavuligerus (29) are all SARP family regulators. The present study has demonstrated that a SARP family regulator, GriR, encoded in the GX biosynthesis gene cluster is the pathway-specific transcriptional activator because (i) transcription of griR was detected before the transcriptional activation of other gri genes, (ii) disruption of griR abolished transcriptional activation of other gri genes and GX production, and (iii) expression of griR from a constitutively active promoter caused overproduction of GX even in the absence of AdpA and in the presence of phosphate at a high concentration. The third observation is important in that both A-factor and phosphate depletion signals are transmitted to the GX biosynthesis genes solely via the griR promoter (Fig. 7). AdpA appears to activate the griR promoter indirectly, because AdpA did not bind the region upstream or downstream of the griR promoter. We assume that a positive transcription factor, the production of which is induced by AdpA, is needed for the transcriptional activation of griR.
The biosynthesis of candicidin in S. griseus IMRU3570 (3) and oxytetracycline in Streptomyces rimosus (20) is controlled by phosphate at the transcriptional level. In addition, phosphate regulation of the actinorhodin (act) and undecylprodigiosin (red) genes in S. coelicolor A3(2) and the pimaricin (pim) genes in Streptomyces natalensis is mediated by a PhoR-PhoP two-component regulatory system (21, 32). In these studies, however, it is not clear whether the phosphate regulation of secondary metabolite formation is mediated only by the transcriptional control of the pathway-specific transcriptional activator genes. Our present study clearly shows that the failure of GX production of the S. griseus wild-type strain in the presence of phosphate at a high concentration is absolutely caused by insufficient transcription of griR. We assume that a positive or negative transcription factor, the production or activity of which is regulated by phosphate, is involved in the control of griR transcription. Further analysis of such a transcription factor controlling griR transcription will reveal a missing link between the PhoR-PhoP two-component regulatory system and the pathway-specific regulators for many secondary metabolites.
GriR binds the direct repeat sequences just on the griC and griJ promoters, indicating that the early active genes (griCDEFG and griJIH), all of which, except for griG, encode the GX biosynthesis enzymes, are directly activated by GriR. On the other hand, GriR did not bind the promoters of griT, griS, and griA, while activation of these three promoters, as well as the griC and griJ promoters, was not observed in the ΔgriR strain. This observation suggests that the late active genes (griT, -S, -A, and -B) are indirectly activated by GriR or activated by GriR in conjunction with some other factor. The molecular mechanisms of activation of these three promoters remain to be elucidated.
GriR was shown to be monomeric by gel filtration chromatography, as is found for a SARP family regulator, DnrI (30). In binding of DnrI to its target site containing two direct repeats, two molecules of DnrI monomer bind its target site in a cooperative manner (30). Three and four molecules of GriR monomer appeared to bind the C1 probe containing three repeats and the J1 probe containing four repeats, respectively. We assume that GriR monomers bind the target DNA in a cooperative manner, because no shift bands other than those probably representing (GriR-H)3-C1 and (GriR-H)4-J1 were detected in the gel mobility shift assay and because a mutation in one of the repeated sequences in J1 abolished the binding of GriR to the mutated J1-M probe. Concerning the positions of binding sites of DnrI and ActII-ORF4 in their respective target promoters, the repeat most proximal to the transcriptional start point is located 8 bp apart from the −10 promoter element (2, 30). The constitution of the GriR binding site with respect to the −10 element in the griC and griJ promoters is the same. In addition, as already pointed out by Wietzorrek and Bibb (40), the distances between the centers of the direct repeats are all 11 bp, as is found for the target sites of DnrI (30) and ActII-ORF4 (2). This means that multiple SARP molecules, including GriR molecules, bind individual repeats in a cooperative way on the same face of the DNA, thus recruiting RNA polymerase to the opposite face.
Enhanced expression of a pathway-specific transcriptional activator gene causes increased production of the respective secondary metabolite. Examples are dnrI for daunorubicin in S. peucetius (34), redD for undecylprodigiosin (37) and actII-orf4 for actinorhodin (8) in S. coelicolor A3(2), ccaR for cephamycin and clavulanic acid in S. clavuligerus (28), and tylR for tylosin in Streptomyces fradiae (33). Furthermore, introduction of actII-orf4 into S. lividans awakens the “sleeping” act genes, resulting in overproduction of actinorhodin (6). In this study, we showed that ectopic expression of griR caused overproduction of GX even under conditions nonpermissive for GX production. Probably, expression of a pathway-specific transcriptional activator gene by using a constitutively active promoter is one of the promising strategies to awaken some sleeping biosynthesis gene cluster, especially in genome mining for undiscovered secondary metabolites.
Supplementary Material
Acknowledgments
We thank K. Ueda for providing pUC-MelG.
This work was supported by grant 03A07002 from the Industrial Technology Research Grant Program in 2003 of the New Energy and Industrial Technology Development Organization of Japan and by a Grant-in-Aid for Scientific Research on Priority Area “Applied Genomics” from Monkasho.
Footnotes
Published ahead of print on 2 March 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Angell, S., E. Schwarz, and M. J. Bibb. 1992. The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol. Microbiol. 6:2833-2844. [DOI] [PubMed] [Google Scholar]
- 2.Arias, P., M. A. Fernández-Moreno, and F. Malpartida. 1999. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J. Bacteriol. 181:6958-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asturias, J. A., P. Liras, and J. F. Martín. 1990. Phosphate control of pabS gene transcription during candicidin biosynthesis. Gene 93:79-84. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingstone, D. O. Moore, J. S. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 5.Beck, E., G. Ludwig, E. A. Auerswald, B. Reiss, and H. Schaller. 1982. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene 19:327-336. [DOI] [PubMed] [Google Scholar]
- 6.Bruheim, P., H. Sletta, M. J. Bibb, J. White, and D. W. Levine. 2002. High-yield actinorhodin production in fed-batch culture by a Streptomyces lividans strain overexpressing the pathway-specific activator gene actII-ORF4. J. Ind. Microbiol. Biotechnol. 28:103-111. [DOI] [PubMed] [Google Scholar]
- 7.Endo, K., K. Kamo, K. Hosono, T. Beppu, and K. Ueda. 2001. Characterization of mutants defective in melanogenesis and a gene for tyrosinase of Streptomyces griseus. J. Antibiot. 54:789-796. [DOI] [PubMed] [Google Scholar]
- 8.Gramajo, H. C., E. Takano, and M. J. Bibb. 1993. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 7:837-845. [DOI] [PubMed] [Google Scholar]
- 9.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 10.Horinouchi, S. 2002. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front. Biosci. 7:d2045-d2057. [DOI] [PubMed] [Google Scholar]
- 11.Kakinuma, S., Y. Takada, H. Ikeda, H. Tanaka, S. Omura, and D. A. Hopwood. 1991. Cloning of large DNA fragments, which hybridize with actinorhodin biosynthesis genes, from kalafungin and nanaomycin A methyl ester producers and identification of genes for kalafungin biosynthesis of the kalafungin producer. J. Antibiot. 44:995-1005. [DOI] [PubMed] [Google Scholar]
- 12.Kato, J., N. Funa, H. Watanabe, Y. Ohnishi, and S. Horinouchi. 2007. Biosynthesis of γ-butyrolactone autoregulators that switch on secondary metabolism and morphological development in Streptomyces. Proc. Natl. Acad. Sci. USA 104:2378-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato, J., Y. Ohnishi, and S. Horinouchi. 2005. Autorepression of AdpA of the AraC/XylS family, a key transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. J. Mol. Biol. 350:12-26. [DOI] [PubMed] [Google Scholar]
- 14.Kelemen, G. H., P. Brian, K. Flärdh, L. Chamberlin, K. F. Chater, and M. J. Buttner. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 180:2515-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieser, H., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 16.Lombó, F., A. F. Braña, C. Méndez, and J. A. Salas. 1999. The mithramycin gene cluster of Streptomyces argillaceus contains a positive regulatory gene and two repeated DNA sequences that are located at both ends of the cluster. J. Bacteriol. 181:642-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Martínez-Hackert, E., and A. M. Stock. 1997. The DNA-binding domain of OmpR: crystal structure of a winged helix transcription factor. Structure 5:109-124. [DOI] [PubMed] [Google Scholar]
- 19.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560. [DOI] [PubMed] [Google Scholar]
- 20.McDowall, K. J., A. Thamchaipenet, and I. S. Hunter. 1999. Phosphate control of oxytetracycline production by Streptomyces rimosus is at the level of transcription from promoters overlapped by tandem repeats similar to those of the DNA-binding sites of the OmpR family. J. Bacteriol. 181:3025-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendes, M. V., S. Tunca, N. Antón, E. Recio, A. Sola-Landa, J. F. Aparicio, and J. F. Martín. 2007. The two-component phoR-phoP system of Streptomyces natalensis: inactivation or deletion of phoP reduces the negative phosphate regulation of pimaricin biosynthesis. Metab. Eng. 9:217-227. [DOI] [PubMed] [Google Scholar]
- 22.Narva, K., and J. Feitelson. 1990. Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2). J. Bacteriol. 172:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh, S. H., and K. F. Chater. 1997. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J. Bacteriol. 179:122-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohnishi, Y., Y. Furusho, T. Higashi, H.-K. Chun, K. Furihata, S. Sakuda, and S. Horinouchi. 2004. Structures of grixazone A and B, A-factor-dependent yellow pigments produced under phosphate depletion by Streptomyces griseus. J. Antibiot. 57:218-223. [DOI] [PubMed] [Google Scholar]
- 25.Ohnishi, Y., and S. Horinouchi. 2004. The A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces. Biofilms 1:319-328. [DOI] [PubMed] [Google Scholar]
- 26.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 27.Ohnishi, Y., H. Yamazaki, J. Kato, A. Tomono, and S. Horinouchi. 2005. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69:431-439. [DOI] [PubMed] [Google Scholar]
- 28.Pérez-Llarena, F. J., P. Liras, A. Rodríguez-García, and J. F. Martín. 1997. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both beta-lactam compounds. J. Bacteriol. 179:2053-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santamarta, I., A. Rodríguez-García, R. Pérez-Redondo, J. F. Martín, and P. Liras. 2002. CcaR is an autoregulatory protein that binds to the ccaR and cefD-cmcI promoters of the cephamycin C-clavulanic acid cluster in Streptomyces clavuligerus. J. Bacteriol. 184:3106-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheldon, P. J., S. B. Busarow, and C. R. Hutchinson. 2002. Mapping the DNA-binding domain and target sequences of the Streptomyces peucetius daunorubicin biosynthesis regulatory protein, DnrI. Mol. Microbiol. 44:449-460. [DOI] [PubMed] [Google Scholar]
- 31.Shinkawa, H., Y. Hatada, M. Okada, H. Kinashi, and O. Nimi. 1995. Nucleotide sequence of a principal sigma factor gene (hrdB) of Streptomyces griseus. J. Biochem. 118:494-499. [DOI] [PubMed] [Google Scholar]
- 32.Sola-Landa, A., R. S. Moura, and J. F. Martín. 2003. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. USA 100:6133-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stratigopoulos, G., N. Bate, and E. Cundliffe. 2004. Positive control of tylosin biosynthesis: pivotal role of TylR. Mol. Microbiol. 54:1326-1334. [DOI] [PubMed] [Google Scholar]
- 34.Stutzman-Engwall, K. J., S. L. Otten, and C. R. Hutchinson. 1992. Regulation of secondary metabolism in Streptomyces spp. and overproduction of daunorubicin in Streptomyces peucetius. J. Bacteriol. 174:144-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki, H., Y. Furusho, T. Higashi, Y. Ohnishi, and S. Horinouchi. 2006. A novel o-aminophenol oxidase responsible for formation of the phenoxazinone chromophore of grixazone. J. Biol. Chem. 281:824-833. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki, H., Y. Ohnishi, Y. Furusho, S. Sakuda, and S. Horinouchi. 2006. Novel benzene ring biosynthesis from C3 and C4 primary metabolites by two enzymes. J. Biol. Chem. 281:36944-36951. [DOI] [PubMed] [Google Scholar]
- 37.Takano, E., H. C. Gramajo, E. Strauch, N. Andres, J. White, and M. J. Bibb. 1992. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol. Microbiol. 6:2797-2804. [DOI] [PubMed] [Google Scholar]
- 38.Tomono, A., Y. Tsai, H. Yamazaki, Y. Ohnishi, and S. Horinouchi. 2005. Transcriptional control by A-factor of strR, the pathway-specific transcriptional activator for streptomycin biosynthesis in Streptomyces griseus. J. Bacteriol. 187:5595-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamazaki, H., A. Tomono, Y. Ohnishi, and S. Horinouchi. 2004. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53:555-572. [DOI] [PubMed] [Google Scholar]
- 40.Wietzorrek, A., and M. J. Bibb. 1997. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 25:1181-1184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.